Submitted:
11 January 2023
Posted:
12 January 2023
You are already at the latest version
Abstract
Keywords:
I. Introduction
II. Related Works
- There are machine-learning-assisted decision support and diagnostic tools (although not widely used or accepted in medical practice) for a non-evasive risk prediction of diabetes in medical patients.
- Deep Learning models have not been used for diabetes risk prediction as much as classical models such as SVM, kNN and regression models.
- The impact of feature engineering on the results of these models needs to be more widely studied.
- Ensemble feature selection method has not been applied yet to diabetes risk prediction.
- Deep belief neural networks and other unsupervised deep learning methods for diabetes risk prediction need more attention.
III. Methods
a. Dataset
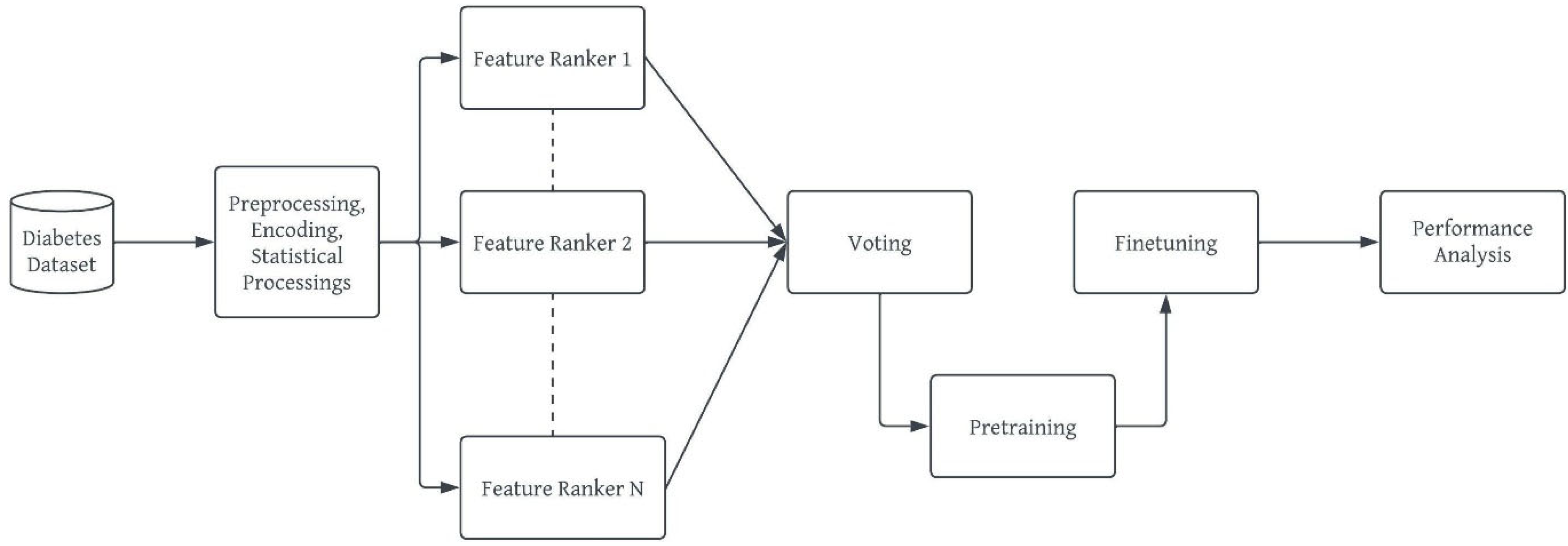
b. Model Development Workflow
- vi.
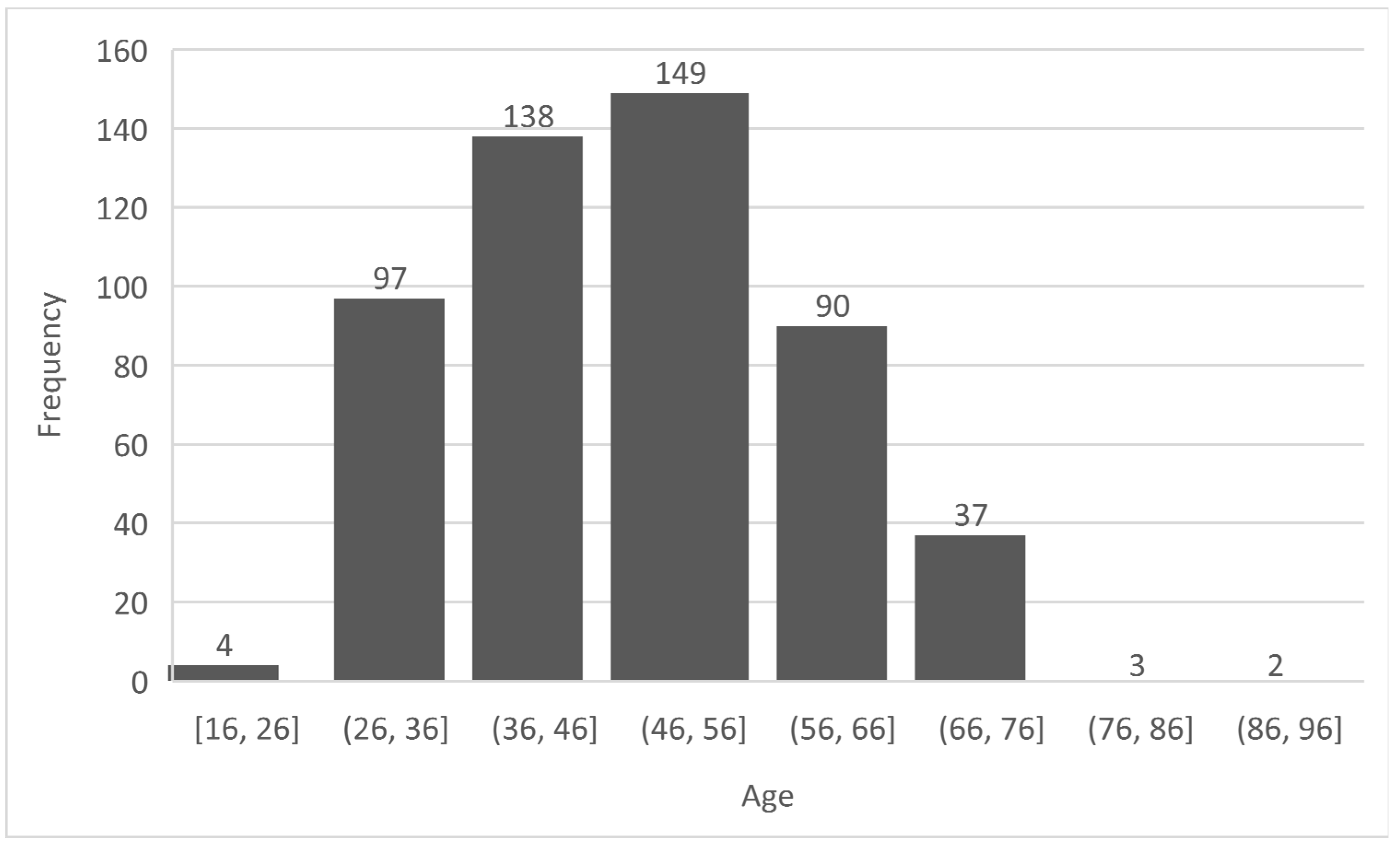
- Preprocessing: This stage ensures that the diabetes dataset to be used is well prepared for the machine learning task [42]. This stage ensures the quality of the dataset in terms of noise and duplicate removal, outlier detection and processing, encoding for a numerical representation of categorical and nominal variables [43]. In the diabetes dataset, all corresponding Yes/No and Male/Female values were replaced with 1/0 respectively. The ages were encoded from 0 – 7 based on the categorization specified in Figure 1 and these values were normalized using the Min-Max normalization [44] to prevent the age column from outweighing other columns during prediction, thereby reducing bias. The output of this stage is a ready dataset for further analyses and model pre-training.
- vi.
- vii. Ensemble Feature Selection: This study uses the ensemble dimensionality reduction framework to select the best feature set for the developed deep learning model while removing redundant features from the dataset. This will avoid misfits, either overfitting or underfitting as well as reduce the curse and complexity of multidimensionality [45,46]. The ensemble selection leverages on the individual strengths of each candidate feature selection method to find the best feature vectors for the deep learning models. The output of this stage is a “project” or subset of the original dataset.
- vi.
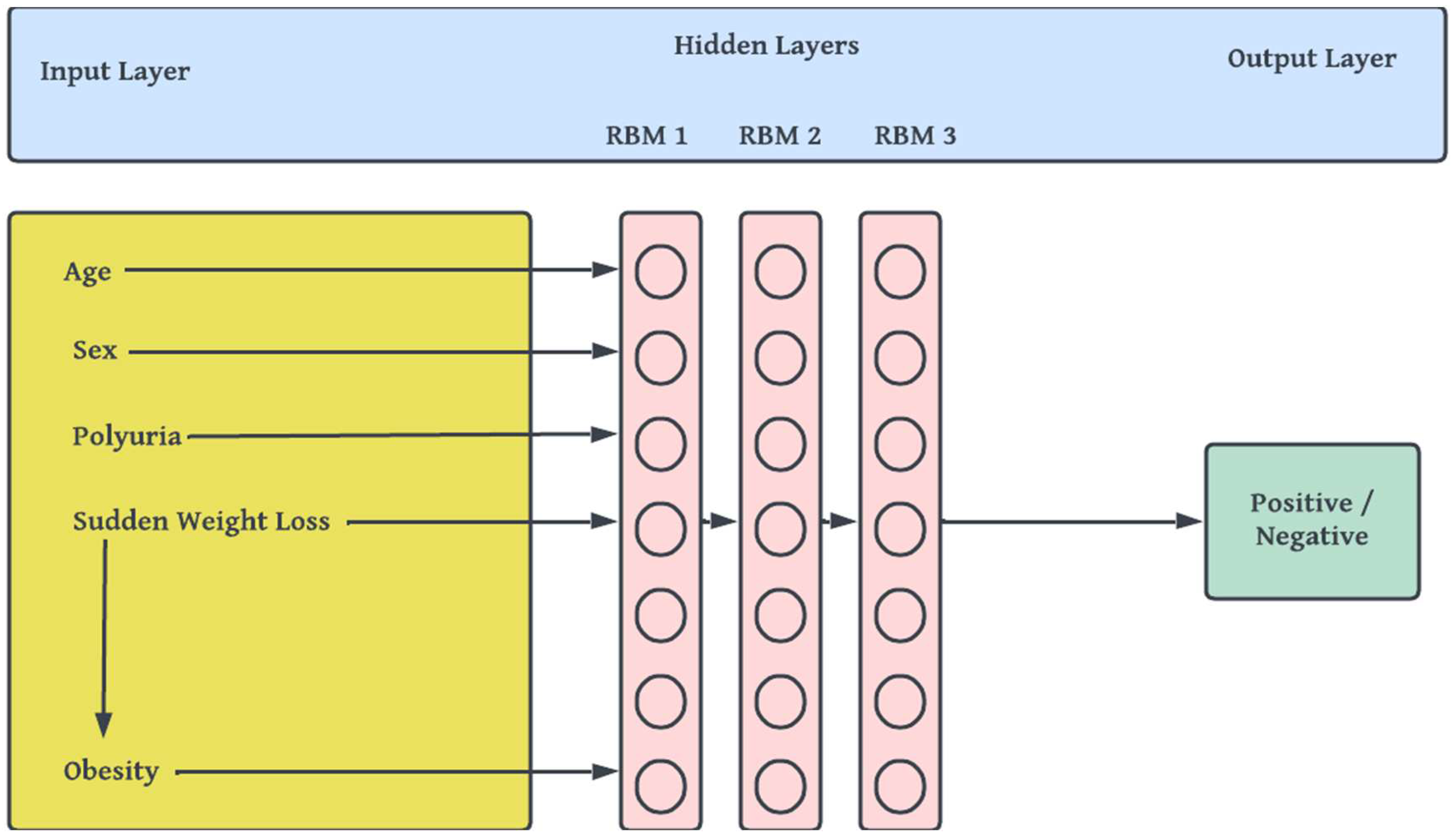
- viii. Building, Pretraining and Finetuning the DBN Model: This step comprises of the actual stacking of Restricted Boltzmann Machines (RBMs) [47] to form a deep net and training. DBN is a generative-graph multi-layered model. The process in which the model is used to predict either in a supervised or unsupervised manner is known as pre-training. Each of the deep – hidden – layers is trained as RBMs. The first stage of training DBN is to train layers sequentially from the bottom visible (observed) layer features. This input layer contains D number of units, where D is input sample dimension. This input layer is fully connected with hidden layers. Each Hidden layer consists of N number of RBM. The output layer consists of one unit which defines the class. The final phase, called fine tuning is to train the second layer based on the results from pre-training step. Finally, the entire hidden layers are learned same way till final hidden layer is reached. The Figure 3 outlines the architecture of model pre-training proposed for our study.
- ix.
- Performance Analysis: Our proposed DBN model for diabetes risk prediction was assessed using F1-Measure, Precision and Recall, wherewhere TP is True Positive, FP is False Positive and FN is False Negative as all obtained from the confusion matrix of the result.
IV. Results and Discussion
V. Conclusion
References
- Preston, E.V., et al., Climate factors and gestational diabetes mellitus risk–a systematic review. Environmental Health, 2020. 19(1): p. 1-19.
- Wang, P., et al., Seasonality of gestational diabetes mellitus and maternal blood glucose levels: evidence from Taiwan. Medicine, 2020. 99(41).
- Boiko, M., R. Ovchinnikova, and A. Shabrina. THE ROLE OF HORMONES IN THE HUMAN BODY. in Чeлoвeк. Oбщecтвo. Kyльтypa. Coциaлизaция. 2019.
- Hauge-Evans, A.C., SUGAR, DOGS, COWS, AND INSULIN—THE STORY OF HOW DIABETES STOPPED BEING DEADLY. Frontiers for young minds, 2021. 9.
- Padhi, S., A.K. Nayak, and A. Behera, Type II diabetes mellitus: A review on recent drug based therapeutics. Biomedicine & Pharmacotherapy, 2020. 131: p. 110708.
- Eizirik, D.L., L. Pasquali, and M. Cnop, Pancreatic β-cells in type 1 and type 2 diabetes mellitus: different pathways to failure. Nature Reviews Endocrinology, 2020. 16(7): p. 349-362.
- 7. Padhi, S., M. Dash, and A. Behera, Nanophytochemicals for the treatment of type II diabetes mellitus: a review. Environmental Chemistry Letters, 2021. 19(6): p. 4349-4373.
- Lee, K.W. , et al., Neonatal outcomes and its association among gestational diabetes mellitus with and without depression, anxiety and stress symptoms in Malaysia: A cross-sectional study. Midwifery, 2020. 81: p. 102586.
- Yang, Q.-Q. , et al., The association between diabetes complications, diabetes distress, and depressive symptoms in patients with type 2 diabetes mellitus. Clinical nursing research, 2021. 30(3): p. 293-301.
- Kopitar, L. , et al., Early detection of type 2 diabetes mellitus using machine learning-based prediction models. Scientific reports, 2020. 10(1): p. 1-12.
- Gadekallu, T.R. , et al., Early detection of diabetic retinopathy using PCA-firefly based deep learning model. Electronics, 2020. 9(2): p. 274.
- Yang, H. , et al., New perspective in diabetic neuropathy: from the periphery to the brain, a call for early detection, and precision medicine. Frontiers in endocrinology, 2020. 10: p. 929.
- Sungheetha, A. and R. Sharma, Design an early detection and classification for diabetic retinopathy by deep feature extraction based convolution neural network. Journal of Trends in Computer Science and Smart technology (TCSST), 2021. 3(02): p. 81-94.
- Tofte, N. , et al., Early detection of diabetic kidney disease by urinary proteomics and subsequent intervention with spironolactone to delay progression (PRIORITY): a prospective observational study and embedded randomised placebo-controlled trial. The lancet Diabetes & endocrinology, 2020. 8(4): p. 301-312.
- Hasan, D.A. , et al. Machine Learning-based Diabetic Retinopathy Early Detection and Classification Systems-A Survey. in 2021 1st Babylon International Conference on Information Technology and Science (BICITS). 2021. IEEE.
- Ben-Israel, D. , et al., The impact of machine learning on patient care: a systematic review. Artificial Intelligence in Medicine, 2020. 103: p. 101785.
- Peiffer-Smadja, N. , et al., Machine learning for clinical decision support in infectious diseases: a narrative review of current applications. Clinical Microbiology and Infection, 2020. 26(5): p. 584-595.
- Bernabe-Ortiz, A. , et al., Diagnostic accuracy of the Finnish Diabetes Risk Score (FINDRISC) for undiagnosed T2DM in Peruvian population. Primary care diabetes, 2018. 12(6): p. 517-525.
- Boulton, A.J. , et al., Comprehensive foot examination and risk assessment: a report of the task force of the foot care interest group of the American Diabetes Association, with endorsement by the American Association of Clinical Endocrinologists. Diabetes care, 2008. 31(8): p. 1679-1685.
- Gray, L. , et al., Implementation of the automated Leicester Practice Risk Score in two diabetes prevention trials provides a high yield of people with abnormal glucose tolerance. Diabetologia, 2012. 55(12): p. 3238-3244.
- Coetzee, A. , et al., The prevalence and risk factors for diabetes mellitus in healthcare workers at Tygerberg hospital, Cape Town, South Africa: a retrospective study. Journal of Endocrinology, Metabolism and Diabetes of South Africa, 2019. 24(3): p. 77–82-77–82.
- El_Jerjawi, N.S. and S.S. Abu-Naser, Diabetes prediction using artificial neural network. International Journal of Advanced Science and Technology, 2018. 121.
- NirmalaDevi, M., S. A. alias Balamurugan, and U. Swathi. An amalgam KNN to predict diabetes mellitus. in 2013 IEEE international conference on emerging trends in computing, communication and nanotechnology (ICECCN). 2013. IEEE.
- Alehegn, M., R. R. Joshi, and P. Mulay, Diabetes Analysis and Prediction Using Random Forest, KNN, Naïve Bayes And J48: An Ensemble Approach. Int. J. Sci. Technol. Res, 2019. 8(9): p. 1346-1354.
- Brown, G.C. , et al., Quality of life associated with diabetes mellitus in an adult population. Journal of Diabetes and its Complications, 2000. 14(1): p. 18-24.
- Tabaei, B.P. and W.H. Herman, A multivariate logistic regression equation to screen for diabetes: development and validation. Diabetes Care, 2002. 25(11): p. 1999-2003.
- Parthiban, G., A. Rajesh, and S. Srivatsa, Diagnosis of heart disease for diabetic patients using naive bayes method. International Journal of Computer Applications, 2011. 24(3): p. 7-11.
- Xu, W. , et al. Risk prediction of type II diabetes based on random forest model. in 2017 Third International Conference on Advances in Electrical, Electronics, Information, Communication and Bio-Informatics (AEEICB). 2017. IEEE.
- Al Jarullah, A.A. Decision tree discovery for the diagnosis of type II diabetes. in 2011 International conference on innovations in information technology. 2011. IEEE.
- Kumari, V.A. and R. Chitra, Classification of diabetes disease using support vector machine. International Journal of Engineering Research and Applications, 2013. 3(2): p. 1797-1801.
- Miotto, R. , et al., Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Scientific reports, 2016. 6(1): p. 1-10.
- Pham, T. , et al., Predicting healthcare trajectories from medical records: A deep learning approach. Journal of biomedical informatics, 2017. 69: p. 218-229.
- Tripathi, G. and R. Kumar. Early prediction of diabetes mellitus using machine learning. in 2020 8th International Conference on Reliability, Infocom Technologies and Optimization (Trends and Future Directions)(ICRITO). 2020. IEEE.
- Parte, R. , et al., Non-invasive method for diabetes detection using CNN and SVM classifier. International journal of research in engineering, science and management, 2019. 2: p. 659-661.
- Swapna, G., K. Soman, and R. Vinayakumar, Diabetes detection using ecg signals: An overview. Deep Learning Techniques for Biomedical and Health Informatics, 2020: p. 299-327.
- Hu, J. , et al., Raman spectrum classification based on transfer learning by a convolutional neural network: Application to pesticide detection. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy, 2022. 265: p. 120366.
- Al-Smadi, M. , et al., A transfer learning with deep neural network approach for diabetic retinopathy classification. International Journal of Electrical and Computer Engineering, 2021. 11(4): p. 3492.
- Spänig, S. , et al., The virtual doctor: an interactive clinical-decision-support system based on deep learning for non-invasive prediction of diabetes. Artificial intelligence in medicine, 2019. 100: p. 101706.
- Nguyen, B.P. , et al., Predicting the onset of type 2 diabetes using wide and deep learning with electronic health records. Computer methods and programs in biomedicine, 2019. 182: p. 105055.
- Ryu, K.S. , et al., A deep learning model for estimation of patients with undiagnosed diabetes. Applied Sciences, 2020. 10(1): p. 421.
- Prabhu, P. and S. Selvabharathi. Deep belief neural network model for prediction of diabetes mellitus. in 2019 3rd international conference on imaging, signal processing and communication (ICISPC). 2019. IEEE.
- Zelaya, C.V.G. Towards explaining the effects of data preprocessing on machine learning. in 2019 IEEE 35th international conference on data engineering (ICDE). 2019. IEEE.
- Deshmukh, D.H., T. Ghorpade, and P. Padiya. Improving classification using preprocessing and machine learning algorithms on NSL-KDD dataset. in 2015 International Conference on Communication, Information & Computing Technology (ICCICT). 2015. IEEE.
- Patro, S. and K.K. Sahu, Normalization: A preprocessing stage. arXiv preprint. arXiv:1503.06462, 2015.
- Saeys, Y., T. Abeel, and Y. Van de Peer. Robust feature selection using ensemble feature selection techniques. in Joint European conference on machine learning and knowledge discovery in databases. 2008. Springer.
- Seijo-Pardo, B. , et al., Ensemble feature selection: homogeneous and heterogeneous approaches. Knowledge-Based Systems, 2017. 118: p. 124-139.
- Zhang, N. , et al., An overview on restricted Boltzmann machines. Neurocomputing, 2018. 275: p. 1186-1199.
- Bonett, D.G. and T.A. Wright, Sample size requirements for estimating Pearson, Kendall and Spearman correlations. Psychometrika, 2000. 65(1): p. 23-28.
- Gómez-Peralta, F. , et al., When does diabetes start? Early detection and intervention in type 2 diabetes mellitus. Revista Clínica Española (English Edition), 2020. 220(5): p. 305-314.
- Gilmer, T.P. and P.J. O'Connor, The growing importance of diabetes screening. 2010, Am Diabetes Assoc. p. 1695-1697.
- Sabariah, M.M.K., S. A. Hanifa, and M.S. Sa'adah. Early detection of type II Diabetes Mellitus with random forest and classification and regression tree (CART). in 2014 International Conference of Advanced Informatics: Concept, Theory and Application (ICAICTA). 2014. IEEE.
| Ref | Techniques/ML Models | Methodology | Major Outcomes | Data Sources |
| [31] | Denosing AE | Normalization, training (704,587), validation (5000) and testing (76,214) | Performance was measured using AUC (0.907) | Mount Sinai Data Warehouse (ICD-9) |
| [32] | Modified Long Short-Term Memory (LSTM), Attention pooling layer | training, validation and testing: 2/3, 1/6 and 1/6 respectively from 53,208 admissions | Study produced maximum accuracy of 79% | EHR data from hospital patients |
| [33] | Restricted Boltzmann machine (RBM) and Recurrent Neural Network (RNN) | Feature selection, Min-Max normalization, train (80%), test (20%) | Sensitivity andprecision: 90.66%,75% respectively | PID Data from the UCI Repository |
| [34] | Modified 1-D CNN and FC layer | The data for training and testing: 15 samples, 10 samples; leave-one out cross-validation | AUC of Type I-Diabetes, Type II - Diabetes, healthy subjects: 0.9659, 0.9625, 0.9644 | Breath samples collected by MOS sensors with 1000-sec intervals |
| [35] | CNN, LSTM, and SVM | Heart rate variability (HRV) data from 71 ECG datasets. 5 fold cross-validation was used. | Validation accuracy of 95.7% was obtained. | ECG data sampled at 500Hz from 40 subjects |
| [36,37,38,39,40] | Deep Multi-Layer Perceptron (DMLP) | Train-test split, data transformation, k-fold cross validation, normalization, feature selection | Maximum accuracy 88.41%, maximum AUC 84.13%, Sensitivity 87.92%, f1 Score 0.808. | PID, Practice Fusion Dataset and HER dataset of |
| [41] | Deep Belief Network | Min-max normalization; feature selection by PCA; pre-training for RBMs; supervised fine-tuning | Sensitivity: 100%, F1 score: 0.808 | Practice Fusion dataset (9948 patients, ICD-9) |
| SN | Attributes | Datatype | Yes (as 1) | No (as 0) |
|---|---|---|---|---|
| 1 | Age | 20 ≤ Age ≤ 100 | ||
| 2 | Sex | Male and Female | Male (328) | Female (192) |
| 3 | Polyuria | Yes/No | 258 | 262 |
| 4 | Polydipsia | Yes/No | 233 | 287 |
| 5 | Sudden Weight Loss | Yes/No | 217 | 303 |
| 6 | Weakness | Yes/No | 305 | 215 |
| 7 | Polyphagia | Yes/No | 237 | 283 |
| 8 | Genital Thrush | Yes/No | 116 | 404 |
| 9 | Visual Blurring | Yes/No | 233 | 287 |
| 10 | Itching | Yes/No | 253 | 267 |
| 11 | Irritability | Yes/No | 126 | 394 |
| 12 | Delayed Healing | Yes/No | 239 | 281 |
| 13 | Partial Paresis | Yes/No | 224 | 296 |
| 14 | Muscle Stiffness | Yes/No | 195 | 325 |
| 15 | Alopecia | Yes/No | 179 | 341 |
| 16 | Obesity | Yes/No | 88 | 432 |
| 17 | Class | Positive/Negative | Positive (320) | Negative (200) |
| SN | Attributes | Chi Square | Mutual Information Gain | Variance Threshold | Voting |
|---|---|---|---|---|---|
| 1 | Age | ✕ | ✓ | ✕ | ✕ |
| 2 | Sex | ✓ | ✓ | ✓ | ✓✓ |
| 3 | Polyuria | ✓ | ✓ | ✓ | ✓✓ |
| 4 | Polydipsia | ✓ | ✓ | ✓ | ✓✓ |
| 5 | Sudden Weight Loss | ✓ | ✓ | ✓ | ✓✓ |
| 6 | Weakness | ✓ | ✓ | ✓ | ✓✓ |
| 7 | Polyphagia | ✓ | ✓ | ✓ | ✓✓ |
| 8 | Genital Thrush | ✕ | ✓ | ✓ | ✓ |
| 9 | Visual Blurring | ✓ | ✕ | ✓ | ✓ |
| 10 | Itching | ✓ | ✕ | ✕ | ✕ |
| 11 | Irritability | ✕ | ✓ | ✓ | ✓ |
| 12 | Delayed Healing | ✓ | ✕ | ✓ | ✓ |
| 13 | Partial Paresis | ✓ | ✓ | ✕ | ✓ |
| 14 | Muscle Stiffness | ✓ | ✓ | ✓ | ✓✓ |
| 15 | Alopecia | ✓ | ✓ | ✓ | ✓✓ |
| 16 | Obesity | ✕ | ✕ | ✕ | ✕ |
| F1-Measure | Recall | Precision | |
|---|---|---|---|
| Full (16) Features | |||
| Deep Belief Networks | 0.87 | 0.66 | 0.80 |
| Decision Tree | 0.72 | 0.62 | 0.72 |
| Random Forest | 0.79 | 0.76 | 0.65 |
| Logistic Regression | 0.86 | 0.59 | 0.67 |
| Support Vector Machine | 0.66 | 0.86 | 0.58 |
| k-Nearest Neighbors | 0.72 | 0.74 | 0.72 |
| All Qualified (13) Features | |||
| Deep Belief Networks | 0.92 | 0.88 | 0.88 |
| Decision Tree | 0.86 | 0.69 | 0.61 |
| Random Forest | 0.77 | 0.72 | 0.70 |
| Logistic Regression | 0.77 | 0.72 | 0.78 |
| Support Vector Machine | 0.89 | 0.69 | 0.68 |
| k-Nearest Neighbors | 0.89 | 0.66 | 0.80 |
| Strongly Qualified (8) Features | |||
| Deep Belief Networks | 1.00 | 0.92 | 1.0 |
| Decision Tree | 0.86 | 0.84 | 0.88 |
| Random Forest | 0.69 | 0.77 | 0.87 |
| Logistic Regression | 0.77 | 0.69 | 0.91 |
| Support Vector Machine | 0.77 | 0.88 | 0.83 |
| k-Nearest Neighbors | 0.86 | 0.88 | 0.91 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





