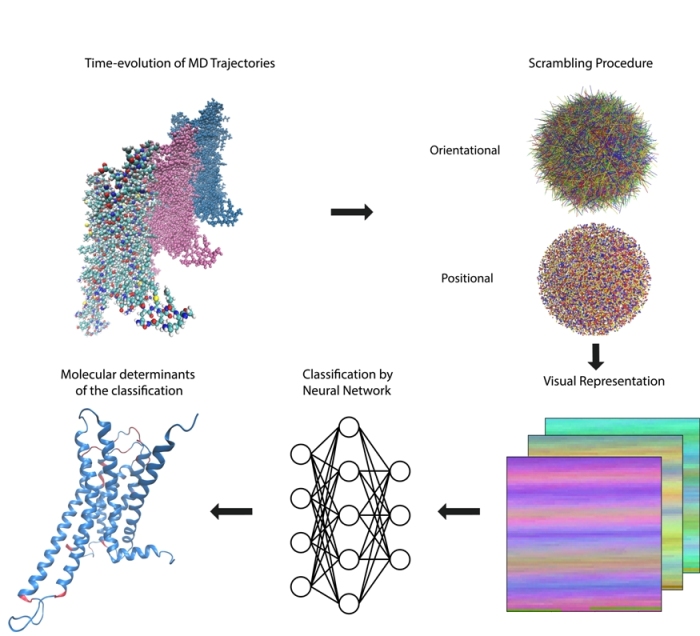
G protein-coupled receptors (GPCRs) play a key role in many cellular signaling mechanisms, and must select among multiple coupling possibilities in a ligand-specific manner in order to carry out a myriad of functions in diverse cellular contexts. Much has been learned about the molecular mechanisms of ligand-GPCR complexes from Molecular Dynamics (MD) simulations. However, to explore ligand-specific differences in the response of a GPCR to diverse ligands, as is required to understand ligand bias and functional selectivity, necessitates creating very large amounts of data from the needed large-scale simulations. This becomes a Big Data problem for the high dimensionality analysis of the accumulated trajectories. Here we describe a new machine learning (ML) approach to the problem that is based on transforming the analysis of GPCR function-related, ligand-specific differences encoded in the MD simulation trajectories into a representation recognizable by state-of-the-art deep learning object recognition technology. We illustrate this method by applying it to recognize the pharmacological classification of ligands bound to the 5-HT2A and D2 subtypes of class A GPCRs from the serotonin and dopamine families. The ML-based approach is shown to perform the classification task with high accuracy, and we identify the molecular determinants of the classifications in the context of GPCR structure and function. This study builds a framework for the efficient computational analysis of MD Big Data collected for the purpose of understanding ligand-specific GPCR activity.