1. Introduction
An essential step in the data preparation phase of MRI-based artificial intelligence (AI) applications and studies is accurately classifying MR images since each image communicates specific anatomical or physiological information [
1]. An example is brain tumor segmentation algorithms requiring information from multiple MR modalities, as distinguishing between healthy brain tissue and tumors is often challenging. However, assuring that the right sequences are used for analysis (classification of sequences) might be a tedious and time-consuming task, especially when dealing with a large amount of data from various sources (multiple scanners, multiple treatment centers) due to possible inconsistent naming schemes. In particular, retrospective data collection yields additional challenges (non-prespecified protocols and sequences).
Gueld et al. demonstrated that classifying medical images based on image metadata (i.e., based on information stored in the DICOM header) is often unreliable [
2]. DICOM tags and the actual examination protocols applied are not always consistently matched. This is mainly done to improve imaging quality, for example, the implementation of different body region imaging protocols due to variabilities and differences among patients' anatomies [
2]. Harvey et al. report data labeling as the costliest part of radiomics studies [
3] and that consistent and unbiased labeling should be performed across the entire dataset to yield robust machine learning models [
3]. However, this can be challenging when large amounts of data are considered. Therefore, automatizing medical image retrieval and classifying data based on the content would be beneficial in terms of time efficiency, accuracy, and, ultimately reproducibility.
Compared to text-based image classification, content-based image classification (CBIC) is independent of inconsistencies between different image sources, is not affected by human error, and is less labor-intensive [
4]. CBIC methods for medical images include the use of traditional classification machine learning techniques such as K-nearest Neighbor (kNN) [
5], support vector machine [
6] (SVM), as well as deep learning methods [
7]. After the success of the deep convolutional neural network (DCNN), AlexNet [
8] in the ImageNet [
9] classification challenge, an increase of interest in DCNN has been seen when dealing with image classification tasks [
10,
11,
12]. In the context of medical image retrieval and classification using DCNNs, four different studies have been identified for the classification of body organs and MR images (Accuracy >90%) [13-16]. A summary of these models can be seen in Supplementary-
Table S1. A limitation of these methods is the inability to deal with the open-set recognition problem, i.e., the failure of a network trained to classify between a specific number of classes to handle unknown classes [
17]. The open-set recognition problem is a common issue when dealing with clinical cohorts since datasets exported from the hospitals' Picture archiving and communication system (PACS) usually include all available medical images and data, resulting in various medical image modalities and sequences.
In this work, we tackle this problem by training a DCNN-based MR image classifier (MR-Class) using a one-vs-all approach. One-vs-all classification is implemented to deal with the open-set recognition problem and thus would enable the handling of unknown classes. A comparison study of the published DCNNs (mentioned above) for medical image classification was first performed to determine the adopted DCNN model. Then, one-vs-all binary class-specific DCNN classifiers were trained to recognize a particular MR image, thus forming MR-Class.
2. Materials and Methods
2.1. Datasets
This study included three datasets: The training/validation cohort (C1) consisted of 320 primary/recurrent high-grade glioma (HGG) patients with a median of 9 image acquisition time points, resulting in 20101 MR images acquired between 2006 and 2018. The dataset was collected retrospectively from 23 scanners at the Heidelberg University Hospital (UKHD). The first testing cohort (C2) consisted of 197 HGG patients, with a median of 7 time points resulting in 11333 images acquired between 2009 and 2017. The dataset was collected retrospectively from 15 different scanners at the UKHD. A public data cohort (C3) was also utilized for the second testing of MR-Class. The data cohort was retrieved from the Cancer Genome Atlas Glioblastoma Multiforme (TCGA-GBM) data collection [
18]. The cohort included scans from 256 GBM patients with a median of 3 time points, resulting in 3522 MR images acquired between 1986 and 2019 and collected from 17 scanners. Patient demographics of all three cohorts can be seen in
Supplementary Table S2.
2.2. MR scans
Multiparametric MRIs (mpMRI) were collected from multiple scanners in all three datasets, resulting in heterogeneous modalities and MR sequence protocols (Supplementary-
Table S3). Conventional multislice (2D) acquired in the axial, sagittal, or coronal plane, as well as 3D scans, are present. The MR sequences found in the cohorts are the widely used sequences for brain tumor imaging [
19] in clinical routines and trials [
20,
21,
22]. All MR images found in the training cohort were included in the training. However, one-vs-all DCNN classifiers were only trained for T1w, contrast-enhanced T1w (T1wce), T2w, T2w fluid-attenuated inversion recovery (FLAIR), apparent diffusion coefficient (ADC), and susceptibility-weighted imaging (SWI). No SWI scans were found in C3. The in-plane resolution ranged from 0.33 x 0.33 to 2 x 2 mm for C1, 0.45 x 0.45 to 1.40 x 1.40 mm for C2 and 0.45 x 0.45 to 1.14 x 1.14 mm for C3. Slice thickness ranged from 0.9 to 7.5 mm in all MR scans. Human experts manually labeled each MR image through an in-house interactive labeling tool. The DICOM attributes "Series Description" (SD) and "Contrast/Bolus Agent" DICOM attribute were then extracted and compared to the derived labels to evaluate the metadata's consistency. Sample images found in the training and testing cohorts are shown in
Figure 1.
2.3. DCNNs comparison study
In the context of medical image retrieval and classification using DCNNs, three different DCNNs are present, i.e., ResNet-18 [
14], Φ-Net [
15], and DeepDicomSort [
16]. Hence, a comparison study was performed where the architecture that showed the highest classification accuracy was adopted in the one-vs-all training approach. Both 2D and 3D ResNet-18 were considered. C1 was used for training, while C2 was for independent testing. C3 was not included in the comparison study as it did not contain all considered MR scans. The comparison study was only performed with the images belonging to one of the six classes considered, resulting in 11246 MR from C1 (8997/80% for training, 2249/20% for validation) and 8326 MR from C2 for testing.
Brief descriptions of the exemplary models behind Φ-Net and DeepDicomSort are given. Visual Geometry Group (VGG) was introduced in 2014 by Simonyan and Zisserman in a paper titled "Very Deep Convolutional Networks for Large-Scale Image Recognition" [
23]. The VGG network architecture is simple, formed by 3x3 convolutional layers stacked on top of each other as depth increases, pooling layers, and fully connected output layers. Residual Networks (ResNet) were introduced in 2015 to deal with the degradation problem, i.e., the degradation of the network accuracy as the depth of the network increases [
24]. Besides the usual DCNN architecture for classification purposes (alternating stack of convolutional, activations, and pooling layers), ResNet introduces skip-connections that skip one or more layers. These skip connections fit the unmodified input from the previous layer to the next layer, preserving the original image signal by performing identity mapping. This results in preserving the norm of the gradient and solving the degradation problem. A softmax layer is appended to the end layer to produce probabilistic predictions of the classes. Schematics of the ResNet and VGG architectures are shown in Supplementary-
Figure S1. Besides the dimensionality increase, no changes were applied to the 3D ResNet-18 architecture. Diagrams and explanations of the architectures of Φ-Net [
14] and DeepDicomSort [
16] are presented in the authors' original papers.
2.3.1. Data preprocessing
Before training, different preprocessing steps were implemented. For the DCNNs trained with Φ-Net and DeepDicomSort, the preprocessing pipelines provided by the authors' GitHub pages were used. As for the 2D and 3D ResNet-18 DCNNs, magnetic field inhomogeneities of the T1w images were first corrected using the N4ITK algorithm [
25]. After reorienting to a common orientation, in-plane cropping was performed to remove background voxels. Then, to account for resolution variability, all MR scans were resampled to a uniform pixel spacing of 2x2 mm
2, and volumes were interpolated to a 2-mm slice thickness. Images were then cropped around the brain into a digital grid of 224x224x224. Padding was performed when the image shape was smaller than the target grid. Lastly, a z-score normalization of the brain voxels was applied to bring all MR images to the same intensity scale. The formula of the Z-score normalization is as follows:
where x is the voxel intensity, μ is the mean of the intensity distribution, and σ is the standard deviation.
2.3.2. DCNNs training and testing.
The 2D and 3D ResNet-18 DCNNs were trained using the deep learning Python library PyTorch (1.7.1) [
26]. A stochastic gradient descent optimizer with a momentum of 0.9 was used with a learning rate scheduler that started with 0.001 and decayed by 0.1 when the training loss did not decrease during three epochs. A categorical cross-entropy loss was considered as the loss function. A learning rate scheduler with a patience number of 3 was used. Early stoppage was performed when no improvement in the loss was observed for five successive epochs. The maximum number of epochs was 100. The batch size was 5 for the 3D ResNet and 50 for the 2D ResNet. The 2D ResNet-18 training included ten slices around the middle slice, extracted from the corresponding preprocessed MR scan acquisition plane. Φ-Net and DeepDicomSort were trained through the training code provided by the authors' GitHub pages. All 4 DCNNs were finally tested on the independent C2, with the 2D DCNNs classifying an MR image as a class through a majority vote (25 slices for DeepDicomSort, 10 slices for the 2D ResNet-18).
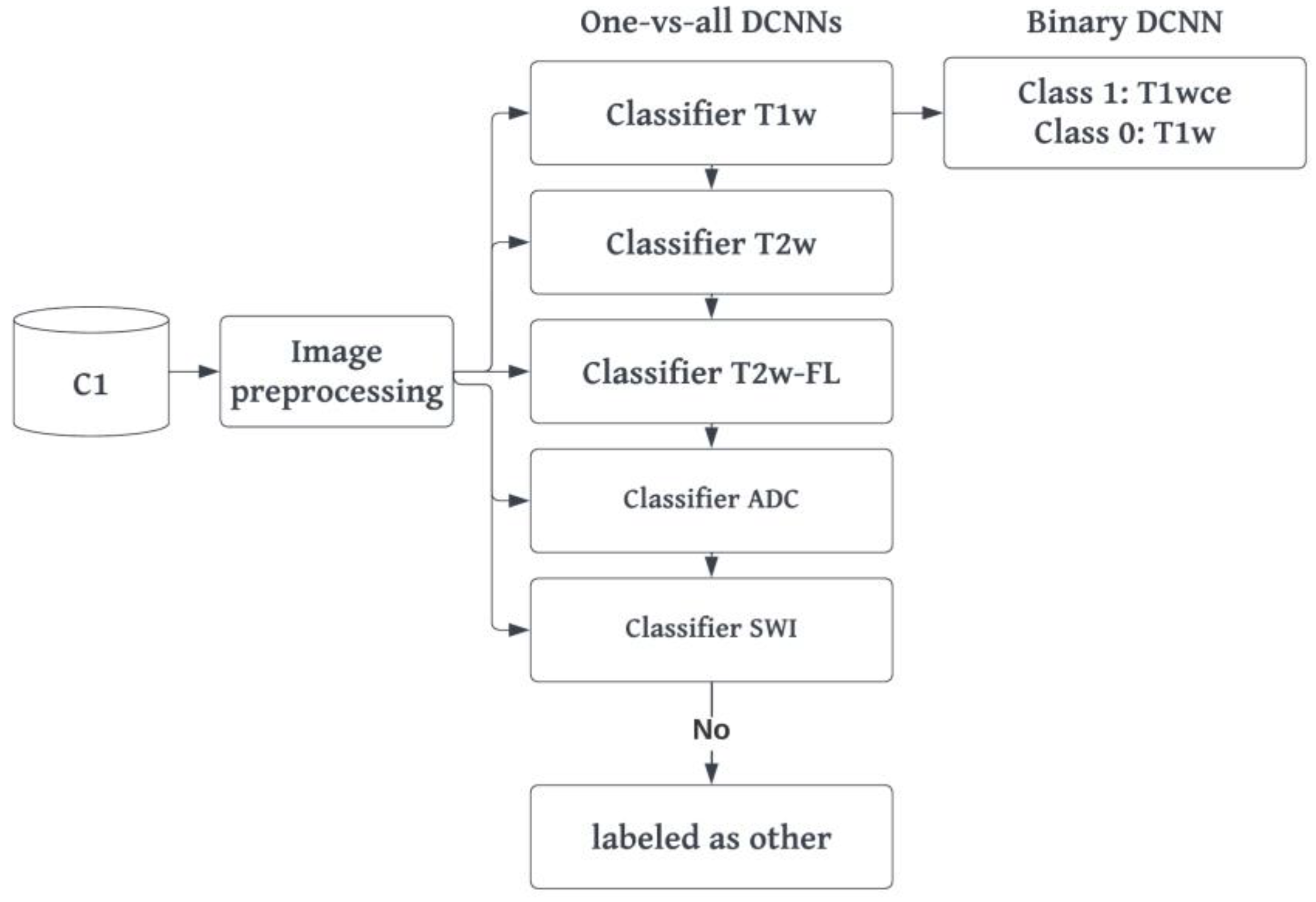
2.4. MR-Class: one-vs-all DCNNs
MR-Class consists of multiple one-vs-all binary classifiers rather than a single multi-class classifier, i.e., a classifier trained to classify all classes (as performed in the comparison study). The intuition behind training multiple one-vs-all DCNN is the open set recognition problem and that training a DCNN image classifier on every possible MR image is cumbersome. The architecture adopted for MR-Class was as of the DCNN, which showed the highest accuracy in the comparison study. The training was performed twice using scans from C1. The first training included all MR images available in the dataset, while the second was performed only with the image volumes of the six considered classes (same images included in the comparison study during training). The latter was performed to obtain a fair comparison of the performance of the one-vs-all dual-class classifiers (MR-Class) against a multi-class DCNN classifier, both trained on the same number of images. Classes for each binary classifier were defined as follows: class 1 included all images corresponding to the targeted class, whereas class 0 contained all remaining images in the dataset. A stratified (by class) 80%-20% dataset split was used for training and validation (
Table 1).
2.4.1. Training and preprocessing
The preprocessing and training approach implemented for the 2D/3D ResNet-18 are likewise applied for the one-vs-all DCNNs. However, further steps were taken to address the imbalanced classes arising from the one-vs-all classification design. First, data augmentation was implemented using the TorchIO python library [
27]. Specifically, the transformations implemented included adding random Gaussian noise, blurring, performing random affine or elastic deformations, and adding random MR motion artifacts like motion, ghosting, or spikes. Second a weighted binary categorical cross-entropy loss was used, where the weights of a class were equal to the size of the largest class divided by the size of that specific class. For example, for the T2w-vs-all DCNN, if class T2w has 1970 and class all has 18131 MR images, the weights would be 9.2 and 1.0, respectively. Finally, the learning rate scheduler was adjusted to decay based on the targeted class training loss instead of the loss of both classes. A summary of the training workflow can be seen in
Figure 2.
2.4.2. Inference and testing.
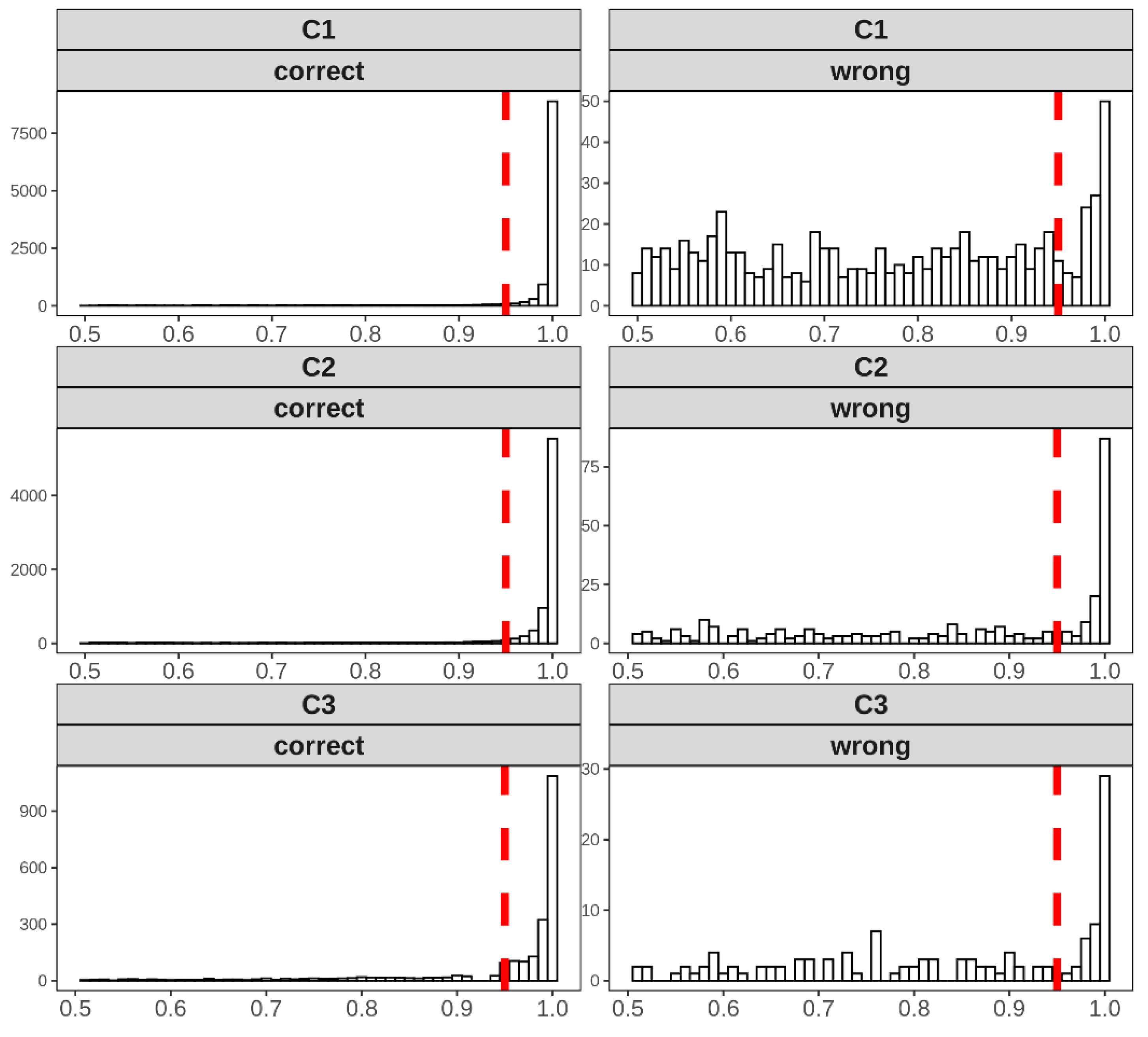
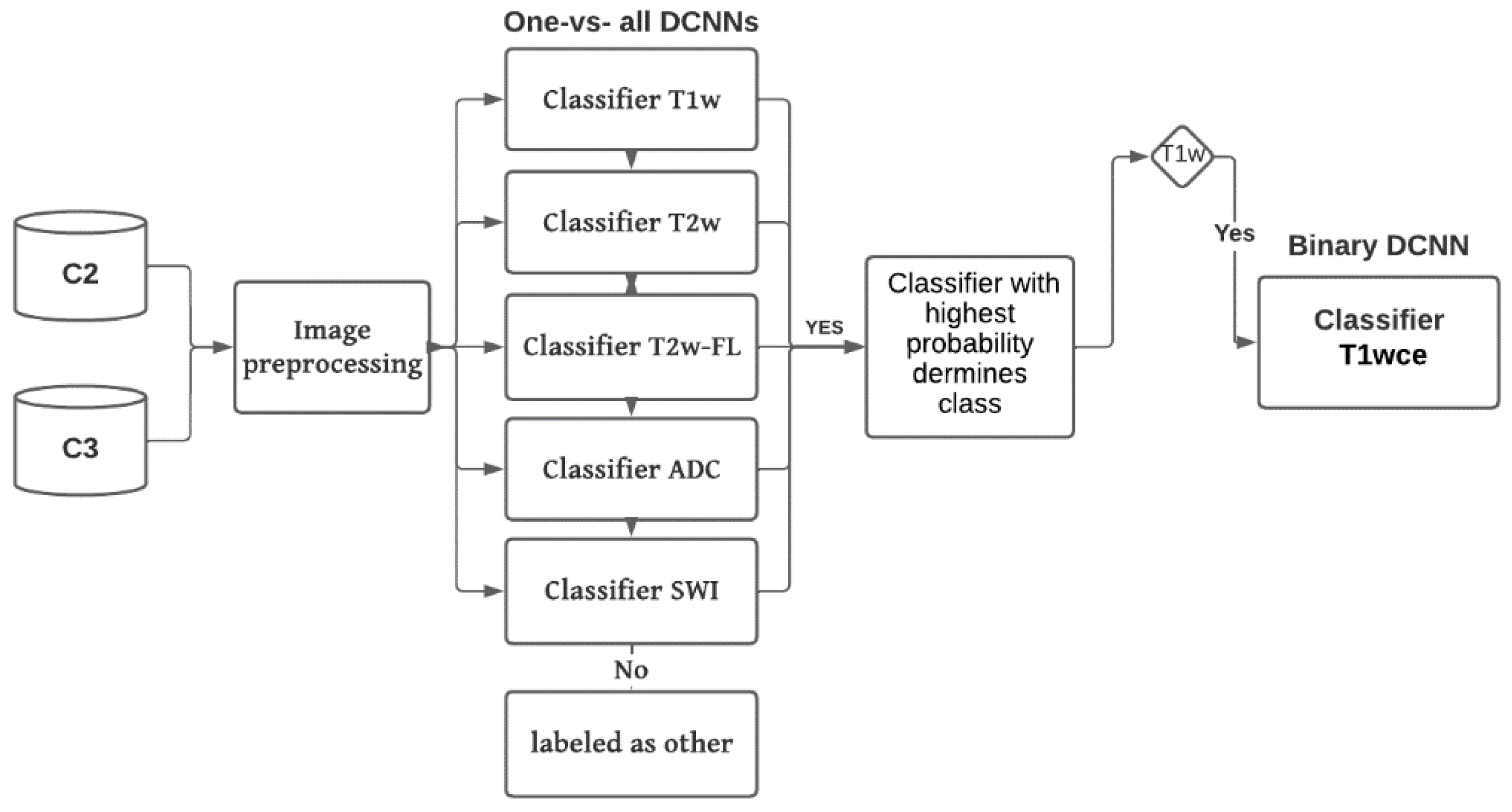
C2 and C3 were used to perform independent testing of MR-Class. In inference mode, the MR images were preprocessed (same as in training) and fed to each DCNN classifier to infer the corresponding class. A classification probability threshold of 0.95 was used. The cutoff threshold value was determined based on the distribution of the probabilities of correct and wrong labeled images when C1 was inferred back to MR-Class (
Figure 5). If an image is labeled by more than one classifier, the classifier with the highest probability determines the class. If none of the classifiers labels an image (i.e., assigned to class 0 by each classifier), it is unclassifiable. The 2D DCNNs classify an MR scan as a class using a majority vote of 10 inferred slices extracted around the middle slice of the corresponding MR acquisition plane.
Figure 3 shows a summary of the inference workflow.
Classifications were compared to ground truth labels, where the number of correct predictions divided by the total number of images derived the accuracy. The 95% confidence interval (CI) was calculated as the Wilson interval [
28]. Classification sensitivity and specificity were calculated to evaluate the performance of each classifier. Lastly, the misclassified images were analyzed to identify the causes of misclassifications.
2.5. MR-Class application: progression-free survival prediction modeling
To demonstrate the applicability of MR-Class in MR-based radiomics applications, Cox proportional hazard models (CPHs) were trained with the T1wce MR sequences of cohort C2 to predict the patients' progression-free survival (PFS) after performing a text-based curation using the DICOM SDs and a content-based curation using MR-Class [
29]. PFS was calculated as the number of days between the beginning of the radiotherapy treatment and disease progression. Progression events were derived from the clinical follow-ups' reports. After performing a series of preprocessing steps on both curated datasets (DICOM SD-based and MR-Class-based curated datasets), radiomics features were calculated automatically from the gross tumor volume (GTV) segmentations extracted from the DICOM RT structure set and the original image, as well as from derived images (Wavelet and Laplacian of Gaussian filtering) from each dataset using Pyradiomics (v 3.0) [
30]
. The image preprocessing diagram is shown in
Supplementary Figure S3. The different feature classes and corresponding feature numbers can be seen in
Supplementary Table S5. A Spearman rank-order correlation coefficient was next used on the total number of features to exclude redundant features (rs>0.80). Three feature selection methods, including a univariate analysis under Cox proportional hazard (CPH) models (P<0.05), a random forest (RF) -based method, and lasso regression, were applied on 1000 random subsamples of the text-based curated and MR-Class curated T1wce datasets (10% left out) separately to identify features correlated to PFS. Significant features identified at least 950 times were selected, and survival analyses were conducted using CPH. Model performances were finally evaluated based on the resampled concordance index (C-I).
3. Results
3.1. Metadata consistency
Between all three datasets, 2704 different DICOM SDs were found (an overview of the number of SDs found for each MR scan is shown in
Supplementary Table S4). 11.4%, 10.6%, and 10.7% of the SDs for C1, C2, and C3, respectively, had misleading or inconsistent entries, not allowing for the proper identification of the MR image class (
Table 2).
3.2. DCNN comparison study
Table 3 summarizes the testing C2 MR scan classification accuracies of all four multi-class DCNN classifiers.
Table 3.
Classification accuracy of the different DCNN architectures in study. T2w-FL: T2w-FLAIR.
Table 3.
Classification accuracy of the different DCNN architectures in study. T2w-FL: T2w-FLAIR.
| |
2D-ResNet |
DeepDicomSort |
Φ-Net |
3D-ResNet |
| T1w |
98.4 |
98.8 |
97.7 |
96.5 |
| T1wce |
97.4 |
95.2 |
97.5 |
96.2 |
| T2w |
98.1 |
97.2 |
96.6 |
97.1 |
| T2w-FL |
99.7 |
99.4 |
96.5 |
98.7 |
| ADC |
99.9 |
99.3 |
98.5 |
99.2 |
| SWI |
98.2 |
98.5 |
97.5 |
98.9 |
| All |
98.6 |
98.1 |
97.4 |
97.8 |
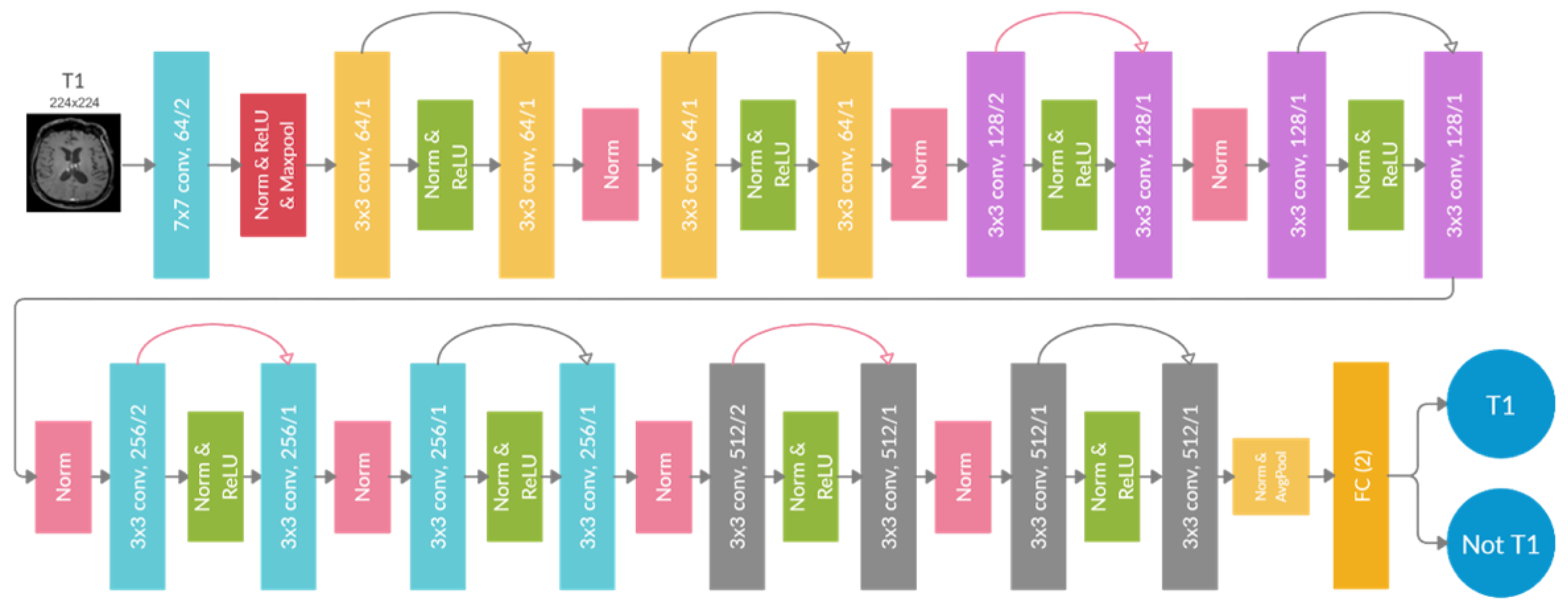
All classifiers achieved high comparable accuracy, with the 2D ResNet-18 having the highest overall accuracy of 98.6%. The training took 18-20 hours for the 3D DCNN (Φ-Net and 3D ResNet-18) and 8-10 hours for the 2D DCNN (DeepDicomSort and 2D ResNet-18) on an Intel Xeon processor with 8 cores and 32 Gb of RAM and a graphics card NVIDIA GeForce GTX 1060 (6 Gb). The average inference time is 0.15 s for a single 2D slice and 4.92 s for a 3D image. Thus, the DCNN one-vs-all architecture implemented in MR-Class was that of the 2D ResNet-18 (a schematic representation is shown in
Figure 4).
3.3. MR-Class: one-vs-all DCNNs
Table 3 summarizes the classification accuracies in the validation sets of all six DCNN classifiers on C1.
Table 3.
Validation classification accuracies of all six binary DCNN classifiers on C1. T2wFL: T2w-FLAIR.
Table 3.
Validation classification accuracies of all six binary DCNN classifiers on C1. T2wFL: T2w-FLAIR.
| Classifier |
Val Acc (%) |
Classifier |
Val Acc (%) |
| T1w-vs-all |
99.1 |
T2wFL-vs-all |
99.4 |
| T1w-vs-T1wce |
97.7 |
ADC-vs-all |
99.6 |
| T2w-vs-all |
99.3 |
SWI-vs-all |
99.7 |
All six classifiers have high validation accuracies, with the lowest at 97.7% for the T1w-vs-T1wce and the highest at 99.7% for the SWI-vs-all and 99.6% for the ADC-vs-all tasks. Passing back the training set dataset I to MR-Class in inference mode, an accuracy of 97.4% [95% CI: 96.2, 98.4] is obtained, i.e., out of 20101 MR scans, MR-Class could not learn 519. As for the multi-class vs multiple binary one-vs-all classification experiment, where only the image volumes of the six considered MR sequences were regarded, the validation accuracy was comparable with 98.6% and 98.1%, respectively.
Distributions of the classification probabilities derived by MR-Class for all 3 cohorts are shown in
Figure 5. Based on C1, a probability cutoff threshold of 0.95 was set for testing MR-Class on C2 and C3.
Figure 5.
Distribution of the probabilities of correct and wrong labeled images for all 3 cohorts in study when inferred to MR-Class. Based on the distributions of C1, a cutoff classification threshold probability of 0.95 was used. Histogram bin width = 0.01.
Figure 5.
Distribution of the probabilities of correct and wrong labeled images for all 3 cohorts in study when inferred to MR-Class. Based on the distributions of C1, a cutoff classification threshold probability of 0.95 was used. Histogram bin width = 0.01.
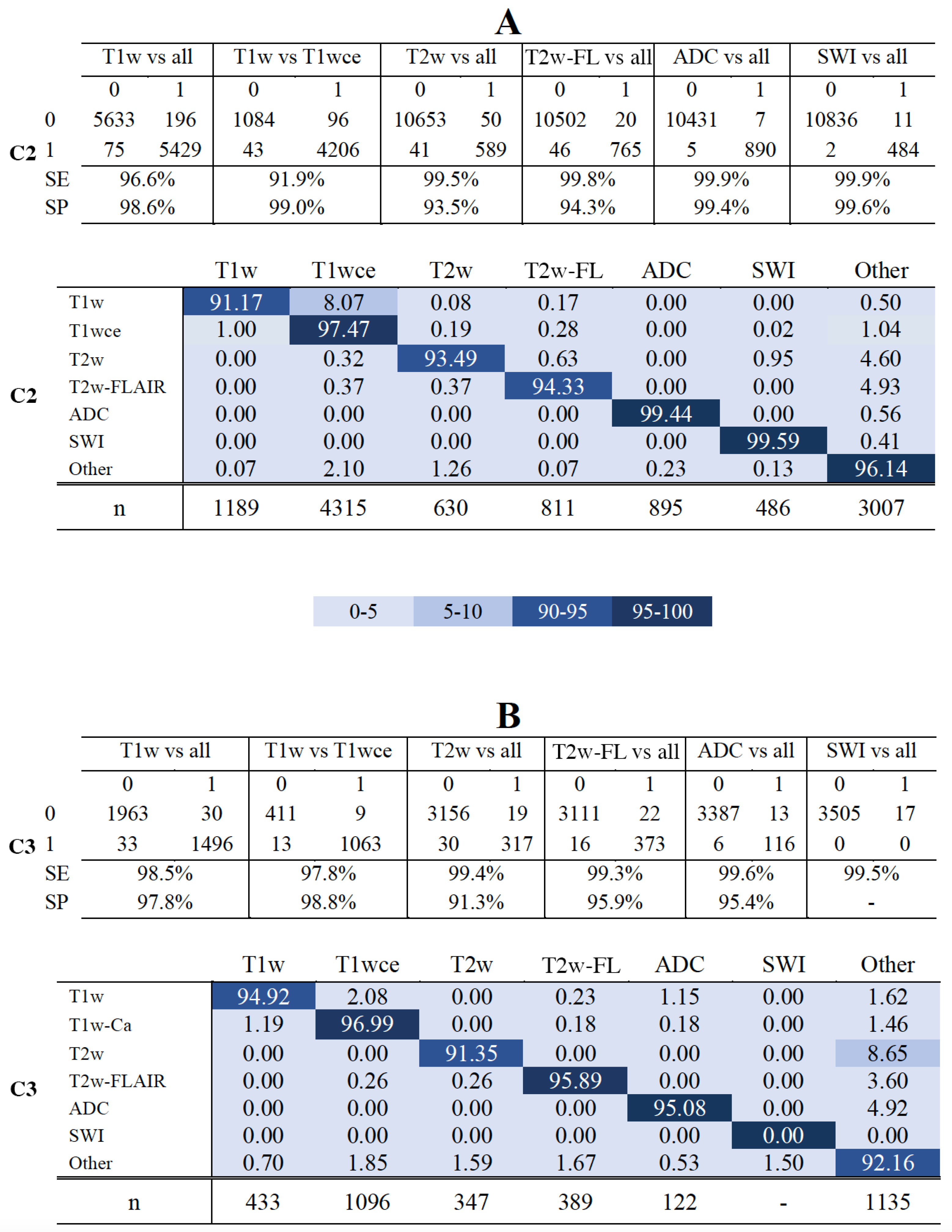
MR-Class's accuracy against the independent C2 was 96.7% [95% CI: 95.8, 97.3], i.e., 424 out of 11333 images were misclassified. All DCNNs had a specificity ranging between 93.5% (T2w-vs-all) and 99.6% (SWI-vs-all). The T1w-vs-T1wce and T1w-vs-all had the lowest sensitivity with 91.9% and 96.6%, while all remaining DCNNs had a high sensitivity (>99%) (Figure 6-A, upper panel). In the multi-class normalized confusion matrix (Figure 6-A, lower panel), it is seen that the classification of T1w is the least reliable, with an accuracy of 91.17%. Against the independent C3, MR-Class achieved an accuracy of 94.4% [95% CI: 93.6, 96.1] with 196 misclassified scans out of 3522. The T1w-vs-T1wce had the lowest sensitivity with 97.4%, while all remaining DCNNs had a sensitivity larger than 98%. Specificity ranged between 91.3% (T2w-vs-all) and 98.8% (T1w-vs-T1wce) (Figure 6-B, upper panel). In the multi-class confusion matrix (Figure 6-D, lower panel), it is seen that the classification of T2w is the least reliable, with an accuracy of 91.35%, with 8.65% classified as "other". Investigations on the misclassified images were performed in the next section.
Figure 6.
Confusion matrices of the 6 DCNNs for C2 (A) and C3 (CB) - The upper panels in A and B show the confusion matrices for datasets C2 and C3. - The lower panels in A and B show MR-Class normalized confusPanelsatrices for datasets C2 and C3, i.e., the percentages (%) of correct classification results per class. SE: sensitivity; SP: specificity. Class ' Other': when none of the DCNNs label an image; n: number of scans per class, T2w-FL: T2w-FLAIR.
Figure 6.
Confusion matrices of the 6 DCNNs for C2 (A) and C3 (CB) - The upper panels in A and B show the confusion matrices for datasets C2 and C3. - The lower panels in A and B show MR-Class normalized confusPanelsatrices for datasets C2 and C3, i.e., the percentages (%) of correct classification results per class. SE: sensitivity; SP: specificity. Class ' Other': when none of the DCNNs label an image; n: number of scans per class, T2w-FL: T2w-FLAIR.
3.4. Analyses of misclassified images.
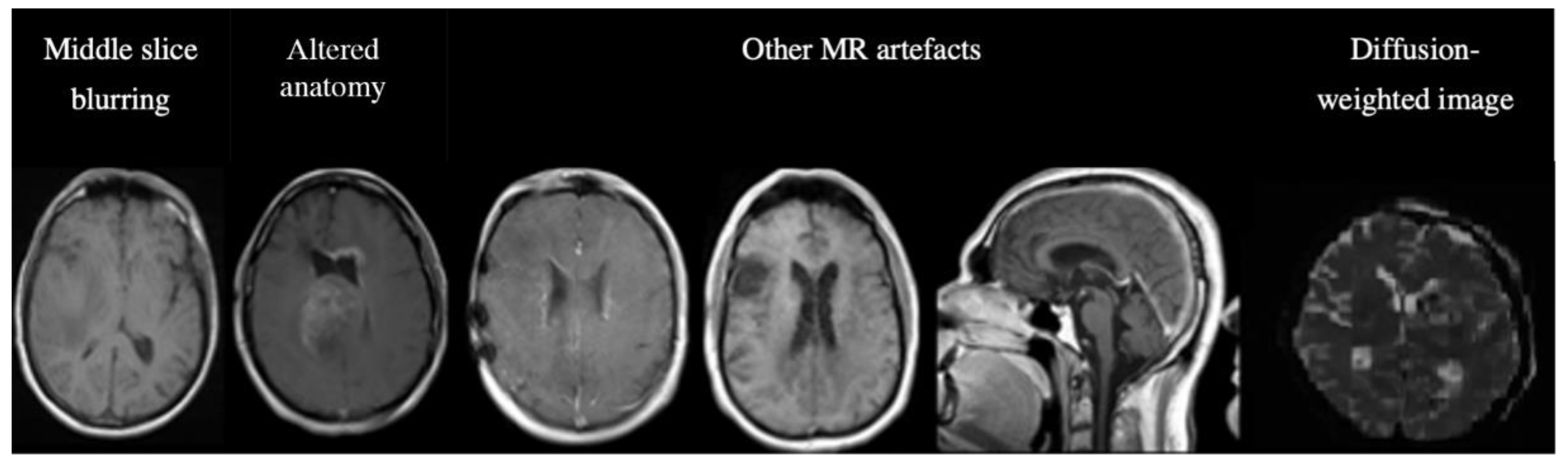
Out of the 14855 inferred images from C2 and C3, MR-Class classified 620 images incorrectly. The misclassifications can be sorted into different categories: MR artifact-middle slice blurring, MR artifacts-other, similar image content for different MR sequences (e.g., a T1w-FLAIR sequence instead of T2w), misclassified diffusion-weighted imaging (DWI) as T2w, and DICOM corrupted scans (sample images shown in Figure 6).
Figure 6.
Examples of misclassified images. The first two images are examples of a misclassified MR, possibly due to blurry images (left) and alterations in expected anatomy (displaced ventricles, large tumor, right). The next three MR images show incorrect predictions due to different MR artifacts (Shading, motion, aliasing). All of these images are falsely classified as "other". The last image is a diffusion-weighted image (DWI), specifically a Trace DWI, misclassified as T2w.
Figure 6.
Examples of misclassified images. The first two images are examples of a misclassified MR, possibly due to blurry images (left) and alterations in expected anatomy (displaced ventricles, large tumor, right). The next three MR images show incorrect predictions due to different MR artifacts (Shading, motion, aliasing). All of these images are falsely classified as "other". The last image is a diffusion-weighted image (DWI), specifically a Trace DWI, misclassified as T2w.
A manual evaluation revealed frequent misclassification (n=122, 19.68%) if the architecture of the ventricles was altered, e.g., displaced by large tumors. This was assessed in detail: we used 122 random, correctly labeled images as a reference group. After manual segmentation of the GTVs and brain, we calculated the Euclidean distance between the brain's center of mass (CoM) and the CoM of the tumor volume. A t-test was then performed between the reference and misclassified CoM distributions. The t-test returned a p-value of 0.04, with a median CoMs distance of 46.15 voxels for the correctly labeled images and 66.31 for the misclassified images. This result shows a statistical difference between the groups, i.e., the further the GTV is from the ventricles, the less likely the image is misclassified. The frequencies of the misclassification categories are shown in
Table 4.
Table 4.
Frequency (n) and percentage (%) of the misclassified images.
Table 4.
Frequency (n) and percentage (%) of the misclassified images.
| Category |
n |
% |
| MR artifact-other |
146 |
26.84 |
| MR artifact-middle slice blurring |
127 |
23.35 |
| Tumor/GTV displacing ventricles |
122 |
22.43 |
| Similar content- different sequence |
80 |
14.71 |
| DWI as T2w |
76 |
13.97 |
| DICOM corrupted images |
69 |
12.68 |
3.5. MR-Class application: progression-free survival prediction modeling
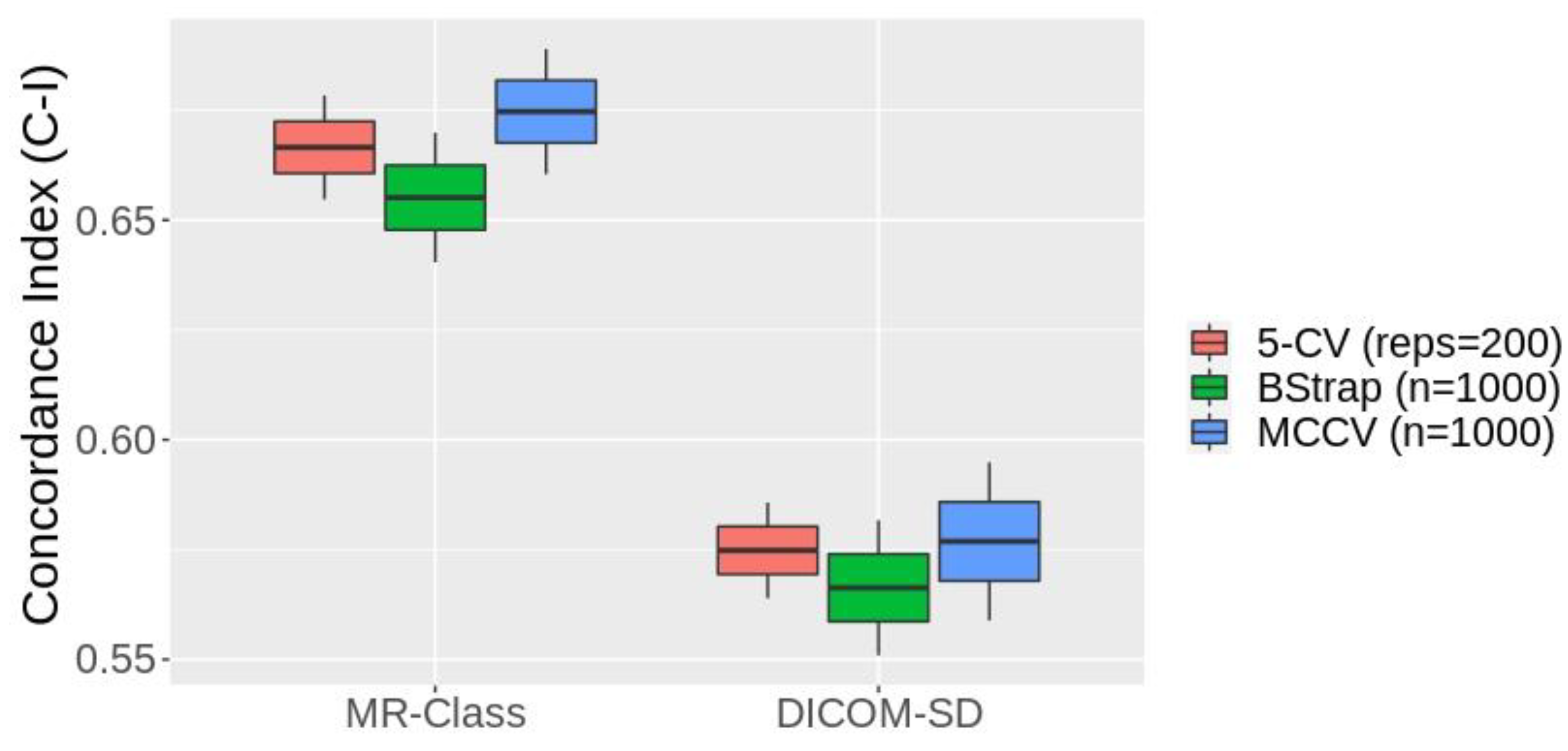
Figure 7 shows the box plots of the 1st - 99th percentiles resulting from the three resampling approaches following the fitting of the PFS CPH models by the radiomics signatures derived from the text-based and MR-Class-based curated datasets. 4 and 2 significant features were identified from the text-based and MR-Class-based curated datasets. The average C-Is across the three different resampling approaches are 0.57 [0.55 0.59] and 0.66 [0.64 0.68] for the DICOM-SD and MR-Class models. The range represents the minimum and maximum C-I achieved. The DICOM SD curated included 7 misclassified T1w and 3 T2w sequences and excluded 10 T1wce images. The MR-Class curated dataset excluded 4 misclassified T1wce images as they were labeled as "other".
4. Discussion
In this manuscript, we present an MRI image sequence classifier, MR-Class, which differentiates between T1w, contrast-enhanced T1w, T2w, T2w-FLAIR, ADC, and SWI while handling unknown classes. Testing was performed on two independent cohorts, where classification accuracies of 96.7% [95% CI: 95.8, 97.3] and 94.4% [95% CI: 93.6, 96.1] were observed. MR-Class consists of 5 one-vs-all DCNNs (one for each class), followed by a binary classifier for T1w images to determine whether a contrast agent was administrated. This design enables MR-Class to handle unknown classes since each DCNN only classifies an image if it belongs to its respective class, and thus an image not labeled by any of the DCNNs is rendered as unknown. To compare the performance of such a design to the basic multi-class classification approach, we performed the multi-class vs multiple dual-class classification experiment. We observed that both methods have comparable classification results (multi-class: 98.6% multiple one-vs-all: 98.1%) in the context of MR brain image classification. However, the latter can deal with the open-set recognition problem, frequently encountered when handling data from clinical cohorts, and thus can help reduce MRI study design times.
MR image DICOM series description (SD) entries usually follow the MR sequence protocol applied. However, they are MR model specific and are sometimes edited by clinical staff. We observed around 10% discrepancies in each cohort when the SD was compared to the manually derived labels. Typical SDs that do not allow for clear MR scan classifications are SDs with only the sequence name, e.g., Spin Echo (SE), or the scan direction, e.g., axial, which can stand for any MR sequence. Typographical errors and empty SD attributes were also observed.
Overall, high accuracies were obtained across all DCNNs in the comparison study. In conjunction with the high performance achieved in literature in medical image classification [
13,
14,
16,
17], it is apparent that DCNNs can learn the intricacies behind different medical image modalities. The 2D ResNet-18 had the best overall accuracy in the DCNN architecture comparison study and thus was the architecture chosen for MR-Class. Furthermore, it was seen that the 2D DCNNs outperformed their 3D counterparts in MR sequence classification. MR scans correctly classified by the 2D DCNNs, while misclassified by the 3D DCNNs, are mainly conventional 2D axial, sagittal, or coronal scans with slice thickness ranging between 5 and 9mm. Scans with a field of view that only encompassed the tumor area were misclassified by both 3D DCNNs (representative images can be seen in Suppl. fig. 3). It is important to note that no data augmentation was performed in the comparison study.
All six one-vs-all classifiers have high validation accuracies, with the lowest being 97.7% for the T1w vs T1wce. After passing the training cohort C1 to MR-Class in inference mode, it was observed that 519 images could not be learned, out of which 336 belonged to class "other", representing 3.8% of the other images used for training. This low error percentage demonstrates that MR-Class can learn to handle different sequences indirectly.
The testing of MR-Class against C2 and C3 yielded an average accuracy of 96.1%, where 620 images (4.2%) were classified incorrectly. Overall, the T2w-vs-all had the worst performance, with a specificity of 93.5% and 91.3% in C2 and C3. This is mostly due to the presence of diffusion-weighted imaging (DWI) sequences (frequently encountered in the datasets), which are inherently a series of T2w sequences. Similarly, C3 included T1w-FLAIR images falsely misclassified as T1w or T2w-FLAIR. It is thus apparent that different sequences with similar content are prone to misclassification by MR-Class. A solution could be to train a subsequent classifier to distinguish between similar sequences, as performed for the T1wce images. Most of the other incorrectly classified images had severe blurring or had other types of MR artifacts. These were observed in a higher prevalence in C3 than in C2. A reason could be the time interval in which the cohort was collected. Most of these classifications were false negatives, i.e., they were labeled as unclassifiable by MR-Class. This can be beneficial for radiomics models since any corrupted image would be automatically disregarded, and all images labeled as a specific class would have similar content. Another subset of the misclassified images showed tumor volumes overlapping the ventricles. Statistical analysis was performed between these misclassified images and a subset of the correctly labeled images, confirming altered anatomy (here: ventricle displacement by large tumors) as a possible reason for misclassification. More detailed analyses are warranted to assess further the impact of surgery on alterations of overall anatomy (i.e., biopsy, partial resection, total resection), as well as on tumors (chemo/radiotherapy) as the latter might, e.g., change the pattern of contrast enhancement.
An essential step in building a radiomics application is to verify the input data labels before training the machine learning model, as inconsistent data can lead to the model drastically failing [
31]. However, this was not performed while building the different survival models to demonstrate the applicability of MR-Class in MR-based radiomics applications. CPHs models were built with the T1wce MR sequences of cohort C2 to predict the patients' PFS after performing a text-based curation using the DICOM SDs and a content-based curation using MR-Class. The MR-Class curated model achieved an average C-I increase of 14.6 %. This is mainly due to the content dissimilarity between the different images in the DICOM SD curated dataset compared to the MR-Class curated dataset.
MR-Class can facilitate the preparation of longitudinal studies for RT treatment assessment as MR data from the three cohorts include scans taken before, after, and throughout the delivery of the RT fractions, which resulted in different tumor volume masses between the different scans, as well as apparent radiation scaring in some of the MR images. Furthermore, the data cohort includes images taken directly after the surgical resection of the tumor, resulting in visible surgical holes and void tumor beds.
The 2D DCNN in this study outperformed their 3D counterparts in classifying MR images. This was mostly due to multiple conventional 2D multislice MR scans acquired in the axial, sagittal, or coronal plane in the 3 cohorts. The classification of MR brain images, as shown by MR-Class and DeepDicomSort, is possible and leads to high classification results. However, the classification of MR sequences of a different entity, e.g., Abdominal and Pelvic MRI, might be more challenging and demand the intrinsic power of 3D DCNN. However, due to the frequent presence of 2D in MR datasets, reconstruction of these low-resolution 2D slices to a high-resolution 3D MR might be a necessary preprocessing step before training. Nevertheless, the one-vs-all classification pipeline implemented in this study on brain MR images can be used for different anatomy sites and other medical image classification problems, for example, the classification of different body parts and organs.
5. Conclusions
MR-Class is a helpful, ready-to-use python tool for the data preparation of MR-based research studies in brain MRI. It eliminates the need to manually sort out the images, a tedious task due to large amounts of data and different naming schemes. Furthermore, since MR-Class classifies images based on the content rather than the metadata, any corrupted image would be automatically disregarded, and all images labeled as a specific class would have related content. Hence, we believe MR-Class is a useful and time-efficient tool for big data MR radiomics-based studies. Future work includes the addition of modalities and sequences to MR-Class for different anatomy sites.
Supplementary Materials
The following supporting information can be downloaded at: www.mdpi.com/xxx/s1. Figure S1: An example of ResNet and VGG architectures with 18 and 16 layers and two output neurons.; Figure S2: Images classified correctly and wrongly by 3D and 2D classifiers; Figure S3: T1wce preprocessing diagram applied before survival prediction modeling; Table S1: state of the art methods specifications; Table S2: Patient demographics; Table S3: MR models found in the cohorts; Table S4: Nr. of series descriptions found for each class; Table S5: The number of shape, first and second-order statistics derived per sequence and calculated on both the original and derived images.
Author Contributions
Conceptualization, Patrick Salome, Francesco Sforazzini, Amir Abdollahi and Maximilian Knoll; Data curation, Patrick Salome and Francesco Sforazzini; Formal analysis, Patrick Salome, Francesco Sforazzini and Maximilian Knoll; Funding acquisition, Jürgen Debus and Amir Abdollahi; Investigation, Gianluca Grugnara, Christel Herold-Mende and Sabine Heiland; Methodology, Patrick Salome and Maximilian Knoll; Resources, Andreas Kudak and Matthias Dostal; Software, Patrick Salome and Francesco Sforazzini; Supervision, Jürgen Debus, Amir Abdollahi and Maximilian Knoll; Visualization, Patrick Salome; Writing – original draft, Patrick Salome; Writing – review & editing, Francesco Sforazzini, Gianluca Grugnara, Christel Herold-Mende, Sabine Heiland, Amir Abdollahi and Maximilian Knoll.
Funding
This study was found by the H2020 MSCA-ITN PREDICT project, Grant Number 766276 and intramural funds of the National Center for Tumor Diseases (NCT) and German Cancer Consortium (DKTK) Radiation Oncology programs.
Institutional Review Board Statement
Ethical approval Institutional Review Board Statement: The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of the Medical Faculty of Heidelberg University (approval number S-540/2010, date of last updated approval: 20 July 2020).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the S-540 study.
Data Availability Statement
Conflicts of Interest
P.S. No relevant relationships. F.S. No relevant relationships. A.K. No relevant relationships. N.B. No relevant relationships. J.D. Grants/contracts from/with Viewray, CRI – The Clinical Research Institute, Accuray International Sarl, RaySearch Laboratories, Vision RT, Merck Serono, Astellas Pharma, AstraZeneca, Siemens Healthcare, Solution Akademie, Ergomed PLC Surrey Research Park, Quintiles, Pharmaceutical Research Associates, Boehringer Ingelheim Pharma & CoKG, PTW-Freiburg Dr. Pychlau, Nanobiotix, Accuray, Varian; participation on a data safety monitoring board or advisory board for Merck Serono. A.A. Predict MarieCurie innovative training network (ITN), in frame of Horizon 2020 from the European Union, Marie Skłodowska-Curie grant agreement No 766276. M.K. No relevant relationships.
References
- Mangrum W, Christianson K, Duncan S, et al. Duke Review of MRI Principles. Mosby. (2012) ISBN:1455700843.
- Gueld MO, Kohnen M, Keysers D, Schubert H, Wein BB, Bredno J, et al. Quality of DICOM header information for image categorization. In: Medical imaging 2002: PACS and integrated medical information systems: design and evaluation. 2002. p. 280–7. [CrossRef]
- Harvey H, Glocker B. A standardized approach for preparing imaging data for machine learning tasks in radiology. In: Artificial Intelligence in Medical Imaging. Springer; 2019. p. 61–72. [CrossRef]
- Ferreira PM, Figueiredo MAT, Aguiar PMQ. Content-Based Image Classification: A Non-Parametric Approach.
- Wagle S, Mangai JA, Kumar VS. An improved medical image classification model using data mining techniques. In: 2013 7th IEEE GCC Conference and Exhibition (GCC). IEEE; 2013. p. 114–8. [CrossRef]
- Varol E, Gaonkar B, Erus G, Schultz R, Davatzikos C. Feature ranking based nested support vector machine ensemble for medical image classification. In: 2012 9th IEEE International Symposium on Biomedical Imaging (ISBI). 2012. p. 146–9. [CrossRef]
- Shen D, Wu G, Suk H-I. Deep learning in medical image analysis. Annu Rev Biomed Eng. 2017;19:221–48. [CrossRef]
- Alom MZ, Taha TM, Yakopcic C, Westberg S, Sidike P, Nasrin MS, et al. The history began from alexnet: A comprehensive survey on deep learning approaches. arXiv Prepr arXiv180301164. 2018. [CrossRef]
- Deng J, Dong W, Socher R, Li L-J, Kai Li, Li Fei-Fei. ImageNet: A large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition. IEEE; 2009. p. 248–55. [CrossRef]
- Liang M, Tang W, Xu DM, Jirapatnakul AC, Reeves AP, Henschke CI, et al. Low-Dose CT Screening for Lung Cancer: Computer-aided Detection of Missed Lung Cancers. Radiology. 2016 Oct;281(1):279–88. [CrossRef]
- Setio AAA, Ciompi F, Litjens G, Gerke P, Jacobs C, van Riel SJ, et al. Pulmonary Nodule Detection in CT Images: False Positive Reduction Using Multi-View Convolutional Networks. IEEE Trans Med Imaging. 2016 May;35(5):1160–9. [CrossRef]
- Kang G, Liu K, Hou B, Zhang N. 3D multi-view convolutional neural networks for lung nodule classification. Deng Y, editor. PLoS One. 2017 Nov 16;12(11): e0188290. [CrossRef]
- Qayyum A, Anwar SM, Awais M, Majid M. Medical image retrieval using deep convolutional neural network. Neurocomputing. 2017; 266:8–20. [CrossRef]
- Ayyachamy S, Alex V, Khened M, Krishnamurthi G. Medical image retrieval using Resnet-18. In: Medical Imaging 2019: Imaging Informatics for Healthcare, Research, and Applications. 2019. p. 1095410. [CrossRef]
- Remedios S, Pham DL, Butman JA, Roy S. Classifying magnetic resonance image modalities with convolutional neural networks. In: Medical Imaging 2018: Computer-Aided Diagnosis. 2018. p. 105752I. [CrossRef]
- van der Voort SR, Smits M, Klein S. DeepDicomSort: An Automatic Sorting Algorithm for Brain Magnetic Resonance Imaging Data. Neuroinformatics. 2021;19(1):159–84. [CrossRef]
- Scheirer WJ, de Rezende Rocha A, Sapkota A, Boult TE. Toward open set recognition. IEEE Trans Pattern Anal Mach Intell. 2012;35(7):1757–72. [CrossRef]
- Scarpace L, Mikkelsen T, Cha S, Rao S, Tekchandani S, Gutman D, et al. Radiology data from the cancer genome atlas glioblastoma multiforme [TCGA-GBM] collection. The Cancer Imaging Archive. Published; 2016. [CrossRef]
- Ellingson BM, Bendszus M, Boxerman J, Barboriak D, Erickson BJ, Smits M, et al. Consensus recommendations for a standardized Brain Tumor Imaging Protocol in clinical trials. Neuro Oncol. 2015;17(9):1188–98. [CrossRef]
- Combs SE, Kieser M, Rieken S, Habermehl D, Jäkel O, Haberer T, et al. Randomized phase II study evaluating a carbon ion boost applied after combined radiochemotherapy with temozolomide versus a proton boost after radiochemotherapy with temozolomide in patients with primary glioblastoma: The CLEOPATRA Trial. BMC Cancer. 2010;10:1–9. [CrossRef]
- Combs SE, Burkholder I, Edler L, Rieken S, Habermehl D, Jäkel O, et al. Randomised phase I/II study to evaluate carbon ion radiotherapy versus fractionated stereotactic radiotherapy in patients with recurrent or progressive gliomas: the CINDERELLA trial. BMC Cancer. 2010;10(1):533. [CrossRef]
- Niyazi M, Adeberg S, Kaul D, Boulesteix AL, Bougatf N, Fleischmann DF, et al. Independent validation of a new reirradiation risk score (RRRS) for glioma patients predicting post-recurrence survival: A multicenter DKTK/ROG analysis. Radiother Oncol. 2018;127(1):121–7. [CrossRef]
- Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv Prepr arXiv14091556. 2014. [CrossRef]
- He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. 2015. [CrossRef]
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29(6):1310–20. [CrossRef]
- Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, et al. PyTorch: An imperative style, high-performance deep learning library. arXiv. 2019;(NeurIPS). [CrossRef]
- Pérez-Garcia F, Sparks R, Ourselin S. TorchIO: a Python library for efficient loading, preprocessing, augmentation and patch-based sampling of medical images in deep learning. Comput Methods Programs Biomed. 2021;106236. [CrossRef]
- Wallis, S. Binomial Confidence Intervals and Contingency Tests: Mathematical Fundamentals and the Evaluation of Alternative Methods. J Quant Linguist. 2013 4 August;20(3):178–208. [CrossRef]
- Lin DY, Wei LJ. The robust inference for the Cox proportional hazards model. Journal of the American statistical Association. 1989;84[408]:1074–8. [CrossRef]
- Van Griethuysen JJ, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. Computational radiomics system to decode the radiographic phenotype. Cancer research. 2017;77[21]:e104–7. [CrossRef]
- Sanders H, Saxe J. Garbage in, garbage out: how purportedly great ML models can be screwed up by bad data. Proceedings of Blackhat. 2017.
- Sforazzini F, Salome P, Kudak A, Ulrich M, Bougatf N, Debus J, et al. pyCuRT: An Automated Data Curation Workflow for Radiotherapy Big Data Analysis using Pythons' NyPipe. Int J Radiat Oncol Biol Phys. 2020;108(3):e772. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).