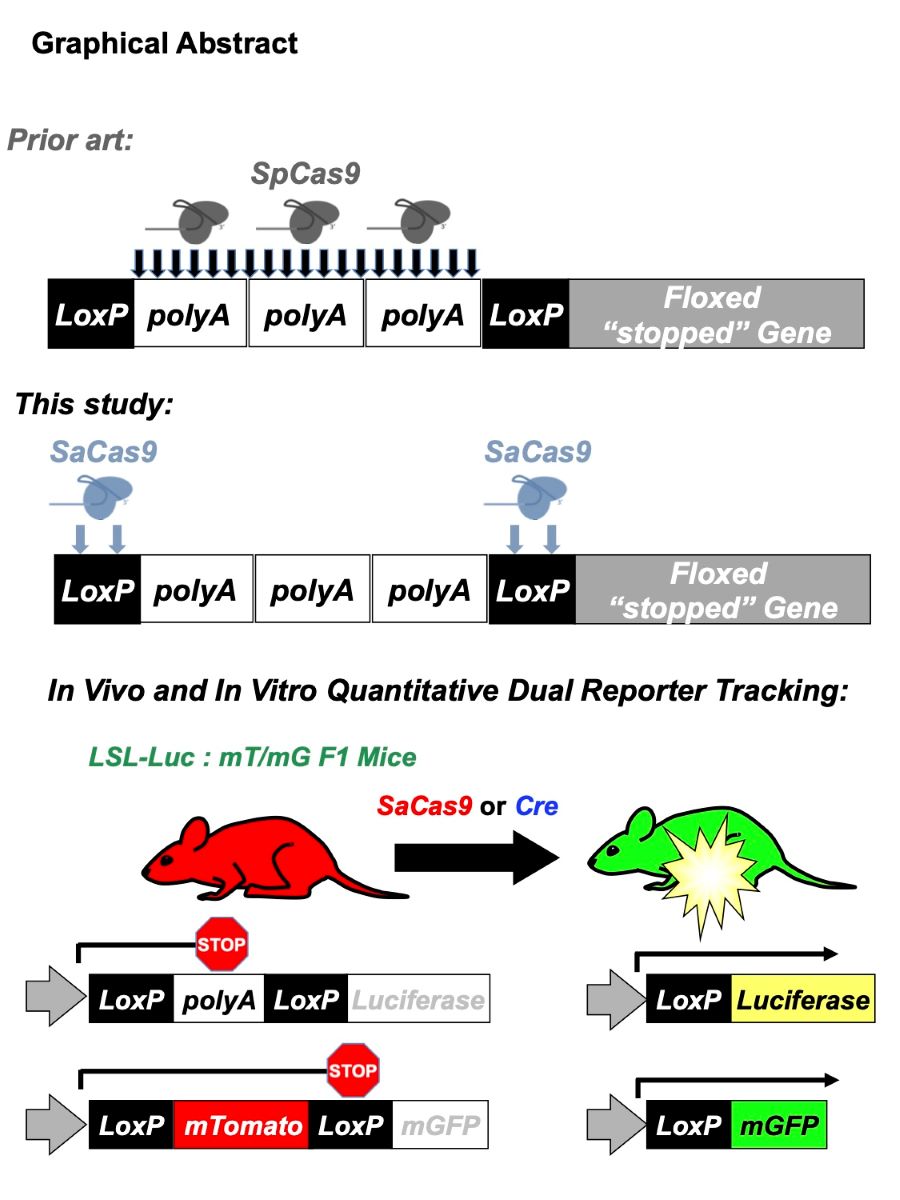
The development of CRISPR associated proteins, such as Cas9, has led to increased accessibility and ease of use in genome editing. However, additional tools are needed to quantify and identify successful genome editing events in living animals. We developed a method to rapidly and quantitatively monitor gene editing activity non-invasively in living animals that also facilitates confocal microscopy and nucleotide level analyses at the end of study. Here we report a new CRISPR “footprinting” approach to activate luciferase and fluorescent proteins in mice as a function of gene editing. This system is based on experience with our prior Cre-detector system and is designed for Cas editors able to target LoxP including gRNAs including SaCas9 and ErCas12a [1, 2]. These CRISPRs cut specifically within LoxP, an approach that is a departure from previous gene editing in vivo activity detection techniques that targeted adjacent stop sequences. In this sensor paradigm, CRISPR activity was monitored non-invasively in living Cre reporter mice (FVB.129S6(B6)-Gt(ROSA)26Sortm1(Luc)Kael/J and Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J, which will be referred to as LSL and mT/mG throughout the paper) after intramuscular or intravenous hydrodynamic plasmid injections, demonstrating utility in two diverse organ systems. The same genome-editing event was examined at the cellular level in specific tissues by confocal microscopy to determine the identity and frequency of successfully genome-edited cells. Further, SaCas9 induced targeted editing at efficiencies that were comparable to Cre recombinase demonstrating high effective delivery and activity in a whole animal. This work establishes genome editing tools and models to track CRISPR editing in vivo non-invasively and to fingerprint the identity of targeted cells. This approach also enables similar utility for any of the thousands of previously generated LoxP animal models.