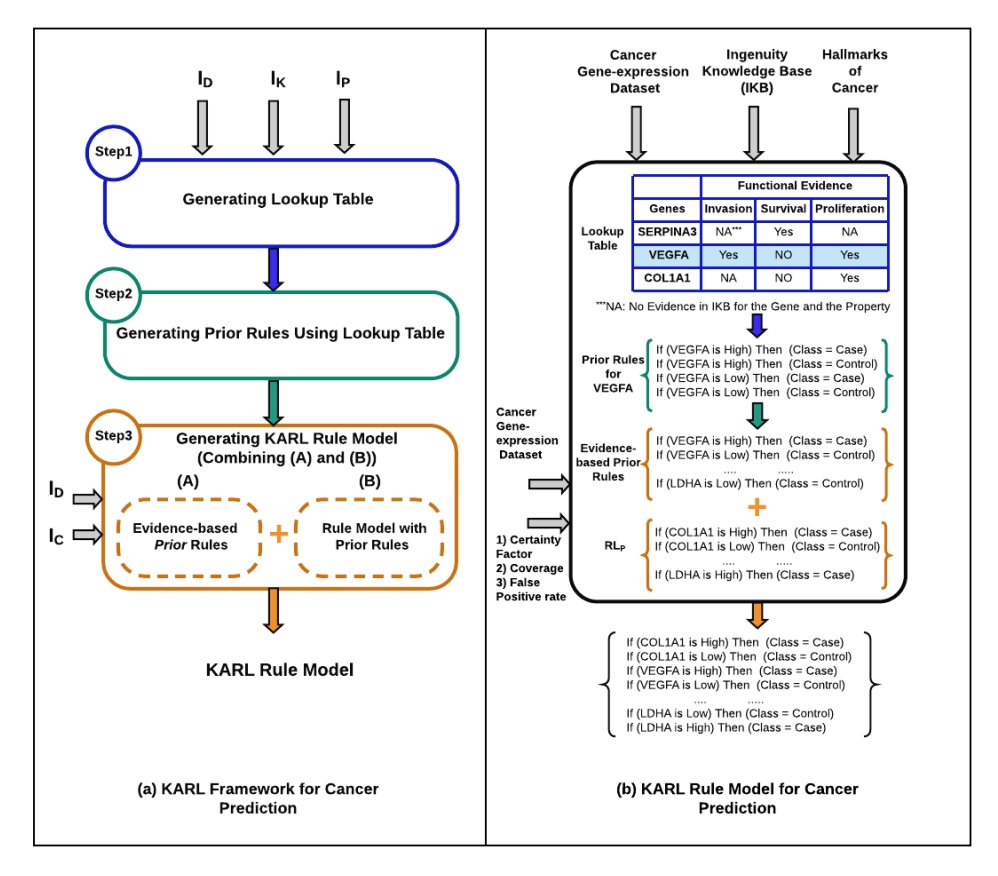
Background: Ongoing molecular profiling studies enabled by advances in biomedical technologies are producing vast amounts of ‘omic’ data for early detection, monitoring, and prognosis of diverse diseases. A major common limitation is the scarcity of biological samples, necessitating integrative modeling frameworks that can make optimal use of available data for disease classification tasks. Related data sets are often available from different studies, but may have been generated using different technology platforms. Thus, there is a critical need for flexible modeling methods that can handle data from diverse sources to facilitate the discovery of robust biomarkers that underlie disease regulatory processes. Results: In this paper, we introduce a novel framework called Knowledge Augmented Rule Learning (KARL), which incorporates two sources of knowledge, domain, and data, for pattern discovery from small and high-dimensional datasets, such as transcriptomic data. We propose KARL as a transfer rule learning framework in which knowledge of the domain is transferred to the learning process on data in order to 1) improve the reliability of the discovered patterns, and 2) study the knowledge of the domain when used along with data for modeling. In this work, we generated KARL models on gene expression datasets for five types of cancer, including brain, breast, colon, lung, and prostate. As our knowledge of the domain, we used the Ingenuity Knowledge Base (IKB) to extract genes related to hallmarks of cancer and annotated these prior relationships before learning classifiers from these datasets. Conclusions: Our results show that KARL produces, on average, rule models that are more robust classifiers than the baseline without such background knowledge, for our tasks of cancer prediction using 25 publicly available gene expression datasets. Moreover, KARL helped us learn insights about previously known relationships in these gene expression datasets, along with new relationships not input as known, to enable informed biomarker discovery for cancer prediction tasks. KARL can be applied to modeling similar data from any other domain and classification task. Future work would involve extensions to KARL to handle hierarchical knowledge to derive more general hypotheses to drive biomedicine.