Submitted:
05 January 2023
Posted:
09 January 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
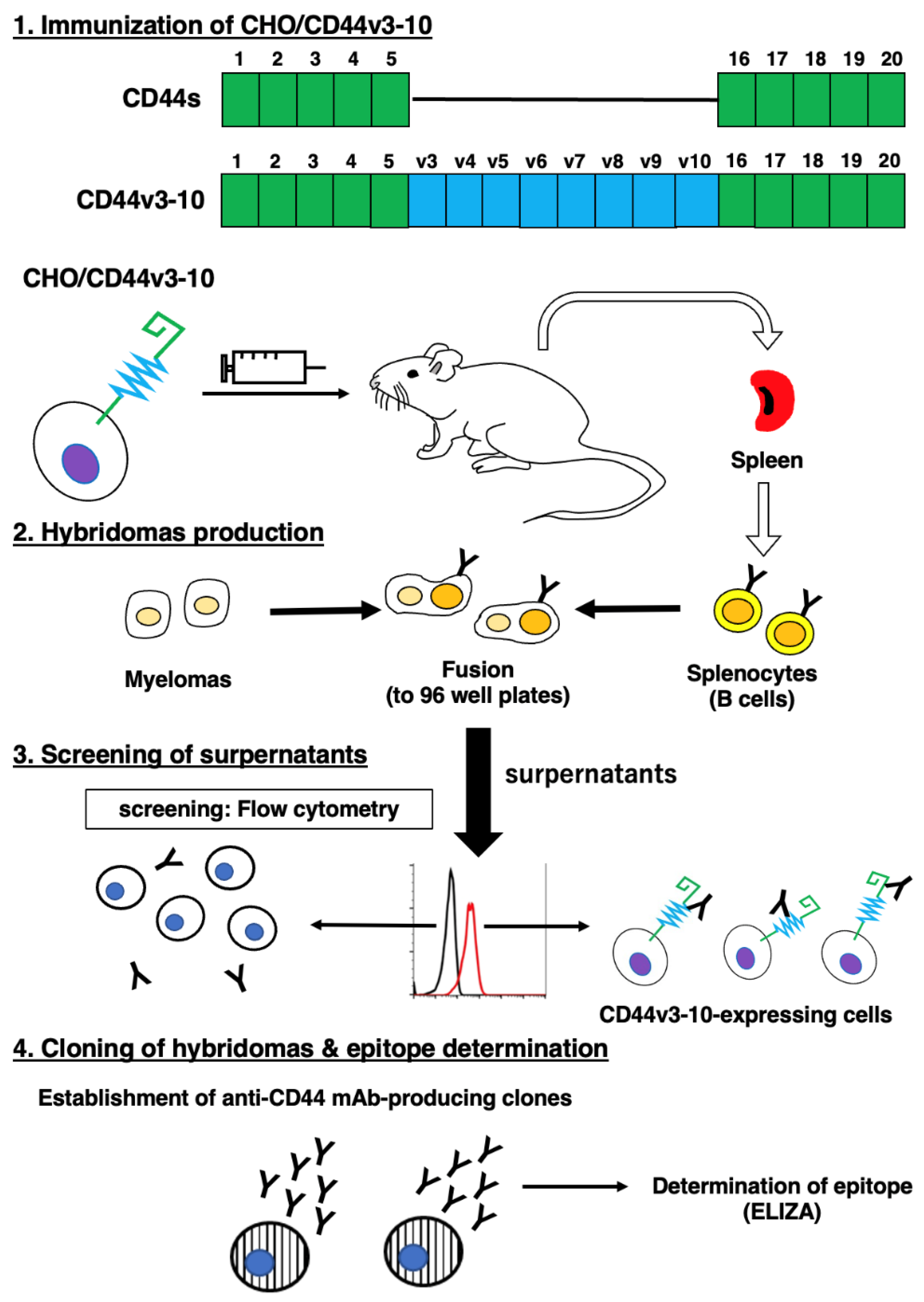
2.1. Establishment of anti-CD44v6 mAb, C44Mab-9
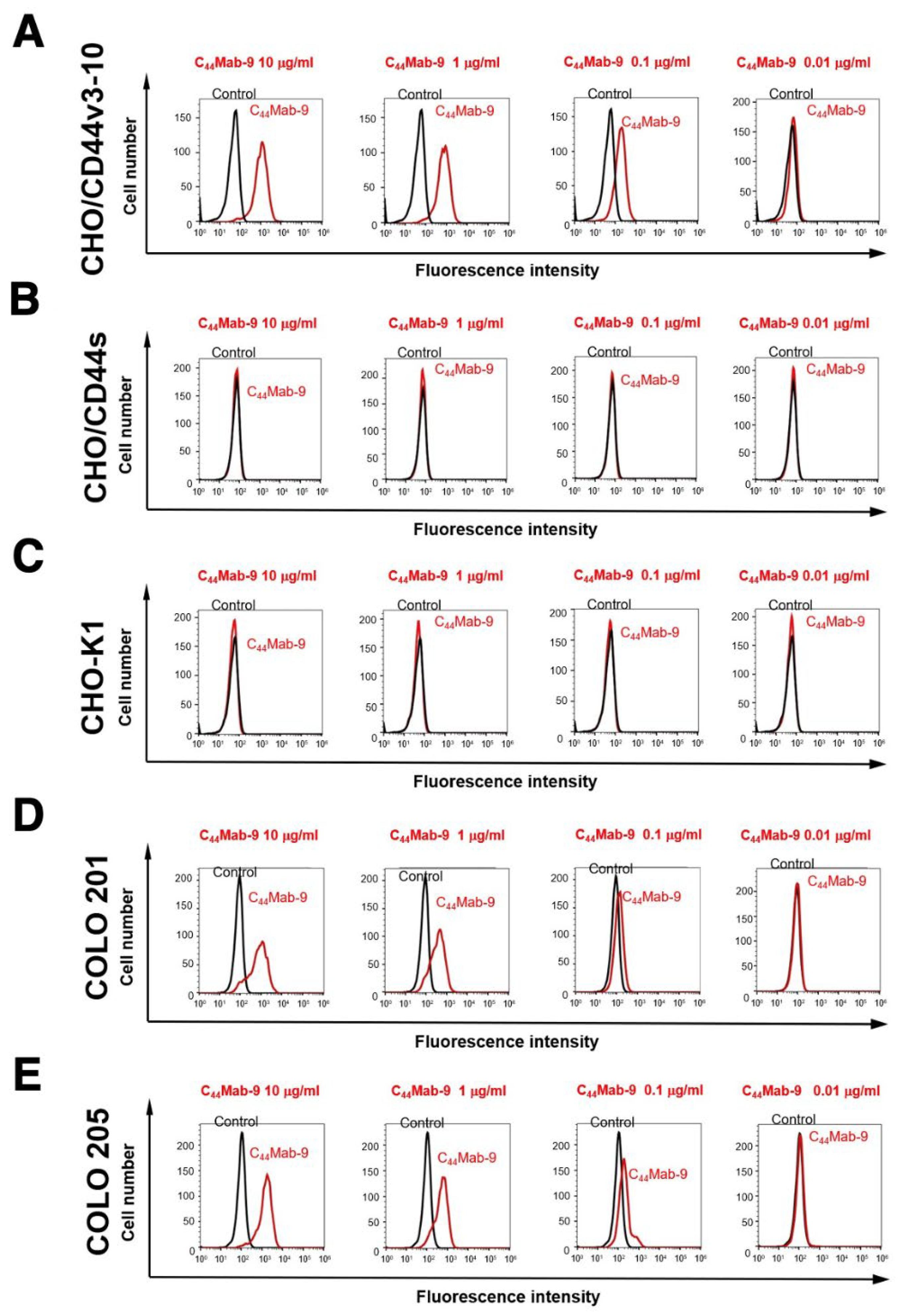
2.2. Flow Cytometric Analysis of C44Mab-9 to CD44-Expressing Cells
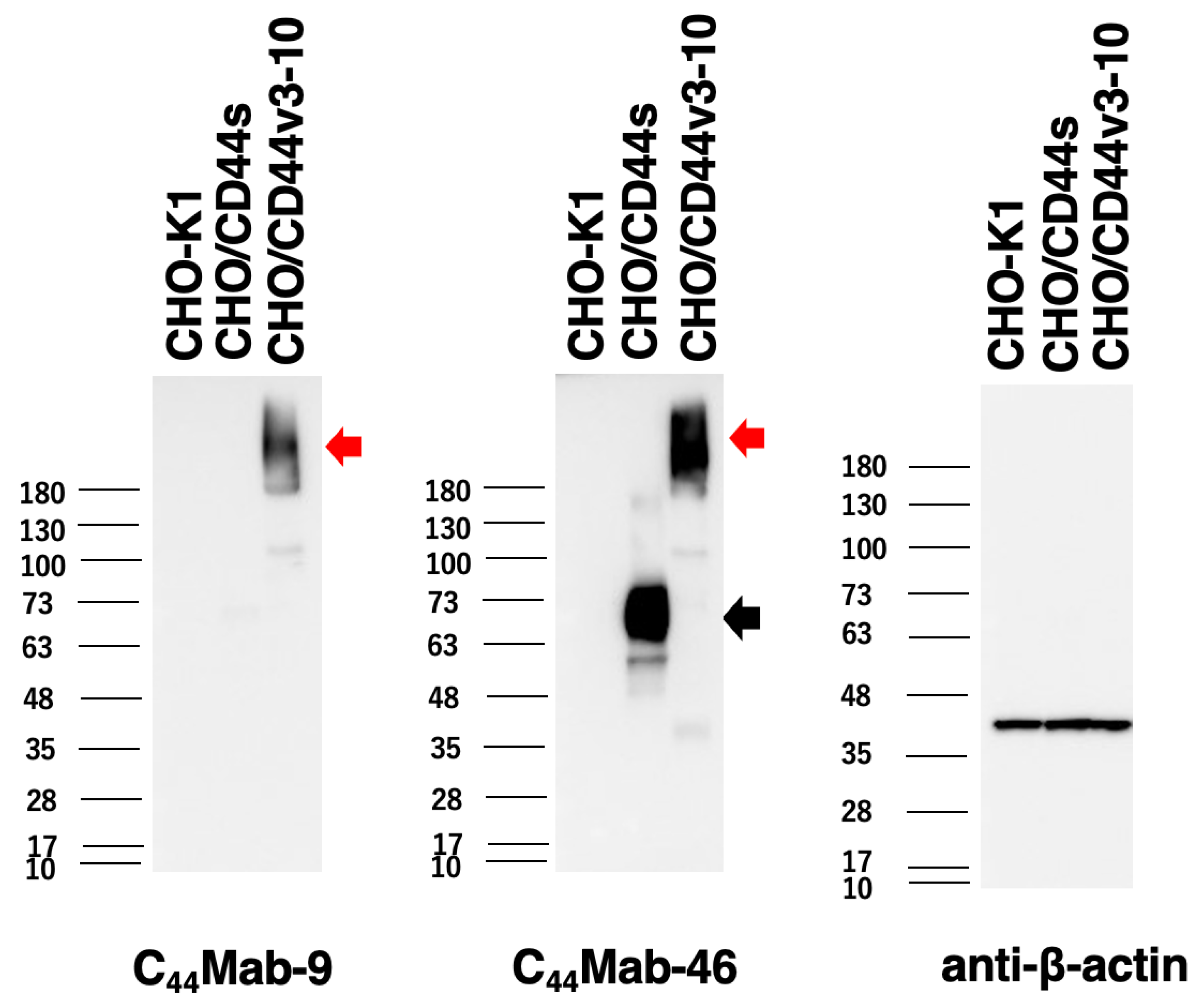
2.3. Western Blot Analysis
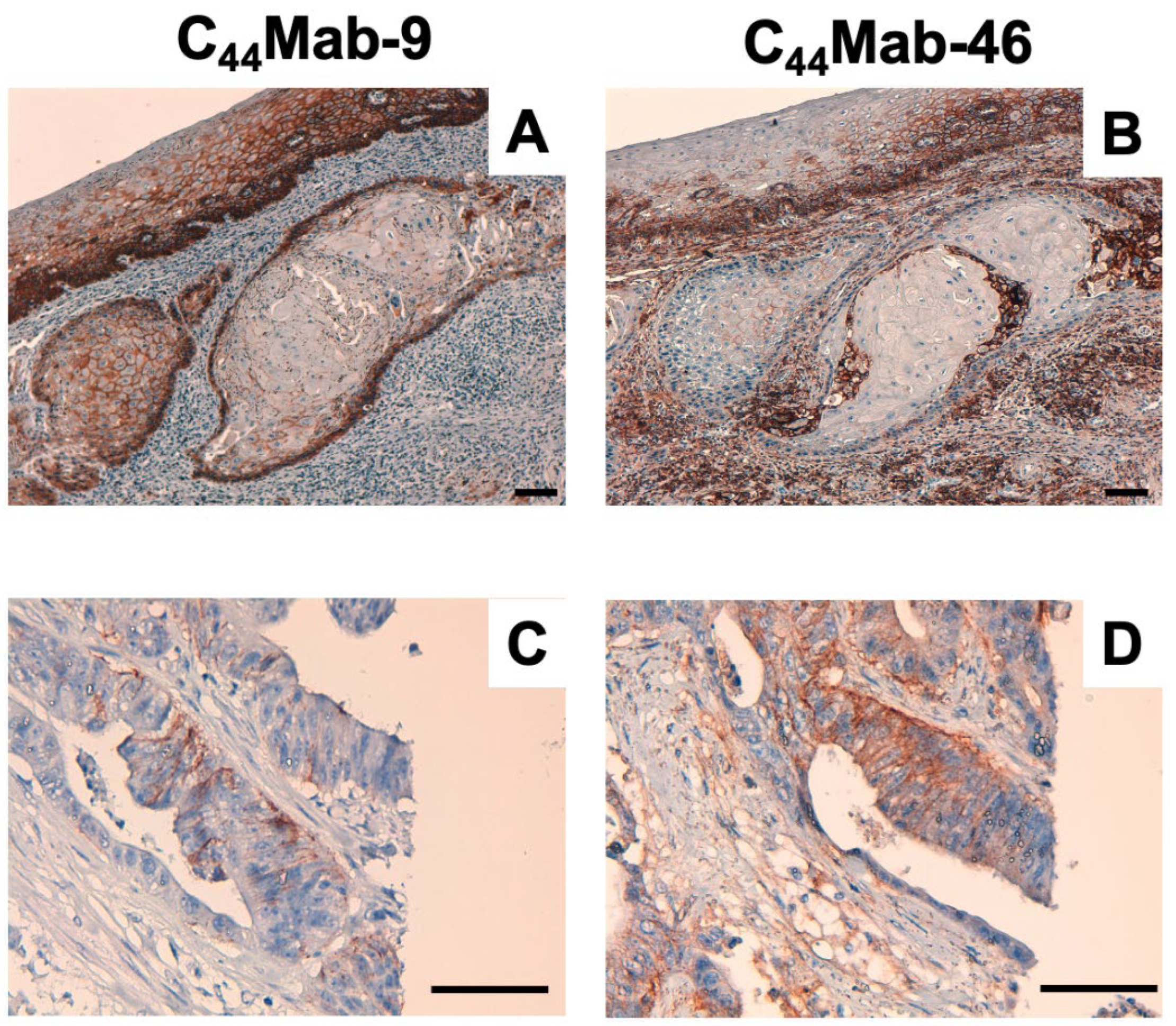
2.4. Immunohistochemical Analysis Using C44Mab-9 against Tumor Tissues
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Hybridoma Production
4.3. ELISA
4.5. Flow Cytometry
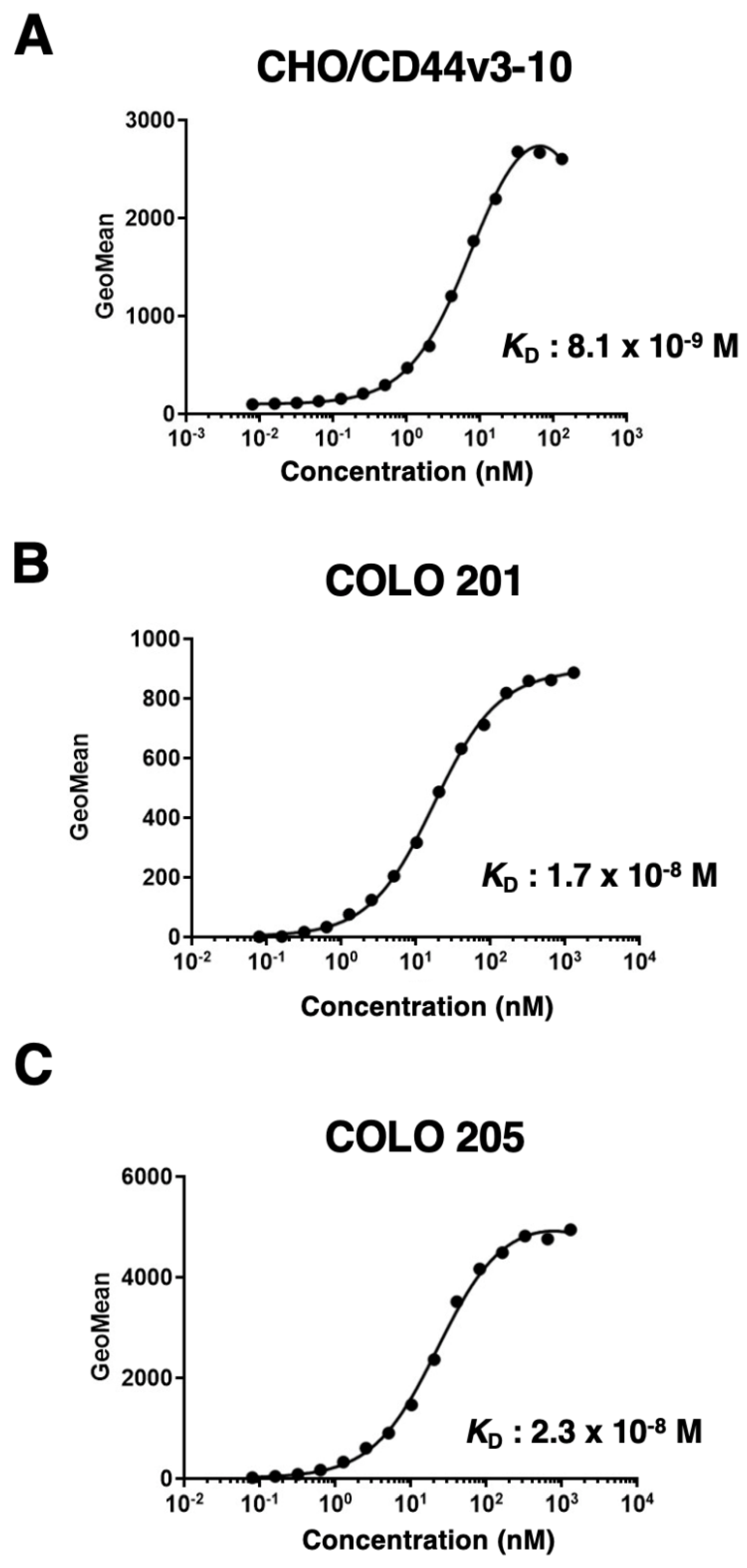
4.6. Determination of Dissociation Constant (KD) by Flow Cytometry
4.7. Western Blot Analysis
4.8. Immunohistochemical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J Clin 2022, 72, 7-33. [CrossRef]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759-767. [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P., et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015, 21, 1350-1356. [CrossRef]
- Puccini, A.; Seeber, A.; Berger, M.D. Biomarkers in Metastatic Colorectal Cancer: Status Quo and Future Perspective. Cancers (Basel) 2022, 14. [CrossRef]
- Zöller, M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer 2011, 11, 254-267. [CrossRef]
- Abbasian, M.; Mousavi, E.; Arab-Bafrani, Z.; Sahebkar, A. The most reliable surface marker for the identification of colorectal cancer stem-like cells: A systematic review and meta-analysis. J Cell Physiol 2019, 234, 8192-8202. [CrossRef]
- Ponta, H.; Sherman, L.; Herrlich, P.A. CD44: from adhesion molecules to signalling regulators. Nat Rev Mol Cell Biol 2003, 4, 33-45. [CrossRef]
- Yan, Y.; Zuo, X.; Wei, D. Concise Review: Emerging Role of CD44 in Cancer Stem Cells: A Promising Biomarker and Therapeutic Target. Stem Cells Transl Med 2015, 4, 1033-1043. [CrossRef]
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J.W. The biology and role of CD44 in cancer progression: therapeutic implications. J Hematol Oncol 2018, 11, 64. [CrossRef]
- Günthert, U.; Hofmann, M.; Rudy, W.; Reber, S.; Zöller, M.; Haussmann, I.; Matzku, S.; Wenzel, A.; Ponta, H.; Herrlich, P. A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell 1991, 65, 13-24. [CrossRef]
- Slevin, M.; Krupinski, J.; Gaffney, J.; Matou, S.; West, D.; Delisser, H.; Savani, R.C.; Kumar, S. Hyaluronan-mediated angiogenesis in vascular disease: uncovering RHAMM and CD44 receptor signaling pathways. Matrix Biol 2007, 26, 58-68. [CrossRef]
- Ma, L.; Dong, L.; Chang, P. CD44v6 engages in colorectal cancer progression. Cell Death Dis 2019, 10, 30. [CrossRef]
- Orian-Rousseau, V.; Morrison, H.; Matzke, A.; Kastilan, T.; Pace, G.; Herrlich, P.; Ponta, H. Hepatocyte growth factor-induced Ras activation requires ERM proteins linked to both CD44v6 and F-actin. Mol Biol Cell 2007, 18, 76-83. [CrossRef]
- Todaro, M.; Gaggianesi, M.; Catalano, V.; Benfante, A.; Iovino, F.; Biffoni, M.; Apuzzo, T.; Sperduti, I.; Volpe, S.; Cocorullo, G., et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell 2014, 14, 342-356. [CrossRef]
- Verel, I.; Heider, K.H.; Siegmund, M.; Ostermann, E.; Patzelt, E.; Sproll, M.; Snow, G.B.; Adolf, G.R.; van Dongen, G.A. Tumor targeting properties of monoclonal antibodies with different affinity for target antigen CD44V6 in nude mice bearing head-and-neck cancer xenografts. Int J Cancer 2002, 99, 396-402. [CrossRef]
- Orian-Rousseau, V.; Ponta, H. Perspectives of CD44 targeting therapies. Arch Toxicol 2015, 89, 3-14. [CrossRef]
- Tijink, B.M.; Buter, J.; de Bree, R.; Giaccone, G.; Lang, M.S.; Staab, A.; Leemans, C.R.; van Dongen, G.A. A phase I dose escalation study with anti-CD44v6 bivatuzumab mertansine in patients with incurable squamous cell carcinoma of the head and neck or esophagus. Clin Cancer Res 2006, 12, 6064-6072. [CrossRef]
- Riechelmann, H.; Sauter, A.; Golze, W.; Hanft, G.; Schroen, C.; Hoermann, K.; Erhardt, T.; Gronau, S. Phase I trial with the CD44v6-targeting immunoconjugate bivatuzumab mertansine in head and neck squamous cell carcinoma. Oral Oncol 2008, 44, 823-829. [CrossRef]
- Yamada, S.; Itai, S.; Nakamura, T.; Yanaka, M.; Kaneko, M.K.; Kato, Y. Detection of high CD44 expression in oral cancers using the novel monoclonal antibody, C(44)Mab-5. Biochem Biophys Rep 2018, 14, 64-68. [CrossRef]
- Goto, N.; Suzuki, H.; Tanaka, T.; Asano, T.; Kaneko, M.K.; Kato, Y. Development of a Novel Anti-CD44 Monoclonal Antibody for Multiple Applications against Esophageal Squamous Cell Carcinomas. Int J Mol Sci 2022, 23. [CrossRef]
- Takei, J.; Asano, T.; Suzuki, H.; Kaneko, M.K.; Kato, Y. Epitope Mapping of the Anti-CD44 Monoclonal Antibody (C44Mab-46) Using Alanine-Scanning Mutagenesis and Surface Plasmon Resonance. Monoclon Antib Immunodiagn Immunother 2021, 40, 219-226. [CrossRef]
- Asano, T.; Kaneko, M.K.; Takei, J.; Tateyama, N.; Kato, Y. Epitope Mapping of the Anti-CD44 Monoclonal Antibody (C44Mab-46) Using the REMAP Method. Monoclon Antib Immunodiagn Immunother 2021, 40, 156-161. [CrossRef]
- Asano, T.; Kaneko, M.K.; Kato, Y. Development of a Novel Epitope Mapping System: RIEDL Insertion for Epitope Mapping Method. Monoclon Antib Immunodiagn Immunother 2021, 40, 162-167. [CrossRef]
- Takei, J.; Kaneko, M.K.; Ohishi, T.; Hosono, H.; Nakamura, T.; Yanaka, M.; Sano, M.; Asano, T.; Sayama, Y.; Kawada, M., et al. A defucosylated antiCD44 monoclonal antibody 5mG2af exerts antitumor effects in mouse xenograft models of oral squamous cell carcinoma. Oncol Rep 2020, 44, 1949-1960. [CrossRef]
- Fox, S.B.; Fawcett, J.; Jackson, D.G.; Collins, I.; Gatter, K.C.; Harris, A.L.; Gearing, A.; Simmons, D.L. Normal human tissues, in addition to some tumors, express multiple different CD44 isoforms. Cancer Res 1994, 54, 4539-4546.
- Heider, K.H.; Sproll, M.; Susani, S.; Patzelt, E.; Beaumier, P.; Ostermann, E.; Ahorn, H.; Adolf, G.R. Characterization of a high-affinity monoclonal antibody specific for CD44v6 as candidate for immunotherapy of squamous cell carcinomas. Cancer Immunol Immunother 1996, 43, 245-253. [CrossRef]
- Heider, K.H.; Mulder, J.W.; Ostermann, E.; Susani, S.; Patzelt, E.; Pals, S.T.; Adolf, G.R. Splice variants of the cell surface glycoprotein CD44 associated with metastatic tumour cells are expressed in normal tissues of humans and cynomolgus monkeys. Eur J Cancer 1995, 31a, 2385-2391. [CrossRef]
- Wang, Z.; Tang, Y.; Xie, L.; Huang, A.; Xue, C.; Gu, Z.; Wang, K.; Zong, S. The Prognostic and Clinical Value of CD44 in Colorectal Cancer: A Meta-Analysis. Front Oncol 2019, 9, 309. [CrossRef]
- Mulder, J.W.; Kruyt, P.M.; Sewnath, M.; Oosting, J.; Seldenrijk, C.A.; Weidema, W.F.; Offerhaus, G.J.; Pals, S.T. Colorectal cancer prognosis and expression of exon-v6-containing CD44 proteins. Lancet 1994, 344, 1470-1472. [CrossRef]
- Wielenga, V.J.; Heider, K.H.; Offerhaus, G.J.; Adolf, G.R.; van den Berg, F.M.; Ponta, H.; Herrlich, P.; Pals, S.T. Expression of CD44 variant proteins in human colorectal cancer is related to tumor progression. Cancer Res 1993, 53, 4754-4756.
- Zlobec, I.; Günthert, U.; Tornillo, L.; Iezzi, G.; Baumhoer, D.; Terracciano, L.; Lugli, A. Systematic assessment of the prognostic impact of membranous CD44v6 protein expression in colorectal cancer. Histopathology 2009, 55, 564-575. [CrossRef]
- Nanashima, A.; Yamaguchi, H.; Sawai, T.; Yasutake, T.; Tsuji, T.; Jibiki, M.; Yamaguchi, E.; Nakagoe, T.; Ayabe, H. Expression of adhesion molecules in hepatic metastases of colorectal carcinoma: relationship to primary tumours and prognosis after hepatic resection. J Gastroenterol Hepatol 1999, 14, 1004-1009. [CrossRef]
- Saito, S.; Okabe, H.; Watanabe, M.; Ishimoto, T.; Iwatsuki, M.; Baba, Y.; Tanaka, Y.; Kurashige, J.; Miyamoto, Y.; Baba, H. CD44v6 expression is related to mesenchymal phenotype and poor prognosis in patients with colorectal cancer. Oncol Rep 2013, 29, 1570-1578. [CrossRef]
- Wang, Z.; Zhao, K.; Hackert, T.; Zöller, M. CD44/CD44v6 a Reliable Companion in Cancer-Initiating Cell Maintenance and Tumor Progression. Front Cell Dev Biol 2018, 6, 97. [CrossRef]
- Baccelli, I.; Schneeweiss, A.; Riethdorf, S.; Stenzinger, A.; Schillert, A.; Vogel, V.; Klein, C.; Saini, M.; Bäuerle, T.; Wallwiener, M., et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol 2013, 31, 539-544. [CrossRef]
- Rupp, B.; Ball, H.; Wuchu, F.; Nagrath, D.; Nagrath, S. Circulating tumor cells in precision medicine: challenges and opportunities. Trends Pharmacol Sci 2022, 43, 378-391. [CrossRef]
- Orian-Rousseau, V.; Chen, L.; Sleeman, J.P.; Herrlich, P.; Ponta, H. CD44 is required for two consecutive steps in HGF/c-Met signaling. Genes Dev 2002, 16, 3074-3086. [CrossRef]
- Nanamiya, R.; Takei, J.; Ohishi, T.; Asano, T.; Tanaka, T.; Sano, M.; Nakamura, T.; Yanaka, M.; Handa, S.; Tateyama, N., et al. Defucosylated Anti-Epidermal Growth Factor Receptor Monoclonal Antibody (134-mG(2a)-f) Exerts Antitumor Activities in Mouse Xenograft Models of Canine Osteosarcoma. Monoclon Antib Immunodiagn Immunother 2022, 41, 1-7. [CrossRef]
- Kawabata, H.; Suzuki, H.; Ohishi, T.; Kawada, M.; Kaneko, M.K.; Kato, Y. A Defucosylated Mouse Anti-CD10 Monoclonal Antibody (31-mG(2a)-f) Exerts Antitumor Activity in a Mouse Xenograft Model of CD10-Overexpressed Tumors. Monoclon Antib Immunodiagn Immunother 2022, 41, 59-66. [CrossRef]
- Kawabata, H.; Ohishi, T.; Suzuki, H.; Asano, T.; Kawada, M.; Suzuki, H.; Kaneko, M.K.; Kato, Y. A Defucosylated Mouse Anti-CD10 Monoclonal Antibody (31-mG(2a)-f) Exerts Antitumor Activity in a Mouse Xenograft Model of Renal Cell Cancers. Monoclon Antib Immunodiagn Immunother 2022, 10.1089/mab.2021.0049. [CrossRef]
- Asano, T.; Tanaka, T.; Suzuki, H.; Li, G.; Ohishi, T.; Kawada, M.; Yoshikawa, T.; Kaneko, M.K.; Kato, Y. A Defucosylated Anti-EpCAM Monoclonal Antibody (EpMab-37-mG(2a)-f) Exerts Antitumor Activity in Xenograft Model. Antibodies (Basel) 2022, 11. [CrossRef]
- Tateyama, N.; Nanamiya, R.; Ohishi, T.; Takei, J.; Nakamura, T.; Yanaka, M.; Hosono, H.; Saito, M.; Asano, T.; Tanaka, T., et al. Defucosylated Anti-Epidermal Growth Factor Receptor Monoclonal Antibody 134-mG(2a)-f Exerts Antitumor Activities in Mouse Xenograft Models of Dog Epidermal Growth Factor Receptor-Overexpressed Cells. Monoclon Antib Immunodiagn Immunother 2021, 40, 177-183. [CrossRef]
- Takei, J.; Ohishi, T.; Kaneko, M.K.; Harada, H.; Kawada, M.; Kato, Y. A defucosylated anti-PD-L1 monoclonal antibody 13-mG(2a)-f exerts antitumor effects in mouse xenograft models of oral squamous cell carcinoma. Biochem Biophys Rep 2020, 24, 100801. [CrossRef]
- Takei, J.; Kaneko, M.K.; Ohishi, T.; Hosono, H.; Nakamura, T.; Yanaka, M.; Sano, M.; Asano, T.; Sayama, Y.; Kawada, M., et al. A defucosylated anti-CD44 monoclonal antibody 5-mG2a-f exerts antitumor effects in mouse xenograft models of oral squamous cell carcinoma. Oncol Rep 2020, 44, 1949-1960. [CrossRef]
- Kato, Y.; Yamada, S.; Furusawa, Y.; Itai, S.; Nakamura, T.; Yanaka, M.; Sano, M.; Harada, H.; Fukui, M.; Kaneko, M.K. PMab-213: A Monoclonal Antibody for Immunohistochemical Analysis Against Pig Podoplanin. Monoclon Antib Immunodiagn Immunother 2019, 38, 18-24. [CrossRef]
- Furusawa, Y.; Yamada, S.; Itai, S.; Sano, M.; Nakamura, T.; Yanaka, M.; Fukui, M.; Harada, H.; Mizuno, T.; Sakai, Y., et al. PMab-210: A Monoclonal Antibody Against Pig Podoplanin. Monoclon Antib Immunodiagn Immunother 2019, 38, 30-36. [CrossRef]
- Furusawa, Y.; Yamada, S.; Itai, S.; Nakamura, T.; Yanaka, M.; Sano, M.; Harada, H.; Fukui, M.; Kaneko, M.K.; Kato, Y. PMab-219: A monoclonal antibody for the immunohistochemical analysis of horse podoplanin. Biochem Biophys Rep 2019, 18, 100616. [CrossRef]
- Furusawa, Y.; Yamada, S.; Itai, S.; Nakamura, T.; Takei, J.; Sano, M.; Harada, H.; Fukui, M.; Kaneko, M.K.; Kato, Y. Establishment of a monoclonal antibody PMab-233 for immunohistochemical analysis against Tasmanian devil podoplanin. Biochem Biophys Rep 2019, 18, 100631. [CrossRef]
- Kato, Y.; Kaneko, M.K.; Kuno, A.; Uchiyama, N.; Amano, K.; Chiba, Y.; Hasegawa, Y.; Hirabayashi, J.; Narimatsu, H.; Mishima, K., et al. Inhibition of tumor cell-induced platelet aggregation using a novel anti-podoplanin antibody reacting with its platelet-aggregation-stimulating domain. Biochem Biophys Res Commun 2006, 349, 1301-1307. [CrossRef]
- Chalise, L.; Kato, A.; Ohno, M.; Maeda, S.; Yamamichi, A.; Kuramitsu, S.; Shiina, S.; Takahashi, H.; Ozone, S.; Yamaguchi, J., et al. Efficacy of cancer-specific anti-podoplanin CAR-T cells and oncolytic herpes virus G47Delta combination therapy against glioblastoma. Mol Ther Oncolytics 2022, 26, 265-274. [CrossRef]
- Ishikawa, A.; Waseda, M.; Ishii, T.; Kaneko, M.K.; Kato, Y.; Kaneko, S. Improved anti-solid tumor response by humanized anti-podoplanin chimeric antigen receptor transduced human cytotoxic T cells in an animal model. Genes Cells 2022, 27, 549-558. [CrossRef]
- Tamura-Sakaguchi, R.; Aruga, R.; Hirose, M.; Ekimoto, T.; Miyake, T.; Hizukuri, Y.; Oi, R.; Kaneko, M.K.; Kato, Y.; Akiyama, Y., et al. Moving toward generalizable NZ-1 labeling for 3D structure determination with optimized epitope-tag insertion. Acta Crystallogr D Struct Biol 2021, 77, 645-662. [CrossRef]
- Kaneko, M.K.; Ohishi, T.; Nakamura, T.; Inoue, H.; Takei, J.; Sano, M.; Asano, T.; Sayama, Y.; Hosono, H.; Suzuki, H., et al. Development of Core-Fucose-Deficient Humanized and Chimeric Anti-Human Podoplanin Antibodies. Monoclon Antib Immunodiagn Immunother 2020, 39, 167-174. [CrossRef]
- Fujii, Y.; Matsunaga, Y.; Arimori, T.; Kitago, Y.; Ogasawara, S.; Kaneko, M.K.; Kato, Y.; Takagi, J. Tailored placement of a turn-forming PA tag into the structured domain of a protein to probe its conformational state. J Cell Sci 2016, 129, 1512-1522. [CrossRef]
- Abe, S.; Kaneko, M.K.; Tsuchihashi, Y.; Izumi, T.; Ogasawara, S.; Okada, N.; Sato, C.; Tobiume, M.; Otsuka, K.; Miyamoto, L., et al. Antitumor effect of novel anti-podoplanin antibody NZ-12 against malignant pleural mesothelioma in an orthotopic xenograft model. Cancer Sci 2016, 107, 1198-1205. [CrossRef]
- Kaneko, M.K.; Abe, S.; Ogasawara, S.; Fujii, Y.; Yamada, S.; Murata, T.; Uchida, H.; Tahara, H.; Nishioka, Y.; Kato, Y. Chimeric Anti-Human Podoplanin Antibody NZ-12 of Lambda Light Chain Exerts Higher Antibody-Dependent Cellular Cytotoxicity and Complement-Dependent Cytotoxicity Compared with NZ-8 of Kappa Light Chain. Monoclon Antib Immunodiagn Immunother 2017, 36, 25-29. [CrossRef]
- Ito, A.; Ohta, M.; Kato, Y.; Inada, S.; Kato, T.; Nakata, S.; Yatabe, Y.; Goto, M.; Kaneda, N.; Kurita, K., et al. A Real-Time Near-Infrared Fluorescence Imaging Method for the Detection of Oral Cancers in Mice Using an Indocyanine Green-Labeled Podoplanin Antibody. Technol Cancer Res Treat 2018, 17, 1533033818767936. [CrossRef]
- Tamura, R.; Oi, R.; Akashi, S.; Kaneko, M.K.; Kato, Y.; Nogi, T. Application of the NZ-1 Fab as a crystallization chaperone for PA tag-inserted target proteins. Protein Sci 2019, 28, 823-836. [CrossRef]
- Shiina, S.; Ohno, M.; Ohka, F.; Kuramitsu, S.; Yamamichi, A.; Kato, A.; Motomura, K.; Tanahashi, K.; Yamamoto, T.; Watanabe, R., et al. CAR T Cells Targeting Podoplanin Reduce Orthotopic Glioblastomas in Mouse Brains. Cancer Immunol Res 2016, 4, 259-268. [CrossRef]
- Kuwata, T.; Yoneda, K.; Mori, M.; Kanayama, M.; Kuroda, K.; Kaneko, M.K.; Kato, Y.; Tanaka, F. Detection of Circulating Tumor Cells (CTCs) in Malignant Pleural Mesothelioma (MPM) with the "Universal" CTC-Chip and An Anti-Podoplanin Antibody NZ-1.2. Cells 2020, 9. [CrossRef]
- Nishinaga, Y.; Sato, K.; Yasui, H.; Taki, S.; Takahashi, K.; Shimizu, M.; Endo, R.; Koike, C.; Kuramoto, N.; Nakamura, S., et al. Targeted Phototherapy for Malignant Pleural Mesothelioma: Near-Infrared Photoimmunotherapy Targeting Podoplanin. Cells 2020, 9. [CrossRef]
- Fujii, Y.; Kaneko, M.; Neyazaki, M.; Nogi, T.; Kato, Y.; Takagi, J. PA tag: a versatile protein tagging system using a super high affinity antibody against a dodecapeptide derived from human podoplanin. Protein Expr Purif 2014, 95, 240-247. [CrossRef]
- Kato, Y.; Kaneko, M.K.; Kunita, A.; Ito, H.; Kameyama, A.; Ogasawara, S.; Matsuura, N.; Hasegawa, Y.; Suzuki-Inoue, K.; Inoue, O., et al. Molecular analysis of the pathophysiological binding of the platelet aggregation-inducing factor podoplanin to the C-type lectin-like receptor CLEC-2. Cancer Sci 2008, 99, 54-61. [CrossRef]
- Kato, Y.; Vaidyanathan, G.; Kaneko, M.K.; Mishima, K.; Srivastava, N.; Chandramohan, V.; Pegram, C.; Keir, S.T.; Kuan, C.T.; Bigner, D.D., et al. Evaluation of anti-podoplanin rat monoclonal antibody NZ-1 for targeting malignant gliomas. Nucl Med Biol 2010, 37, 785-794. [CrossRef]
- Itai, S.; Ohishi, T.; Kaneko, M.K.; Yamada, S.; Abe, S.; Nakamura, T.; Yanaka, M.; Chang, Y.W.; Ohba, S.I.; Nishioka, Y., et al. Anti-podocalyxin antibody exerts antitumor effects via antibody-dependent cellular cytotoxicity in mouse xenograft models of oral squamous cell carcinoma. Oncotarget 2018, 9, 22480-22497. [CrossRef]
| Peptide | Coding exon | Sequence | C44Mab-9 |
|---|---|---|---|
| CD44p21–40 | 2 | QIDLNITCRFAGVFHVEKNG | – |
| CD44p31–50 | 2 | AGVFHVEKNGRYSISRTEAA | – |
| CD44p41–60 | 2 | RYSISRTEAADLCKAFNSTL | – |
| CD44p51–70 | 2 | DLCKAFNSTLPTMAQMEKAL | – |
| CD44p61–80 | 2/3 | PTMAQMEKALSIGFETCRYG | – |
| CD44p71–90 | 2/3 | SIGFETCRYGFIEGHVVIPR | – |
| CD44p81–100 | 3 | FIEGHVVIPRIHPNSICAAN | – |
| CD44p91–110 | 3 | IHPNSICAANNTGVYILTSN | – |
| CD44p101–120 | 3 | NTGVYILTSNTSQYDTYCFN | – |
| CD44p111–130 | 3/4 | TSQYDTYCFNASAPPEEDCT | – |
| CD44p121–140 | 3/4 | ASAPPEEDCTSVTDLPNAFD | – |
| CD44p131–150 | 4 | SVTDLPNAFDGPITITIVNR | – |
| CD44p141–160 | 4 | GPITITIVNRDGTRYVQKGE | – |
| CD44p151–170 | 4/5 | DGTRYVQKGEYRTNPEDIYP | – |
| CD44p161–180 | 5 | YRTNPEDIYPSNPTDDDVSS | – |
| CD44p171–190 | 5 | SNPTDDDVSSGSSSERSSTS | – |
| CD44p181–200 | 5 | GSSSERSSTSGGYIFYTFST | – |
| CD44p191–210 | 5 | GGYIFYTFSTVHPIPDEDSP | – |
| CD44p201–220 | 5 | VHPIPDEDSPWITDSTDRIP | – |
| CD44p211–230 | 5/v3 | WITDSTDRIPATSTSSNTIS | – |
| CD44p221–240 | v3 | ATSTSSNTISAGWEPNEENE | – |
| CD44p231–250 | v3 | AGWEPNEENEDERDRHLSFS | – |
| CD44p241–260 | v3 | DERDRHLSFSGSGIDDDEDF | – |
| CD44p251–270 | v3/v4 | GSGIDDDEDFISSTISTTPR | – |
| CD44p261–280 | v4 | ISSTISTTPRAFDHTKQNQD | – |
| CD44p271–290 | v4 | AFDHTKQNQDWTQWNPSHSN | – |
| CD44p281–300 | v4 | WTQWNPSHSNPEVLLQTTTR | – |
| CD44p291–310 | v4 | PEVLLQTTTRMTDVDRNGTT | – |
| CD44p301–320 | v4/v5 | MTDVDRNGTTAYEGNWNPEA | – |
| CD44p311–330 | v5 | AYEGNWNPEAHPPLIHHEHH | – |
| CD44p321–340 | v5 | HPPLIHHEHHEEEETPHSTS | – |
| CD44p331–350 | v5/v6 | EEEETPHSTSTIQATPSSTT | – |
| CD44p341–360 | v5/v6 | TIQATPSSTTEETATQKEQW | – |
| CD44p351–370 | v6 | EETATQKEQWFGNRWHEGYR | + |
| CD44p361–380 | v6 | FGNRWHEGYRQTPREDSHST | – |
| CD44p371–390 | v6 | QTPREDSHSTTGTAAASAHT | – |
| CD44p381–400 | v6/v7 | TGTAAASAHTSHPMQGRTTP | – |
| CD44p391–410 | v6/v7 | SHPMQGRTTPSPEDSSWTDF | – |
| CD44p401–420 | v7 | SPEDSSWTDFFNPISHPMGR | – |
| CD44p411–430 | v7 | FNPISHPMGRGHQAGRRMDM | – |
| CD44p421–440 | v7/v8 | GHQAGRRMDMDSSHSTTLQP | – |
| CD44p431–450 | v8 | DSSHSTTLQPTANPNTGLVE | – |
| CD44p441–460 | v8 | TANPNTGLVEDLDRTGPLSM | – |
| CD44p451–470 | v8/v9 | DLDRTGPLSMTTQQSNSQSF | – |
| CD44p461–480 | v9 | TTQQSNSQSFSTSHEGLEED | – |
| CD44p471–490 | v9 | STSHEGLEEDKDHPTTSTLT | – |
| CD44p481–500 | v9/v10 | KDHPTTSTLTSSNRNDVTGG | – |
| CD44p491–510 | v10 | SSNRNDVTGGRRDPNHSEGS | – |
| CD44p501–520 | v10 | RRDPNHSEGSTTLLEGYTSH | – |
| CD44p511–530 | v10 | TTLLEGYTSHYPHTKESRTF | – |
| CD44p521–540 | v10 | YPHTKESRTFIPVTSAKTGS | – |
| CD44p531–550 | v10 | IPVTSAKTGSFGVTAVTVGD | – |
| CD44p541–560 | v10 | FGVTAVTVGDSNSNVNRSLS | – |
| CD44p551–570 | v10/16 | SNSNVNRSLSGDQDTFHPSG | – |
| CD44p561–580 | v10/16 | GDQDTFHPSGGSHTTHGSES | – |
| CD44p571–590 | 16 | GSHTTHGSESDGHSHGSQEG | – |
| CD44p581–600 | 16/17 | DGHSHGSQEGGANTTSGPIR | – |
| CD44p591–606 | 17 | GANTTSGPIRTPQIPEAAAA | – |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





