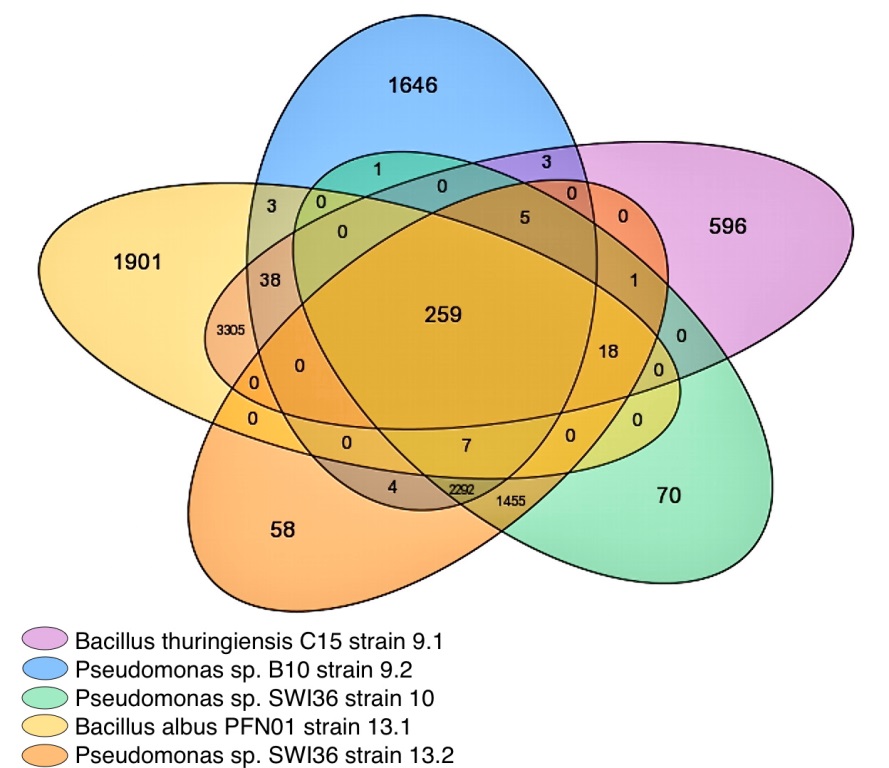
Global use of single-use non-biodegradable plastics, like bottles made of polyethylene tereph-thalate (PET), have contributed to catastrophic levels of plastic pollution. Fortunately, microbi-al communities are adapting to assimilate plastic waste. Previously, our work showed a full consortium of five bacteria capable of synergistically degrading PET. Using omics approaches we identified key genes implicated in PET degradation within the consortium’s pangenome and transcriptome. This analysis led to the discovery of a novel PETase, EstB, discovered to hydrolyze oligomer BHET, and polymer PET. Besides genes implicated in PET degradation, many other biodegradation genes were discovered. Over 200 plastic and plasticizer degrada-tion related genes were discovered through the Plastic Microbial Biodegradation Database (PMBD). Diverse carbon source utilization was observed by a microbial community-based as-say, which paired with an abundant number of plastic and plasticizer degrading enzymes in-dicates a promising possibility for mixed plastic degradation. Using RNAseq differential analysis, several genes were predicted to be involved in PET degradation including aldehyde dehydrogenases and several classes of hydrolases. Active transcription of PET monomer me-tabolism was also observed, including the generation of polyhydroxyalkanoate (PHA) bi-opolymers. These results present an exciting opportunity for the bio-recycling of mixed plastic waste with upcycling potential.