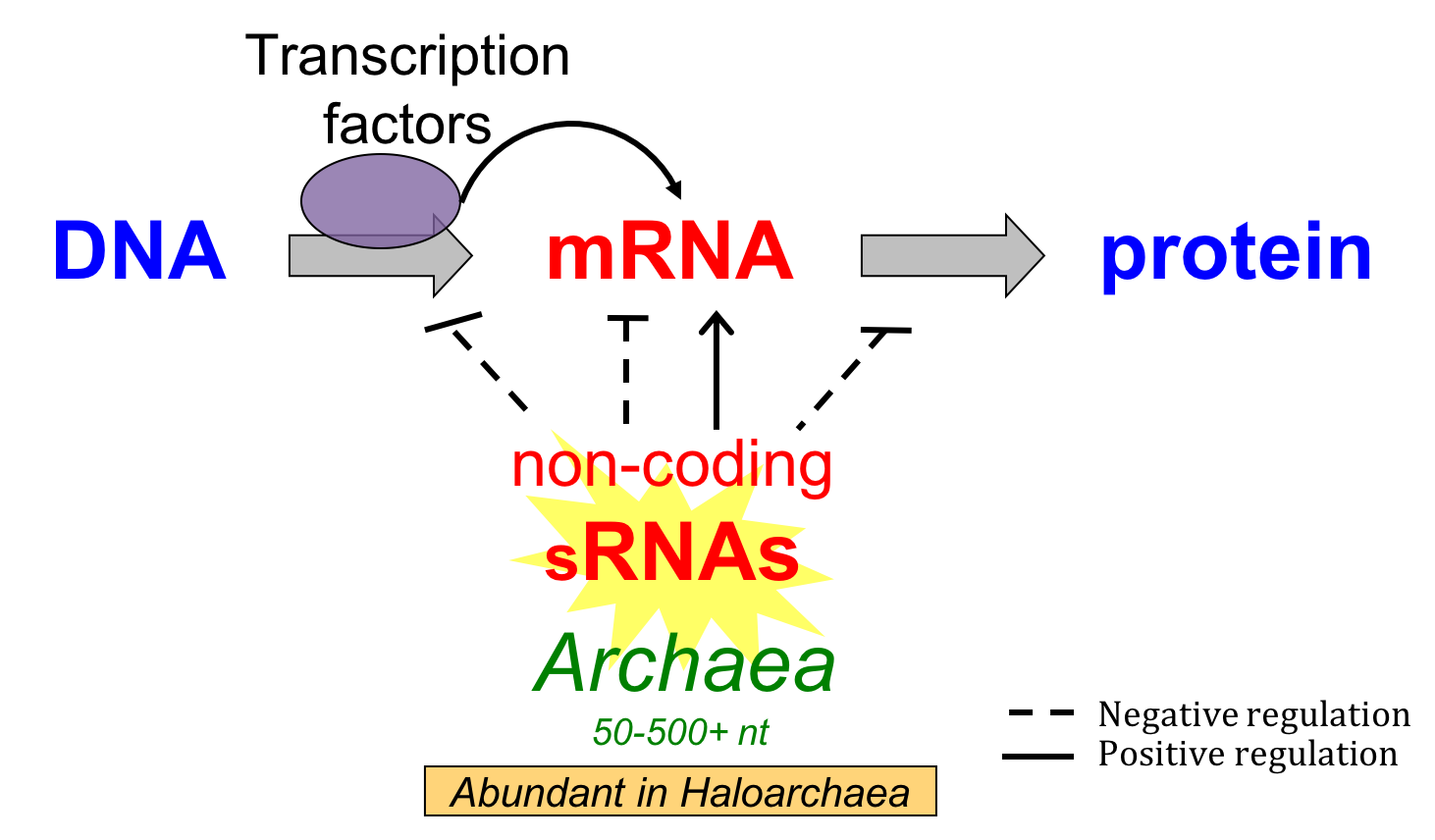
Small non-coding RNAs (sRNAs) are ubiquitously found in the three domains of life playing large-scale roles in gene regulation, transposable element silencing, and defense against foreign elements. While a substantial body of experimental work has been done to uncover function of sRNAs in Bacteria and Eukarya, the functional roles of sRNAs in Archaea are still poorly understood. Recently, high throughput studies using RNA-sequencing revealed that sRNAs are broadly expressed in the Archaea, comprising thousands of transcripts within the transcriptome during non-challenged and stressed conditions. Antisense sRNAs, which overlap a portion of a gene on the opposite strand (cis-acting), are the most abundantly expressed non-coding RNAs and they can be classified based on their binding patterns to mRNAs (3’ UTR, 5’ UTR, CDS-binding). These antisense sRNAs target many genes and pathways, suggesting extensive roles in gene regulation. Intergenic sRNAs are less abundantly expressed and their targets are difficult to find because of a lack of complete overlap between sRNAs and target mRNAs (trans-acting). While many sRNAs have been validated experimentally, a regulatory role has only been reported for very few of them. Further work is needed to elucidate sRNA-RNA binding mechanisms, the molecular determinants of sRNA-mediated regulation, whether protein components are involved, and how sRNAs integrate with complex regulatory networks.