1. Introduction
Strongyloides spp. (Nematode, Strongyloididae) or threadwormis, a chronic causative infectious agent, is one of the neglected tropical infectious pathogens. This causes a serious public health issue due to the worm’s capacity to complete its entire lifecycle in the human host through autoinfection [1]. It can eventually lead to a hyper-infection syndrome in immunosuppressed patients, which can be fatal [1,2,3,4].
People acquire strongyloidiasis through penetration of the infective filariform larvae (L3) during contact contaminated soil, such as agricultural cultivation and recreation activities [1,4,5]. The clinical symptoms of strongyloidiasis range from absence of symptoms to gastrointestinal symptoms,e.g., abdominal pain, diarrhea, and colitis and urticaria from the penetration of the larvae through the skin causingchronic inflammatory response. Disseminated infections with mortality rates as high as 87% can occur specially among immunocompromised patients [8,9].
Diagnosis of strongyloidiasis is traditionally based on wet mount microscopy to detect rhabditiform/filariform of Strongyloides spp. larvae in faeces [7,10]. However, a low parasite load in patients with chronic infection affects the sensitivity of microscopic diagnosis. Serological methods such as Enzyme-linked immunosorbent assay (ELISA) have been used to detect human immunoglobulin G (IgG) against secreted antigen of filariform larvae during migration in the blood vessels. This is one of the most widely used techniques for the diagnosis of strongyloidiasis in a chronically infected population, with the sensitivity ranging from 73% to 100% [7,11,12]. However, this technique lacks specificity due to the cross-reactivity with antigen of other helminths, such as those that cause filariasis, schistosomiasis, ascariasis, hookworm, toxocariasis, and fascioliasisamong others [7,13,14]. Recently, molecular techniques have been developed for the detection and differentiation of Strongyloides spp. in fecal samples which are highly sensitive and more cost-effective for developing countries where Strongyloides spp are endemic [15,16,17,18].
Verweij JJ (2009), and Stefanie Krammeet al. (2011) have developed a real-time PCR method targeting the small subunit of the rRNA (18S and 28S) gene for specific detection of Strongyloides spp. DNA in fecal samples [19,20]. It was introduced as a promising alternative diagnostic approach that could determine species components, monitor the true prevalence and intensity of Strongyloides spp. infection in community [18,21].
Strongyloidiasis was firstly discovered in 1876 in the stools of French soldiers on duty in Southern Vietnam who had severe diarrhea, which was later proved to be endemic in Vietnam [7,22]. A recent meta-analysis reviewed the prevalence of Strongyloides spp. infection, from 0.2 to 2.5% in the northern and 1.19% in the southern, using stool examination and 7.6% using sera-immunological tests [22,23,24]. However, studies on species distinction of Strongyloides spp. infecting human are still limited.
Duc Hoa is a rural district of Long An Province in the Mekong Delta region. The district shares its borders with the administrative units such as Trang Bang District of Tay Ninh Province and Cu Chi District, Hoc Mon District of Ho Chi Minh city [25]. There has not been any research on strongyloidiasis or the prevalence of dominant Strongyloides species in this region. This study aims to determine the species of Strongyloides that infect the resident population in Duc Hoa district, Long Anprovince, Southern Vietnam.
2. Materials and Methods
2.1. Study Design, Area and Population
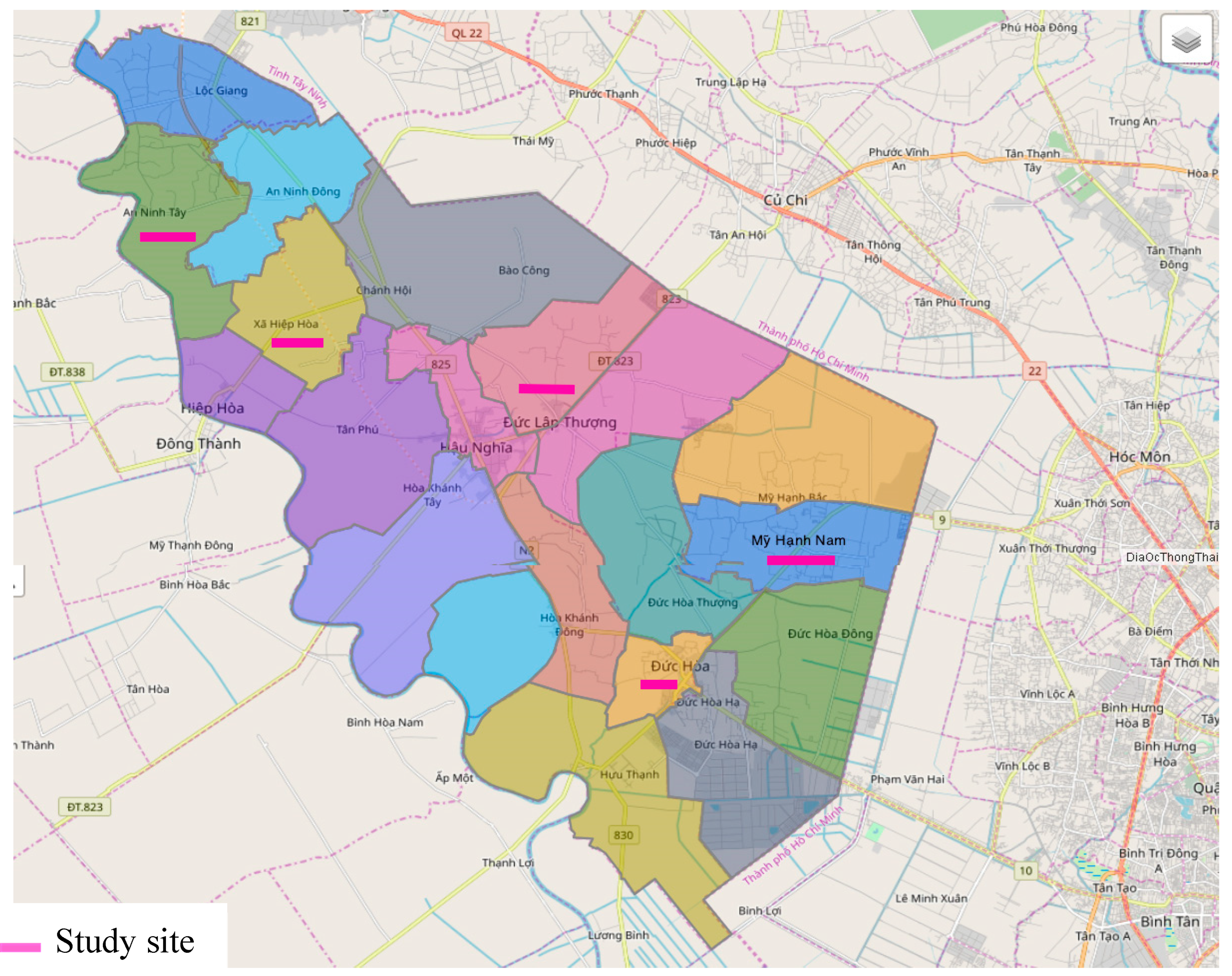
A cross-sectional study was carried out from July, 2017 to November, 2018 among the general population in four communes and one town in Duc Hoa district, Long An province, Vietnam. The study sites were randomly selected from the list of 17 communes and 3 towns of Duc Hoa (total number of residences: 245,617) (
Figure 1)
Selection criteria: The study was conducted on the whole population, regardless of gender, race, over six-month residence at the study sites, and fluency in Vietnamese language. The subjects joined voluntarily and learned about this study before signing their consent to participate. Moreover, the study participants had not taken any deworming medication for 6 weeks. A total number of 1,190 participants distributed in 4 communes and 1 town in Duc Hoa satisfied the research criteria and were recruited in this study.
Sample containers were distributed one day before collection. The participants' fecal samples were colected in the wide-mouth screw-capped plastic containers, pre-labelled with the general information about the participants such as name, ID code of site, sex, and age. Stool samples were stored at the temperature of 25oC and transported to the Laboratory of Medical Parasitology, Pham Ngoc Thach University of Medicine within two hours of sampling. All samples were stored at 2 - 8°C and processed within 48 hours for microscopic examination. Molecular detection was carried out at laboratory of molecular biology, Pham Ngoc Thach University of Medicine, Hochiminh city, Vietnam.
2.2. Laboratory Detection of Strongyloides in Fecal Samples
Stool samples were checked in terms of the quality, volume, sampling time, research location in the laboratory and each was divided into two halves. The first half was tested by direct wet mount smear for microscopicmethod to detect motile protozoan trophozoites, helminth’s eggs and larvae under a light microscope [26,27]. The second half was adapted from the Harada-Mori filter paper culture (HMFPC) to identify Strongyloides spp. and hookworm larvae [28,29]. In short, each sample was tested in duplicate; and if the result was positive in either or both of the methods above, it was considered a positive result. This procedure complianced the guideline on detection of Strongyloides in fecal samples developed by the Laboratory of Medical Parasitology and Laboratory of Molecular Biology, Pham Ngoc Thach University of Medicine, Hochiminh city, Vietnam
Morphological characteristics of all larval forms of Strongyloides spp. development stages were photographed in details by using Dp26 camera connected to Olympus BX53 microscope system, at magnifications of 100, 200 and 400 times. The pictures were analyzed using CellSens 5.0 software. Worms/larvae were measured in terms of body length, body width, esophageal length, abdominal cavity length, tail shape and transverse size at the tip of the tail. The developmental stages are classified based on the open shape of the mouth, esophagus, genital spines, ovaries, and intestines [26,30,31]. The Strongyloides spp. larvae were washed 3 times with distilled water and preserved in 75% ethanol solution. Each sample (2 larvae/100 µl) had its own code and was kept at ambient temperature for DNA extraction.
2.3. A RealTime PCR Molecular Detection of Strongyloides Spp.
Approximately 500 µl of the 2nd stage Strongyloidesspp larvae (2 larvae/100 µl), preserved in 75% ethanol solution, were used for DNA extraction by using the High Pure PCR Template Preparation Kit (Product No: 11796828001, Roche Life Science, USA) in accordance with the manufacturer’s instructions. The concentration and purity of the extracted DNA were determined by using an Amersham Pharmacia's GeneQuant pro spectrophotometer (Amersham Pharmacia Biotech). Each DNA sample was measured 3 times, and an average was calculated for evaluation in accordance with the company's regulations. The criteria for evaluating the purity of DNA samples after extraction were based on the OD260/OD280 ratio. The DNA sample was considered suitable for realtime PCR assay when the ratio was in the range of 1.6-2.0. Samples were stored at -40C for further experiments.
A realtime PCR targeting the 18Sribosomal RNA gene was developed for identification of the genus
Strongyloides including species such as
Strongyloidesstercoralis and
Strongyloidesrati. The assay in this study used three sets of species-specific primers that obtained from previous studies [19,20,32] and performed in two separate reactions in the thermal cycler Applied Biosystems 7500 Real-Time PCR (Applied Biosystems, USA). The primers sequence details were described in
Table 1.
The optimized realtime PCR mixture that was used in a 25 µL reaction was characterized as follows: 12.5 μL qPCR Master mix (Absolute®QPCR, Aligen-Abgen, UK), 0.6 μl (300nmol) each of primers (IDT, Singapore), 0.6 μL (200nmoL) each of labelled probes (IDT, Singapore), 5 μL of extracted DNA template from larvae and deionized water. The condition for realtime PCR involved denaturation at 95 oC for 15 minutes and was followed by 40 cycles at 95 oC for 15s and the final one at 60 oC for 1 minute.
The positive control DNA for Strongyloides stercoralis was obtained from the larvae culture medium and negative control was deionized water without DNA. Amplification, detection and data analysis were performed in StratageneMx3005P system (Agilent Technologies Division, Germany). Fluorescence was measured at the end of each extension step and the threshold cycle or Cut-off value of each positive samples were calculated.
2.4. Nested PCR and Sequencing
18S rDNA gene of 70 positive samples was amplified by a nested-polymerase chain reaction (nested-PCR). Primers were designed for 18S ribosomal DNA small subunit based on the Strongyloides spp. sequence deposited in GenBank (accession numbers: AJ417023) [33]. All PCR products were purified and sent to the First BASE Laboratories-Axil Scientific, Malaysia for sequencing.
2.5. Data analysis
Data were analyzed using the statistical package for social sciences (SPSS) software for windows version 20.0 (SPSS, IL, USA). Frequencies were expressed as percentages.
DNA sequences were edited by using the Bioedit v.2.6 program [34]. Multiple sequence alignments were performed with Clustal W method and databases (
http://www.ncbi.nlm.nih.gov/). Phylogenetic tree of identified
Strongyloides spp. was was constructed by using MEGA v6.0 with the neighbourjoining (NJ) cluster algorithm and Tamura-3-Parameter. Bootstrap with 1000 replications was utilized for determining the topology reliability of the tree.
3. Results
3.1. Microscopic Diagnosis
A total of 79 samples among 1,190 stool samples (6.6%) were found to be positive for the larvae Strongyloides spp. using both techniques. The direct smear microscopy detected 58.2% of total cases, much lower than the mHMFPC technique (93.7%).
3.2. Morphological Characteristics of Strongyloidesspp Larvae Stages
All of the 1st stage larvae had pointed tail with an average body length of 279.9 µm and the esophageal length was 27.1% of the body's. When cultured at day 3, the 2nd larvae stage had slender shape, with the tip of the tail blunt or notched. Free-living male of
Strongyloides spp. showed a pointed tail at an average length of 778.8 µm. In contrast, free-living female of
Strongyloides spp. had an average length of 916.7µL with the vulva located near the middle of the body (
Table 2).
3.3. Realtime PCR Technique
70 of the 79 extracted DNA samples (88.6%) had the OD260/280 ratio, greater than 1.7 and passed the quality needed for realtime PCR. Nine others were detected in DNA negative sample.
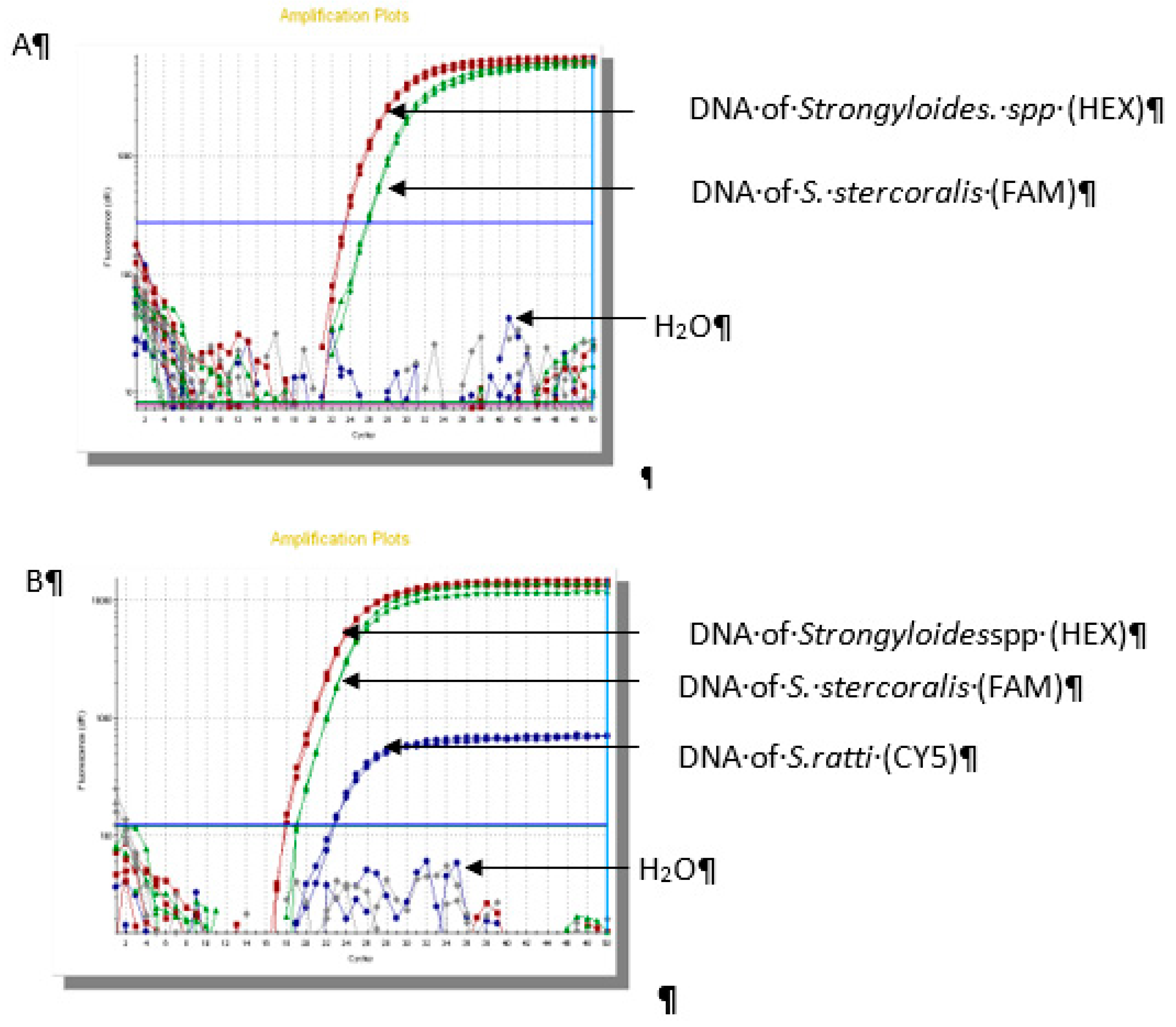
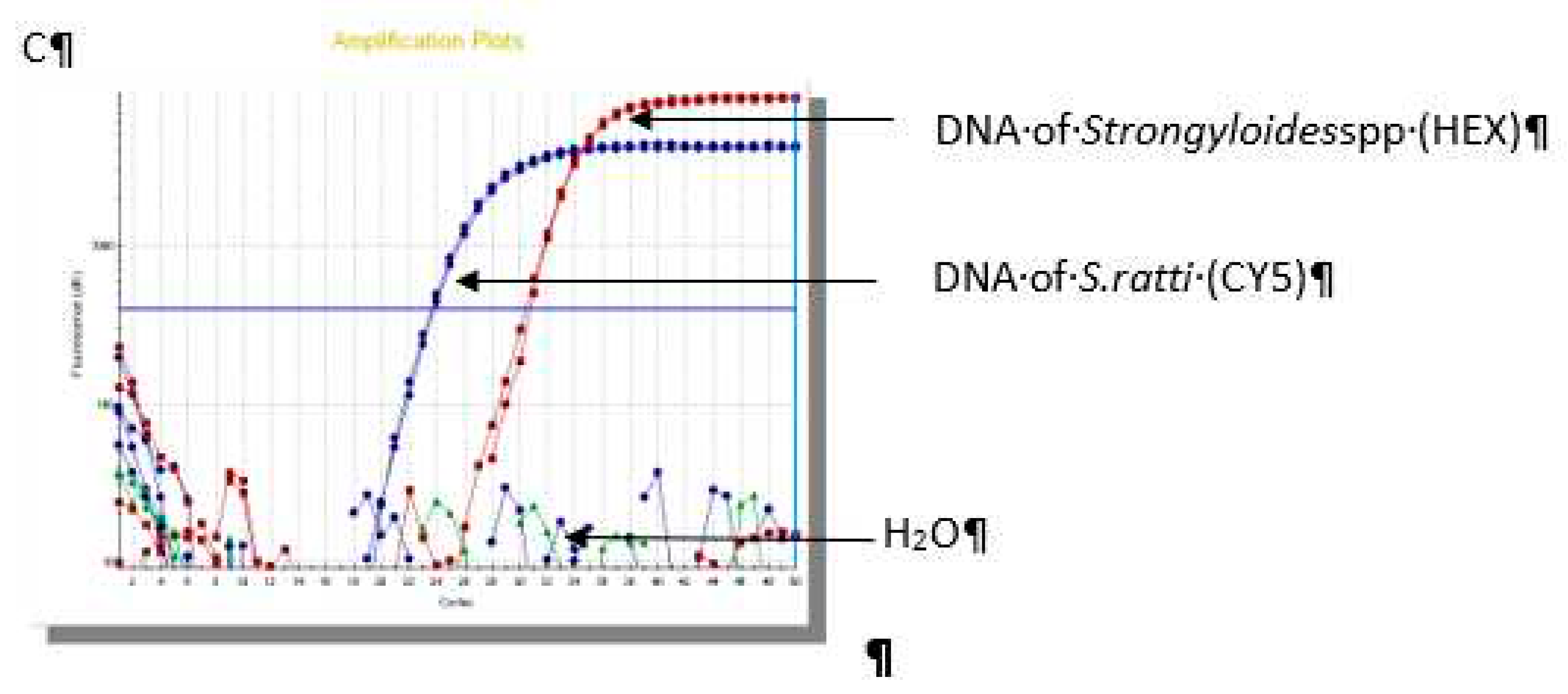
The results of real-time PCR assay showed that 66/70 (94.2%) samples were positive for
Strongyloidesstercolaris, 2/70 (2.9%) samples were
Strongyloides ratti and 2/70 (2.9%) samples were co-infection (
Figure 2).
3.4. Nested - PCR and Sequencing
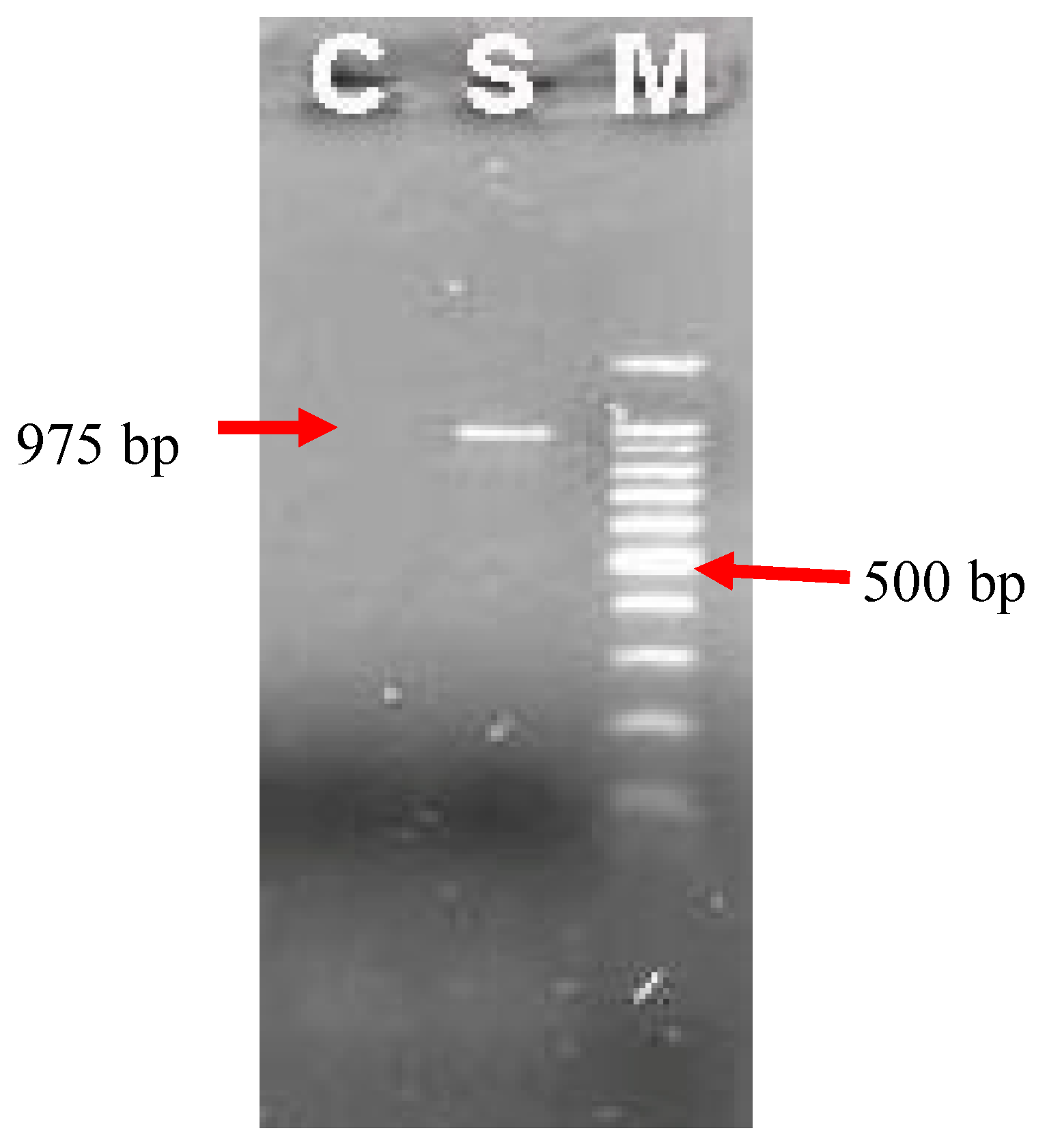
A total number of 14 samples were tested by nested PCR, including 10 out of 66 of
Strongyloidesstercolaris and 4 samples of
Strogyloides ratti DNA. The result product was electrophoresed on agarose gel shown in
Figure 3 with the expected size (975 bp). No DNA product was detected in negative sample.
Figure 2.
Result of real-time PCR in identifiication of Strongyloidesspp.(A): Sample No.1 S. stercoralis; (B): Sample No.25 coinfection of S. stercoralis - S. ratti; (C): Sample No.54 S. ratti.
Figure 2.
Result of real-time PCR in identifiication of Strongyloidesspp.(A): Sample No.1 S. stercoralis; (B): Sample No.25 coinfection of S. stercoralis - S. ratti; (C): Sample No.54 S. ratti.
All 14 nested-PCR products were purified, sequenced and compared with the 18S rRNA of the
Strongyloides spp. deposited in Genbank by using BLAST program. The result showed that the genetic homologysequence at AJ417023 location of
Strongyloides spp. dominated in Duc Hoa. While the sequence homology elsewhere (AB453329.1, AB923889.1, LN609412.1, AB923888.1, LL999065.1, LM528082.1, LL999063.1, MK369923.1 and LL999126.1) was 93-100% for
S. stercoralis and more than 98% for
S.ratti (
Table 3).
Figure 3.
Electrophoresis products of nested PCR on agarose gel 1.5%.M: scale of DNA 100 bp; C: Negative control (H2O); S: DNA sample of Strongyloidesspp.
Figure 3.
Electrophoresis products of nested PCR on agarose gel 1.5%.M: scale of DNA 100 bp; C: Negative control (H2O); S: DNA sample of Strongyloidesspp.
4. Discussion
Investigating the prevalence and identifying the species components of Strongyloidesspp among the residents of Duc Hoa are of high priority due to the special ecological context of this location and suitable condition for distribution and transmission of this intestinal nematode [23,24,25,35].
In this study, 6.6% (79/1,190) participants were confirmed with a gastrointestinal strongyloidiasis, however, simple wet mount microscopic technique was only able to detect 58.2% (46/79). It suggested that the detection rate of direct smear preparation technique for strongyloidiasis was quite low. Therefore, it is not recommended as the main diagnostic test for routine screening of Strongyloidesspp infection in community because of its low sensitivity.
On the other hand, modified Harada-Mori filter paper culture (mHMFPC) was able to detect more Strongyloidesspp larvae than wet mount (93.7% vs 58.2%). This result was higher than 78.4% in a study at Cu Chi district (2005) [36] and 47.8% by Rayzan H. Z et al (2012) [15] in Egypt. There were still 5 out of 79 (6.3%) cases with negative results in the mHMFPC techniquebut positive for the direct smear technique. Therefore, it is suggested that both should be used in field studies as the combination would increase detection rates [37].
The 1st stage larvae morphology in this study tended to be longer than in the studies by Grove DI (1989) [30] and Prayong R.(2013) [31], while the horizontal size of larvae was similar with these studies. The average length of the esophagus was 75.7 µm, with the bulb forms; and the average ratio compared to the body length was 27.1%, completely consistent with the structure of stage 1stlarvae.The buccal cavity of 1st stage of strongyloidesspp larvae had an average length of 4.4 µm, the min - max was 3.9 µm - 5.3 µm. This was an important structure to distinguish with 1st stage hookworm larvae that had long buccal cavity.This is consistent with the findings by authors Grove D. I (1989) [30], Hong TT (2017) [38] and Prayong R. (2013) [31] with the mean of buccal cavity of 4-8 µm. From the above results, it would be confirmed that all surveyed larvae in this study were 1st stage Strongyloidesspp larvae.
The 2nd stage larve of Strongyloides spp. had a body length of 576.4 µm in average, the average horizontal size was 16.9 µm. The results were consistent with those by Grove D.I (1989) [30], Prayong R. (2013) [31], which reported between 450 - 600 µm, horizontally slender than 1st stage larvae. The larvae had a cylindrical esophagus with an average length of 244.7 µm, and had a body length ratio of 42.5%. In addition, none of the larvae had pointed tail, in fact, 86.1% of them had notched tail. This is entirely consistent with the structure of the 2nd stage larvae with cylindrical esophagus which covers one-third of the body length.
Only 8 adults of Strongyloides spp., including 5 males and 3 females, were collected and morphologically characterized in this study. The average length of the male’s body was 778.8 µm, and the average horizontal size was 45.1 µm. Although this result was higher than that reported by Prayong R. (2013) of appropriately700 µm [31], it is completely consistent with those by Grove D.I. (1989) [30] from 700 to 900 µm. They had an average esophageal length of 131.3 µm, and reached a ratio of 17% of the larval body length.The average spicule of adult male worm was 33.4 µm. In addition, it was also shown that the female worm was about 916.7 µm x 46.2 µm. This result was within the threshold but at a low level compared to the reported by Grove D.I. (1989) [30]. Free-living females had a longer esophagus than males (130.6µm vs. 131.3µm). This resulted in a smaller esophagus-to-body length ratio in females, reaching 14.3% compared to 17% in males. The two branches of the uterus, filled with eggs, were located symmetrically across the vulva. Distinguishing Strongyloidesspieces could only be based on morphology. This was quite challenging because the internal organs of this species were similar in shape and too small in size [30]. For the above reasons, in terms of morphology corresponding to the design in this study, only 1st, 2nd stage larvae, male and female adults of Strongyloidesspp were identified.
In this study, 70 extracted DNA samples of Strongyloidesspp isolates, originally retrieved from human, were characterized by real-time PCR amplification of the 18S, 28S region of the rDNA gene. The result showed that S. stercoralis accounted for 97.1%, of which 2.9% was co-infected with S. ratti.
The dominant S. stercoralis in this study was consistent with those in the studies by N.V.DE (2017) [39] and D.T. Hong (2019) [40] (97.1% vs. 100%, respectively), although the authors did not use the same realtime PCR technique. Moreover, in this study the causative agent of S.ratti, a common gastro-intestinal parasite of the rat, has been detected by molecular biology. It means that this species could be a human pathogen and it could not be differentiated due to the fact that morphological structure of Strongloidesspieces larvae was almost unidentical.
Among 70 DNA samples characterized, 10 positive samples with S. stercoralis and 4 samples with S. ratti were randomly choosen to be amplfied by the nested-PCR. All PCR results yielded an identical pattern, with a visible fragment size of 975-bp. For all 14 isolates, nucleotide sequencing was achieved; and comparison with other human pathogen Strongyloidesspp available sequences in GenBank was performed, using BLAST program. The results confirmed the identity of 12/14 sequences as S. stercoralis with high level of similarity (91.3% - 100%) and over 98% for S. ratti. Between the two co-infection samples S. stercoralis and S. ratti (samples 25 and 65), the higher similarity belonged to S. stercoralisspecies. This result could be explained by the highert number of S. stercoralis pathogens in the sample, or the S. ratti gene segment not being amplified through reaction.
The similarity of S. stercoralis sequence was recorded in comparision with the reference sequence in Genbankin the study by De NV (2017) [39] and Hong DT (2019) [40]. However, both authors experimented in small groups, consisting of only 2 and 7 samples respectively, thus it is inevitable that the rate of variation would be lower.
On the other hand, the phylogenetic tree showed the S. ratti species in the study completely closed to the species originating from the Rattus novegicus and it was consistent with the findings by Polanco Campo L F. (2018) [41] in Brazil. Thus, with the identity of new species - S. ratti, this study provided solid evidence to confirm the presence of S. ratti as a causative infectious disease on human at the molecular level.
This study has three main limitations. Firstly, the study sites were not representative for the ecological environment of Long An province as well as the Southern provinces in Vietnam. The second limitation is the small number of participants and positive samples, thus it is necessary for further studies to be conducted in different areas in Vietnam, in order to compare with the results of this study. Last but not least, the 18S and 28S rDNA gene would help to differentiate Strongyloides species, however, it is required other specific genetic markers such as cytochrome C oxidase subunit 1 (Cox1) gene to be used to describe the phylogenetic tree of Strongyloides spp. more specifically.
In conclusion, the study shows that it is not possible to make distinction among different Strongyloides species’ larvae by performing morphologically identification because some larvae may be zoonostic pathogen. A molecular amplification of small subunit ribosome DNA and following sequence analysis is a suitable method for discrimination of Strongyloides spp. isolated from faecal samples. It is also the first report of zoonosis Strongyloides ratti infection in human in Vietnam.
Author Contributions
Conceptualization, Huong Nguyen-Thu and Vinh Le Duc; Data curation, Toan Nguyen Minh and Lang Ngo Van; Formal analysis, Thach Nguyen Kim and Quang Huynh Hong; Funding acquisition, Hanh Tran Thi Duc and Quang Huynh Hong; Investigation, Vinh Le Duc; Methodology, Huong Nguyen Thu and Vinh Le Duc; Project administration, Binh Do Nhu and Hanh Tran Thi Duc; Resources, Vinh Le Duc and Thach Nguyen Kim; Software, Toan Nguyen Minh and Lang Ngo Van; Supervision, Huong Nguyen-Thu; Validation, Quang Huynh Hong; Visualization, Binh Do Nhu and Huong Nguyen-Thu; Writing – original draft, Huong Nguyen Thu, Vinh Le Duc and Thach Nguyen Kim; Writing – review & editing, Huong Nguyen Thu, Hanh Tran Thi Duc and Binh Do Nhu. All authors have read and agreed to the published version of the manuscript.
Funding
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Ethical Statement
All people described in this research were signed written informed consent for the publication of the case details, and the protocol was approved by the Ethical Review Committee of National Institute of Malariology, Parasitology and Entomology (No. 1796/QD-VSR, date 18/11/2019). The study was in line with the Declaration of Helsinki. Written informed consent was signed by all participants after full explanation. In addition, the study was also approved by the local government of Duc Hoa district, Long An province, Vietnam.
Informed Consent Statement
All people described in this research were signed written informed consent for the publication of the case details.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request. If you have concerns about sharing the data, please contact nth14@huph.edu.vn and ducvinh@pnt.edu.vn.
Acknowledgments
We thank you all the staffs at the Laboratory of Medical Parasitology and Laboratory of Molecular Biology, Pham Ngoc Thach University of Medicine, Hochiminh city, Vietnam, for supporting this study. We are grateful to all the people and local authorities of Duc Hoa district, Long An province, Vietnam for their enthusiastic participation in the study.
Disclosure
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- Page, W.; Judd, J.A.; Bradbury, R.S. The unique life cycle of Strongyloides stercoralis and implications for public health action. Trop. Med. Infect. Dis. 2018, 3, 53. [Google Scholar] [CrossRef]
- WHO. Strongyloidiasis; In. World Health Organization: Geneva, 2019. [Google Scholar]
- Mora Carpio, A.L.M.M. Strongyloides Stercoralis (Strongyloidiasis) [Updated 2019 Dec 25]; StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2019. [Google Scholar]
- Schär, F.; Trostdorf, U.; Giardina, F.; Khieu, V.; Muth, S.; Marti, H.; Vounatsou, P.; Odermatt, P. Strongyloides stercoralis: global distribution and risk factors. PLOS Negl. Trop. Dis. 2013, 7, e2288. [Google Scholar] [CrossRef]
- Mai, T.X.D.T.; Tuan, P.A. Medical Parasitology; In: Medical Publishing House, 2010; pp. 202–210. [Google Scholar]
- Lindo, J.F.; Lee, M.G. Strongyloides stercoralis and S. fulleborni. In Principles and practice of clinical parasitology; John Wiley & Sons Ltd Chichester, 2001; pp. 485–490. [Google Scholar]
- Siddiqui, A.A.; Berk, S.L.; Ericsson, C.D.; Steffen, R. Diagnosis of Strongyloides stercoralis infection. Clin. Infect. Dis. 2001, 33, 1040–1047. [Google Scholar] [CrossRef]
- Feely, N.M.; Waghorn, D.J.; Dexter, T.; Gallen, I.; Chiodini, P. Strongyloides stercoralis hyperinfection: difficulties in diagnosis and treatment. Anaesthesia 2010, 65, 298–301. [Google Scholar] [CrossRef] [PubMed]
- Marcos, L.A.; Terashima, A.; DuPont, H.L.; Gotuzzo, E. Strongyloides hyperinfection syndrome: an emerging global infectious disease. Trans. R. Soc. Trop. Med. Hyg. 2008, 102, 314–318. [Google Scholar] [CrossRef]
- de Kaminsky, R.G. Evaluation of three methods for laboratory diagnosis of Strongyloides stercoralis infection. J. Parasitol. 1993, 79, 277–80. [Google Scholar] [CrossRef]
- Conway, D.; Atkins, N.; Lillywhite, J.; Bailey, J.; Robinson, R.; Lindo, J.; Bundy, D.; Bianco, A. Immunodiagnosis of Strongyloides stercoralis infection: a method for increasing the specificity of the indirect ELISA. Trans. R. Soc. Trop. Med. Hyg. 1993, 87, 173–176. [Google Scholar] [CrossRef] [PubMed]
- Bisoffi, Z.; Buonfrate, D.; Sequi, M.; Mejia, R.; Cimino, R.O.; Krolewiecki, A.J.; Albonico, M.; Gobbo, M.; Bonafini, S.; Angheben, A.; et al. Diagnostic accuracy of five serologic tests for Strongyloides stercoralis infection. PLOS Negl. Trop. Dis. 2014, 8, e2640. [Google Scholar] [CrossRef] [PubMed]
- Koosha, S.; Fesharaki, M.; Rokni, M.B. Comparison of enzyme-linked immunosorbent assay and indirect immunofluorescence assay in the diagnosis of human strongyloidiasis. Indian J. Gastroenterol. 2004, 23, 214–6. [Google Scholar]
- Levenhagen, M.A.; Costa-Cruz, J.M. Update on immunologic and molecular diagnosis of human strongyloidiasis. Acta Trop. 2014, 135, 33–43. [Google Scholar] [CrossRef]
- Rayan, H.Z.; Soliman, R.H.; Galal, N.M. Detection of Strongyloides stercoralis in fecal samples using conventional parasitological techniques and real-time PCR: a comparative study. Parasitol United J. 2012, 5, 27–34. [Google Scholar]
- Buonfrate, D.; Requena-Mendez, A.; Angheben, A.; Cinquini, M.; Cruciani, M.; Fittipaldo, A.; Giorli, G.; Gobbi, F.; Piubelli, C.; Bisoffi, Z. Accuracy of molecular biology techniques for the diagnosis of Strongyloides stercoralis infection—A systematic review and meta-analysis. PLOS Neglected Trop. Dis. 2018, 12, e0006229. [Google Scholar] [CrossRef] [PubMed]
- Sitta, R.B.; Malta, F.M.; Pinho, J.R.; Chieffi, P.P.; Gryschek, R.C.B.; Paula, F.M. Conventional PCR for molecular diagnosis of human strongyloidiasis. Parasitology 2014, 141, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Buonfrate, D.; Formenti, F.; Perandin, F.; Bisoffi, Z. Novel approaches to the diagnosis of Strongyloides stercoralis infection. Clin. Microbiol. Infect. 2015, 21, 543–552. [Google Scholar] [CrossRef]
- Kramme, S.; Nissen, N.; Soblik, H.; Erttmann, K.; Tannich, E.; Fleischer, B.; Panning, M.; Brattig, N. Novel real-time PCR for the universal detection of Strongyloides species. J. Med Microbiol. 2011, 60, 454–458. [Google Scholar] [CrossRef] [PubMed]
- Verweij, J.J.; Canales, M.; Polman, K.; Ziem, J.; Brienen, E.A.; Polderman, A.M.; van Lieshout, L. Molecular diagnosis of Strongyloides stercoralis in faecal samples using real-time PCR. Trans. R. Soc. Trop. Med. Hyg. 2009, 103, 342–346. [Google Scholar] [CrossRef]
- Jaleta, T.G.; Lok, J.B. Advances in the Molecular and Cellular Biology of Strongyloides spp. Curr. Trop. Med. Rep. 2019, 6, 161–178. [Google Scholar] [CrossRef] [PubMed]
- Diep, N.T.N.; Thai, P.Q.; Trang, N.N.M.; Jäger, J.; Fox, A.; Horby, P.; Phuong, H.V.M.; Anh, D.D.; Mai, L.T.Q.; VAN Doorn, H.R.; et al. Strongyloides stercoralis seroprevalence in Vietnam. Epidemiology Infect. 2017, 145, 3214–3218. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Cheong, F.W.; Liew, J.W.K.; Lau, Y.L. Seroprevalence of fascioliasis, toxocariasis, strongyloidiasis and cysticercosis in blood samples diagnosed in Medic Medical Center Laboratory, Ho Chi Minh City, Vietnam in 2012. Parasites Vectors 2016, 9, 486. [Google Scholar] [CrossRef]
- Van De, N.; Minh, P.N.; Van Duyet, L.; Mas-Coma, S. Strongyloidiasis in northern Vietnam: epidemiology, clinical characteristics and molecular diagnosis of the causal agent. Parasites Vectors 2019, 12, 515. [Google Scholar] [CrossRef]
- Hicks, N. Agricultural Extension Officers in Two Districts of Long An Province; local government in Vietnam: Beyond Hanoi, 2004; Volume 229. [Google Scholar]
- Organization, W.H. Basic laboratory methods in medical parasitology; World Health Organization: 1991.
- Organization, W.H. Bench aids for the diagnosis of intestinal parasites; World Health Organization: 2019.
- Hotez, P.; Hawdon, J.; Cox, N.; Schad, G.; Richards, F. An apparatus for modified Harada-Mori cultures of third-stage hookworm larvae. J Helminthol Soc Wash. 1990, 57, 167–169. [Google Scholar]
- Khanna, V.; Tilak, K.; Prakash, P.Y.; Mukhopadhyay, C. Modified agar plate culture method for culture of Strongyloides stercoralis. Trop. Parasitol. 2015, 5, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Grove, D.I. Strongyloidiasis: a major roundworm infection of man; Taylor and Francis: London - Newyork – Philadenphia, 1989. [Google Scholar]
- Radomyos, P. Atlas of medical parasitology, 9 ed.; Medical Media: Bangkok, 2013. [Google Scholar]
- Watts, M.R.; James, G.; Sultana, Y.; Ginn, A.N.; Outhred, A.C.; Kong, F.; Verweij, J.J.; Iredell, J.R.; Chen, S.C.; Lee, R. A loop-mediated isothermal amplification (LAMP) assay for Strongyloides stercoralis in stool that uses a visual detection method with SYTO-82 fluorescent dye. Am. J. Trop. Med. Hyg. 2014, 90, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Kousha, S.; Kazemi, B.; Bonyadi, F. The 18S Ribosomal DNA sequence of Strongyloides stercoralis in Iran. 2009.
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: an important software for molecular biology. GERF Bull Biosci. 2011, 2, 60–61. [Google Scholar]
- Schär, F.; Giardina, F.; Khieu, V.; Muth, S.; Vounatsou, P.; Marti, H.; Odermatt, P. Occurrence of and risk factors for Strongyloides stercoralis infection in South-East Asia. Acta Trop. 2016, 159, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Vinh, T.D.H.L. Investigation of Strongyloides spp. ifection by improved faecal testing methods in Phu My Hung commune, Cu Chi district, Ho Chi Minh City. Hochiminh Med. J. 2005, 9, 101–105. [Google Scholar]
- Pocaterra, L.A.; Ferrara, G.; Peñaranda, R.; Rojas, E.; Pérez-Chacón, G.; Hernán, A.; Certad, G.; Goldstein, C.; Núñez, L. Improved detection of Strongyloides stercoralis in modified agar plate cultures. Am. J. Trop. Med. Hyg. 2017, 96, 863–865. [Google Scholar] [CrossRef] [PubMed]
- TT, H. Medical Parasitology (Strongyloidiasis); Medical Publishing House, 2017.
- De NV, M.P. Clinical manifestations, subclinical characterization and identification of Strongyloides stercoralis in humans. J. Malar. Parasite Dis. Control. 2017, 2, 15–20. [Google Scholar]
- al HDe. Identification of Strongyloides stercoralis larvae collected from patients by molecular technique based on 18S gene sequence. Paper presented at: National conference of parasitology 2019.
- Campo-Polanco, L.; Sarmiento, J.M.H.; Mesa, M.A.; Franco, C.J.V.; López, L.L.; Botero, L.E.; Builes, L.A.G. Strongyloidiasis in humans: diagnostic efficacy of four conventional methods and real-time polymerase chain reaction. Rev. Soc. Bras. Med. Trop. 2018, 51, 493–502. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).