Submitted:
28 January 2026
Posted:
28 January 2026
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
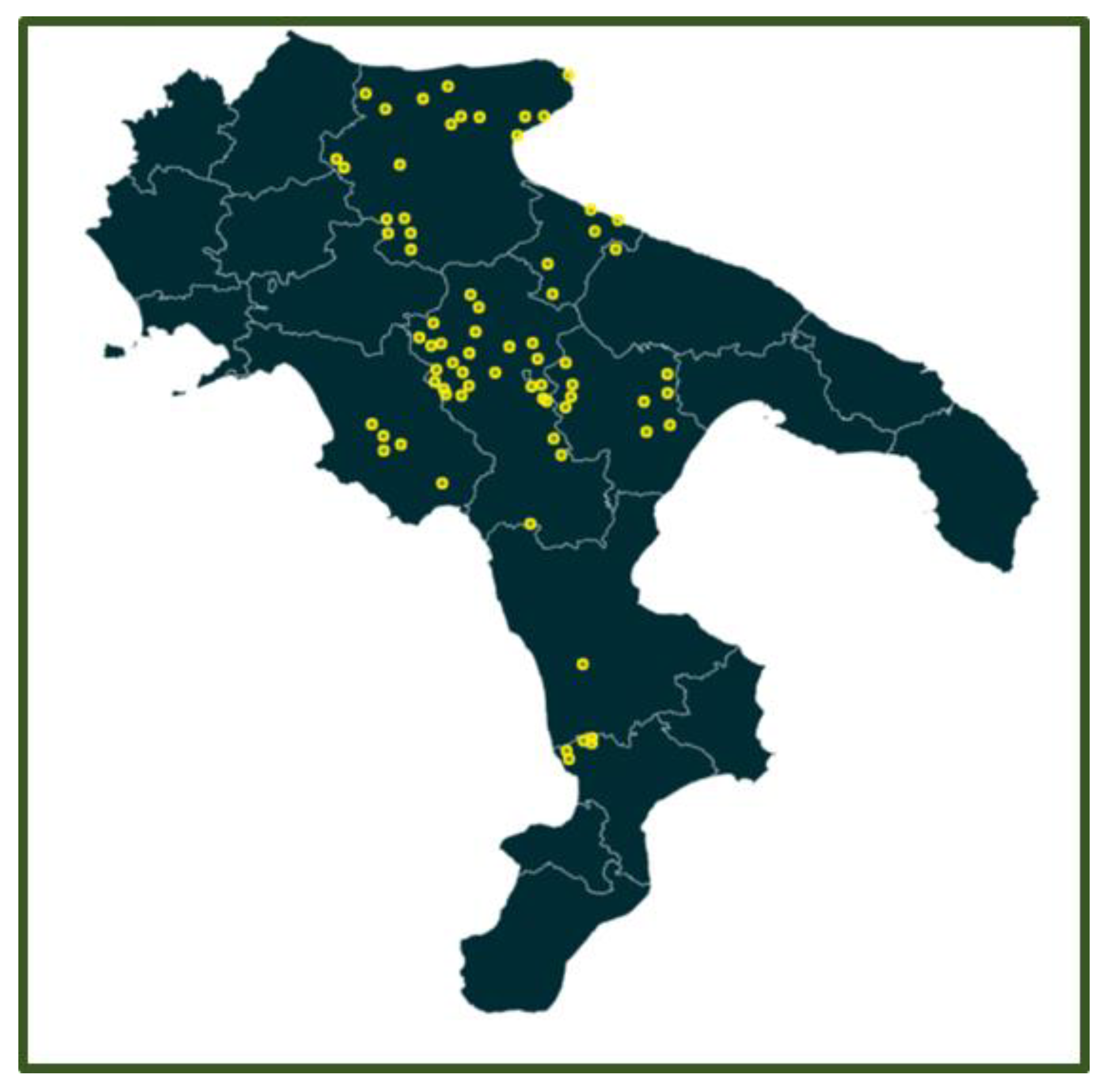
2.1. Serological Investigation
2.2. Nucleic Acid Extraction
2.3. Molecular Detection of Influenza Viruses
2.4. Molecular Detection of Flaviviruses
2.5. Molecular Detection of PCV2
3. Results
4. Discussion
5. Conclusions
Funding
References
- Pittiglio, C.; Khomenko, S.; Beltran-Alcrudo, D. Wild boar mapping using population-density statistics: From polygons to high resolution raster maps. PLoS One 2018, 13. [Google Scholar]
- Lewis, J.S.; Farnsworth, M.L.; Burdett, C.L.; Theobald, D.M.; Gray, M.; Miller, R.S. Biotic and abiotic factors predicting the global distribution and population density of an invasive large mammal. Sci Rep. 2017, 7. [Google Scholar] [CrossRef]
- Torres, R.T.; Fernandes, J.; Carvalho, J.; Cunha, M.V.; Caetano, T.; Mendo, S.; et al. Wild boar as a reservoir of antimicrobial resistance. Vol. 717, Science of the Total Environment. Elsevier B.V.; 2020.
- Massei, G.; Kindberg, J.; Licoppe, A.; Gačić, D.; Šprem, N.; Kamler, J.; et al. Wild boar populations up, numbers of hunters down? A review of trends and implications for Europe. Pest Manag Sci. 2015, 71, 492–500. [Google Scholar] [CrossRef]
- Bieber, C.; Ruf, T. Population dynamics in wild boar Sus scrofa: Ecology, elasticity of growth rate and implications for the management of pulsed resource consumers. Journal of Applied Ecology 2005, 42, 1203–1213. [Google Scholar] [CrossRef]
- Ruiz-Fons, F.; Segalés, J.; Gortázar, C. A review of viral diseases of the European wild boar: Effects of population dynamics and reservoir rôle. Veterinary Journal 2008, 176, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Rosell, C.; Navàs, F.; Romero, S.; De Dalmases, I. ACTIVITY PATTERNS AND SOCIAL ORGANIZATION OF WILD BOAR (Sus scrofa, L.) IN A WETLAND ENVIRONMENT: PRELIMINARY DATA ON THE EFFECTS OF SHOOTING INDIVIDUALS. [CrossRef]
- Vicente, J.; Ruiz-Fons, F.; Vidal, D.; Höfle, U.; Acevedo, P.; Villanúa, D.; et al. Serosurvey of Aujeszky’s disease virus infection in European wild boar in Spain. Veterinary Record 2005, 156, 408–412. [Google Scholar] [CrossRef]
- Mostafa, A.; Naguib, M.M.; Nogales, A.; Barre, R.S.; Stewart, J.P.; García-Sastre, A.; et al. Avian influenza A (H5N1) virus in dairy cattle: origin, evolution, and cross-species transmission. Vol. 15, mBio. American Society for Microbiology; 2024.
- Graziosi, G.; Lupini, C.; Catelli, E.; Carnaccini, S. Highly Pathogenic Avian Influenza (HPAI) H5 Clade 2.3.4.4b Virus Infection in Birds and Mammals. Animals 2024, 14. [Google Scholar] [CrossRef]
- Liang, Y. Pathogenicity and virulence of influenza. Vol. 14, Virulence. Taylor and Francis Ltd.; 2023.
- Abdelwhab, E.M.; Mettenleiter, T.C. Zoonotic Animal Influenza Virus and Potential Mixing Vessel Hosts. Viruses 2023, 15. [Google Scholar] [CrossRef]
- AbuBakar, U.; Amrani, L.; Kamarulzaman, F.A.; Karsani, S.A.; Hassandarvish, P.; Khairat, J.E. Avian Influenza Virus Tropism in Humans. Viruses 2023, 15. [Google Scholar] [CrossRef]
- Ferrara, G.; Longobardi, C.; D’ambrosi, F.; Amoroso, M.G.; D’alessio, N.; Damiano, S.; et al. Aujeszky’s disease in south-Italian wild boars (Sus Scrofa): A serological survey. Animals 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Veljović, L.; Paunović, M.; Glišić, D.; Šolaja, S.; Zurovac Sapundžić, Z.; Maletić, J.; et al. Wild Mammals as Sentinels for West Nile Virus Circulation: Evidence from Serbia. Pathogens 2025, 14. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Melo, N.; González-Barrio, D.; Pinto, M.V.; Ruiz-Fons, F. Aujeszky’s disease in hunted wild boar (Sus scrofa) in the iberian peninsula. J Wildl Dis. 2021, 57, 543–552. [Google Scholar]
- Ciarello, F.P.; Moreno, A.; Miragliotta, N.; Antonino, A.; Fiasconaro, M.; Purpari, G.; et al. Aujeszky’s disease in hunting dogs after the ingestion of wild boar raw meat in Sicily (Italy): clinical, diagnostic and phylogenetic features. BMC Vet Res. 2022, 18. [Google Scholar] [CrossRef]
- Amoroso, M.G.; Serra, F.; Esposito, C.; D’alessio, N.; Ferrara, G.; Cioffi, B.; et al. Prevalence of infection with porcine circovirus types 2 and 3 in the wild boar population in the campania region (Southern italy). Animals 2021, 11. [Google Scholar] [CrossRef]
- Fanelli, A.; Pellegrini, F.; Camero, M.; Catella, C.; Buonavoglia, D.; Fusco, G.; et al. Genetic Diversity of Porcine Circovirus Types 2 and 3 in Wild Boar in Italy. Animals 2022, 12. [Google Scholar] [CrossRef]
- Gillespie, J.; Opriessnig, T.; Meng, X.J.; Pelzer, K.; Buechner-Maxwell, V. Porcine circovirus type 2 and porcine circovirus-associated disease. Journal of Veterinary Internal Medicine 2009, 23, 1151–1163. [Google Scholar] [CrossRef]
- Cillis, G.; Statuto, D.; Picuno, P. Historical gis as a tool for monitoring, preserving and planning forest landscape: A case study in a mediterranean region. Land 2021, 10. [Google Scholar] [CrossRef]
- Poonsuk, K.; Cheng, T.Y.; Ji, J.; Zimmerman, J.; Giménez-Lirola, L. Detection of porcine epidemic diarrhea virus (PEDV) IgG and IgA in muscle tissue exudate (“meat juice”) specimens. Porcine Health Manag 2018, 4. [Google Scholar]
- Patel, P.; Landt, O.; Kaiser, M.; Faye, O.; Koppe, T.; Lass, U.; et al. Development of one-step quantitative reverse transcription PCR for the rapid detection of flaviviruses. Virol J. 2013, 10. [Google Scholar] [CrossRef]
- Zmijewski, T.; Modzelewska-Kapituła, M. The influence of age and sex on carcass characteristics and chemical composition of the longissimus thoracis et lumborum muscle in wild boars (Sus scrofa). Arch Anim Breed 2021, 64. [Google Scholar] [CrossRef]
- Pandit, P.S.; Doyle, M.M.; Smart, K.M.; Young, C.C.W.; Drape, G.W.; Johnson, C.K. Predicting wildlife reservoirs and global vulnerability to zoonotic Flaviviruses. Nat Commun. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Ojeyinka, O.T.; Omaghomi, T.T. Wildlife as sentinels for emerging zoonotic diseases: A review of surveillance systems in the USA. World Journal of Advanced Research and Reviews 2024, 21, 768–776. [Google Scholar] [CrossRef]
- Petruccelli, A.; Zottola, T.; Ferrara, G.; Iovane, V.; Di Russo, C.; Pagnini, U.; et al. West nile virus and related flavivirus in european wild boar (Sus scrofa), latium region, Italy: A retrospective study. Animals 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Pepin, K.M.; Pedersen, K.; Wan, X.F.; Cunningham, F.L.; Webb, C.T.; Wilber, M.Q. Individual-Level Antibody Dynamics Reveal Potential Drivers of Influenza A Seasonality in Wild Pig Populations. In Integrative and Comparative Biology; Oxford University Press, 2019; pp. 1231–1242. [Google Scholar]
- De Marco, M.A.; Cotti, C.; Raffini, E.; Frasnelli, M.; Prosperi, A.; Zanni, I.; et al. Long-Term Serological Investigations of Influenza A Virus in Free-Living Wild Boars (Sus scrofa) from Northern Italy (2007–2014). Microorganisms 2022, 10. [Google Scholar]
- Delogu, M.; Cotti, C.; Vaccari, G.; Raffini, E.; Frasnelli, M.; Nicoloso, S.; et al. Serologic and virologic evidence of influenza a viruses in wild boars (Sus scrofa) from two different locations in Italy. J Wildl Dis. 2019, 55, 158–163. [Google Scholar]
- Ferrara, G; Longobardi, C; D’ambrosi, F; Amoroso, MG; D’alessio, N; Damiano, S; et al. Aujeszky’s disease in south-Italian wild boars (Sus Scrofa): A serological survey. Animals 2021, 11. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




