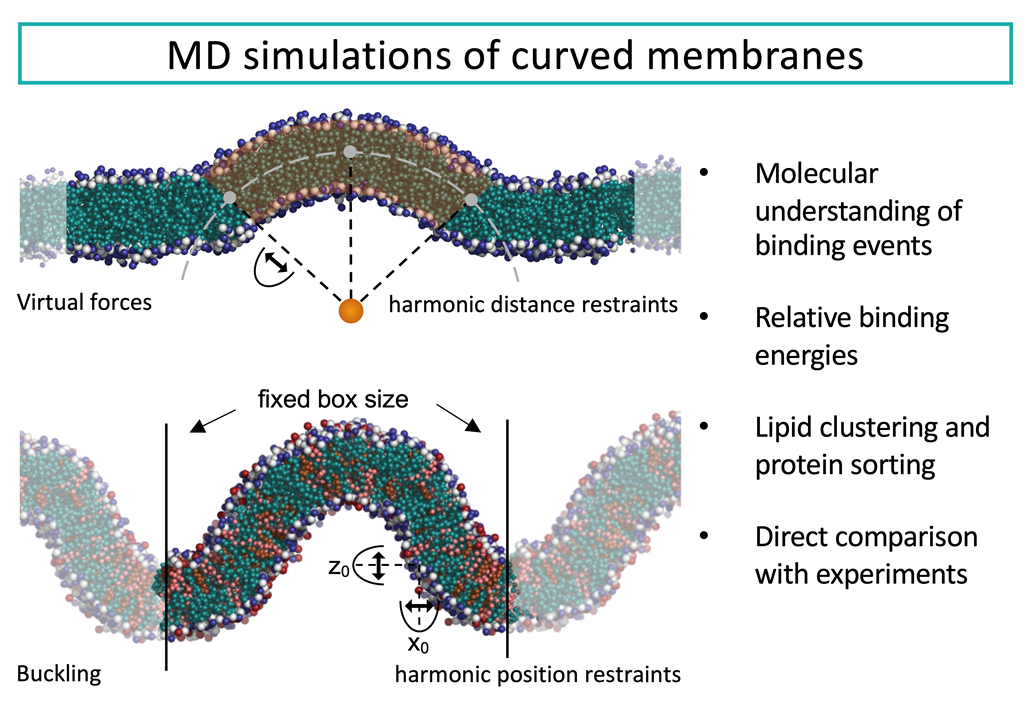
Eukaryotic cells contain membranes with various curvatures, from the near-plane plasma membrane to the highly curved membranes of organelles, vesicles, and membrane protrusions. These curvatures are generated and sustained by curvature-inducing proteins, peptides, and lipids, and describing these mechanisms is an important scientific challenge. In addition to that, some molecules can sense membrane curvature and a thereby be trafficked to specific locations. The description of curvature-sensing is another fundamental challenge. Curved lipid membranes and their interplay with membrane-associated proteins can be investigated with molecular dynamics (MD) simulations, given the right tools. Various methods for simulating curved membranes with MD are discussed here, including tools for setting up simulation of vesicles, and methods for sustaining membrane curvature. The latter are divided into methods that exploit scaffolding virtual beads, methods that use curvature-inducing molecules, and methods applying virtual forces. The variety of simulation tools allow the researcher to closely match the conditions of experimental studies of membrane curvatures.