Submitted:
09 January 2026
Posted:
09 January 2026
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Proposed System
- The absence of explicit background suppression at the input level, and
- The decoupling of feature learning and classification, which prevents end-to-end optimization and limits task-driven feature adaptation.
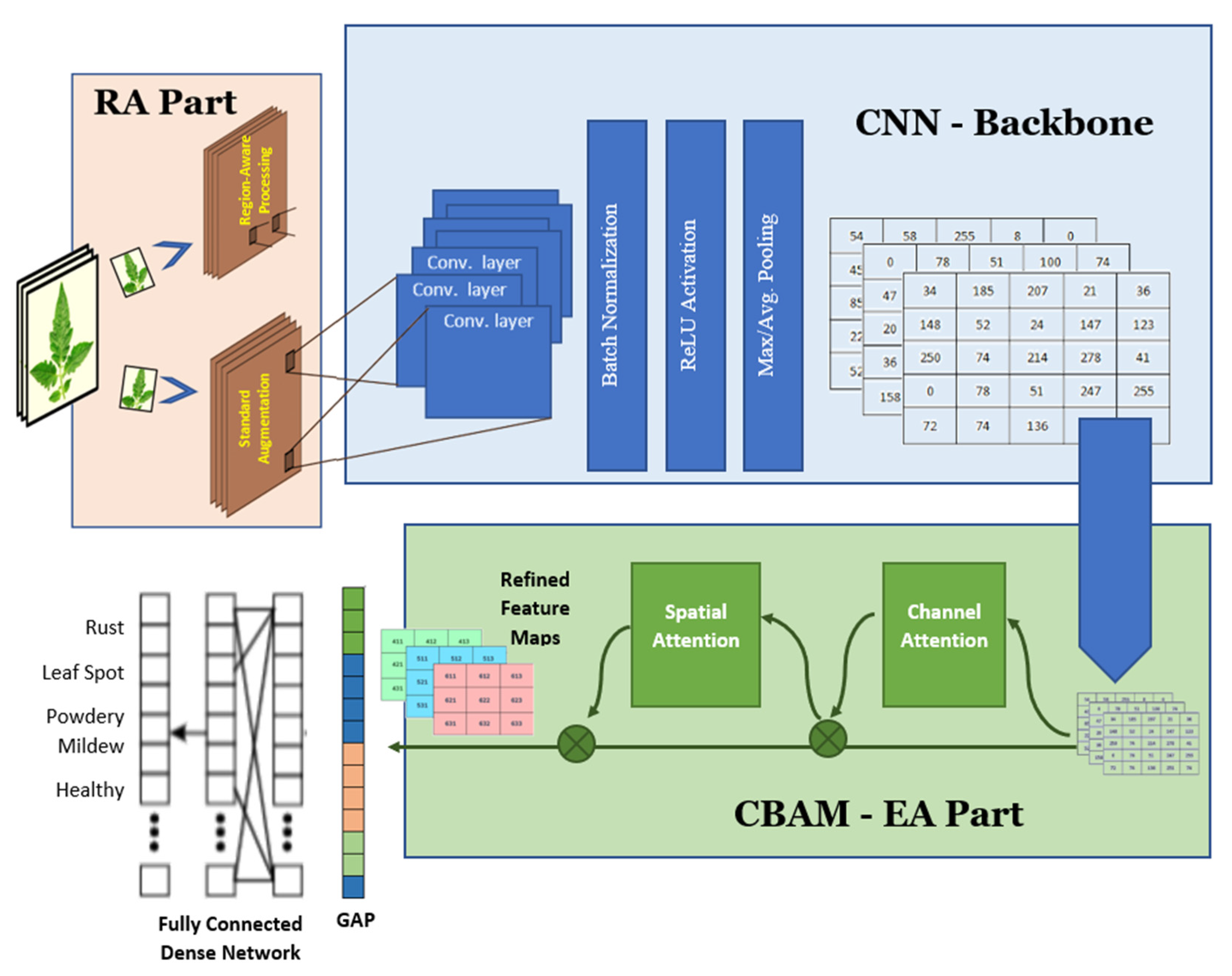
2.1. REA-CNN: Architecture and Learning Framework
2.2. Stage I – Region-Aware Processing (RA)
- A.
- Contrast normalization: First, contrast normalization is performed using min–max normalization:
- B.
- Color normalization and brightness/saturation adjustment: Next, color normalization and brightness–saturation adjustment is applied to improve chromatic consistency and highlight disease-relevant patterns:
- C.
- Output of Region-Aware Processing: The final output of this stage, IENH constitutes a refined, leaf-centric, and visually enhanced image representation. As illustrated in Figure 2, this pipeline includes foreground–background separation, background suppression, and final image enhancement, producing an input that emphasizes disease-relevant visual features while minimizing environmental noise. This enhanced representation is then forwarded to the CNN backbone for robust feature extraction.
2.3. Stage II – CNN Backbone (Feature Extraction)
- A.
- Convolutional Feature Extraction: At the lth convolutional layer, the feature maps are computed as:
- B.
- Batch Normalization: To stabilize training and accelerate convergence, batch normalization is applied to the convolutional outputs:
- C.
- Nonlinearity and Pooling: Nonlinear activation is introduced using the Rectified Linear Unit (ReLU):
- D.
- Output of CNN Backbone: After multiple stacked convolutional blocks, the CNN backbone produces a high-level feature tensor:
2.4. Stage III – Enhanced Attention via CBAM (EA)
- A.
- Channel Descriptor Generation: Global information is first aggregated from the feature tensor using both global average pooling, GAP(·) and global max pooling, GMP(·):
- B.
- Channel Weight Estimation: The pooled descriptors are passed through a shared multi-layer perceptron (MLP) to compute channel attention weights:
- C.
- Channel-wise Feature Refinement: The original feature tensor is refined by channel-wise multiplication:
2.5. Spatial Attention Mechanism
- A.
- Spatial Descriptor Computation: Spatial descriptors are generated by applying average and max pooling across the channel dimension:
- B.
- Spatial Attention Map Generation: The pooled feature maps are concatenated and passed through a convolutional layer to compute the spatial attention map:
- C.
- Spatial Feature Refinement: The final attention-enhanced feature tensor is obtained as:
- D.
- Output of Enhanced Attention Stage: The output FEA represents an attention-refined feature tensor that integrates both channel-wise and spatial importance. By explicitly modelling what features and where they matter, the CBAM module significantly improves feature discriminability and robustness, particularly for visually similar plant diseases. This refined representation is subsequently forwarded to the feature aggregation and classification stages.
2.6. Stage IV – Feature Aggregation
2.7. Stage V – Classification Head
2.8. End-to-End Learning Strategy
- The classification objective directly influences feature learning,
- The attention weights are dynamically adapted based on task relevance, and
- The convolutional filters learn representations that are aligned with disease-specific visual patterns.
- Explicit background suppression reduces input entropy
- Attention enhances semantic and spatial discriminability
- GAP improves generalization and parameter efficiency
- Fully end-to-end differentiability ensures robustness under domain shift
2.9. Comparison with Previous Hybrid Models
- REA-CNN does not freeze feature extractors
- The classifier influences feature learning directly
- Attention mechanisms adapt dynamically during training
3. Experimental Setup
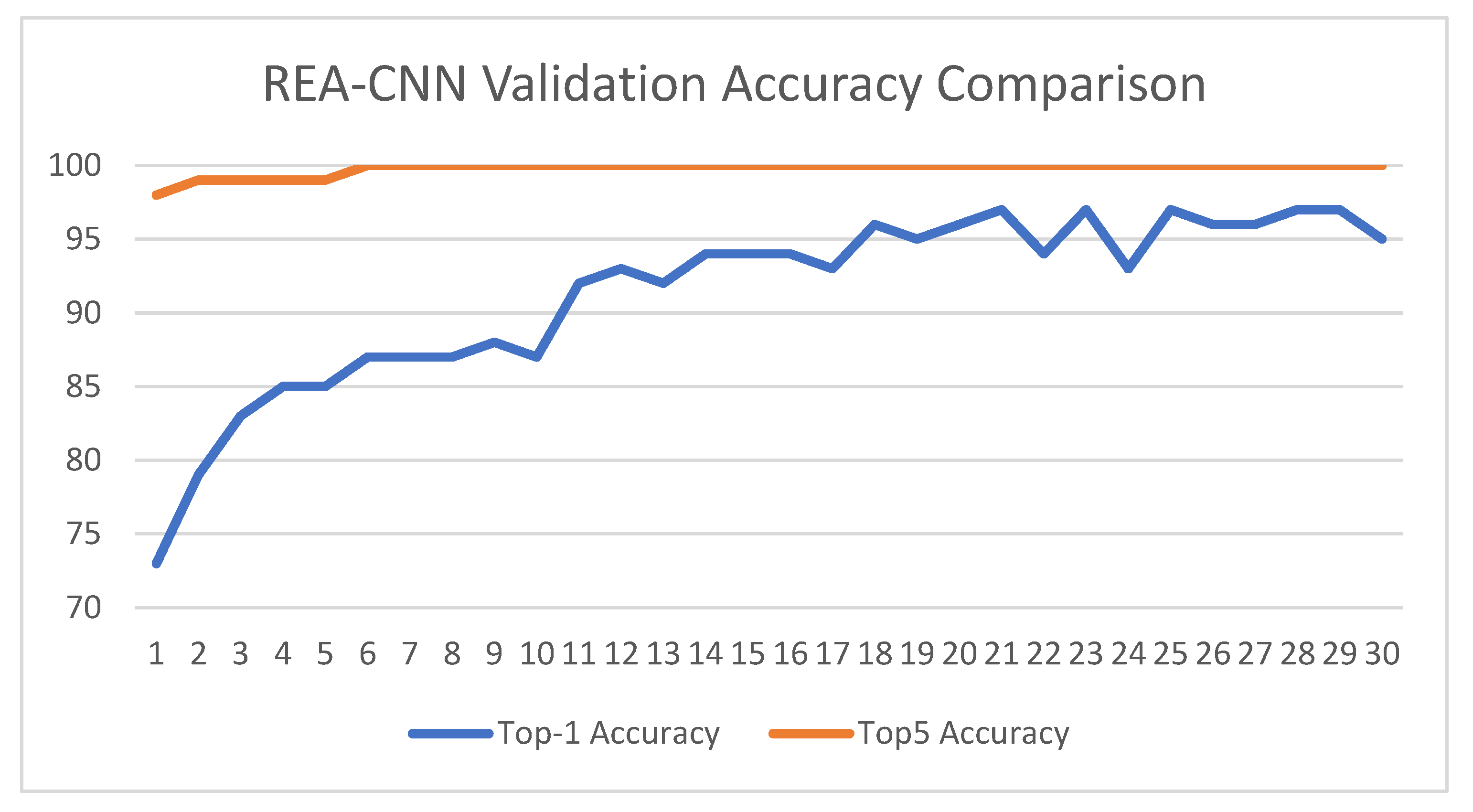
3.1. Evaluation Metrics
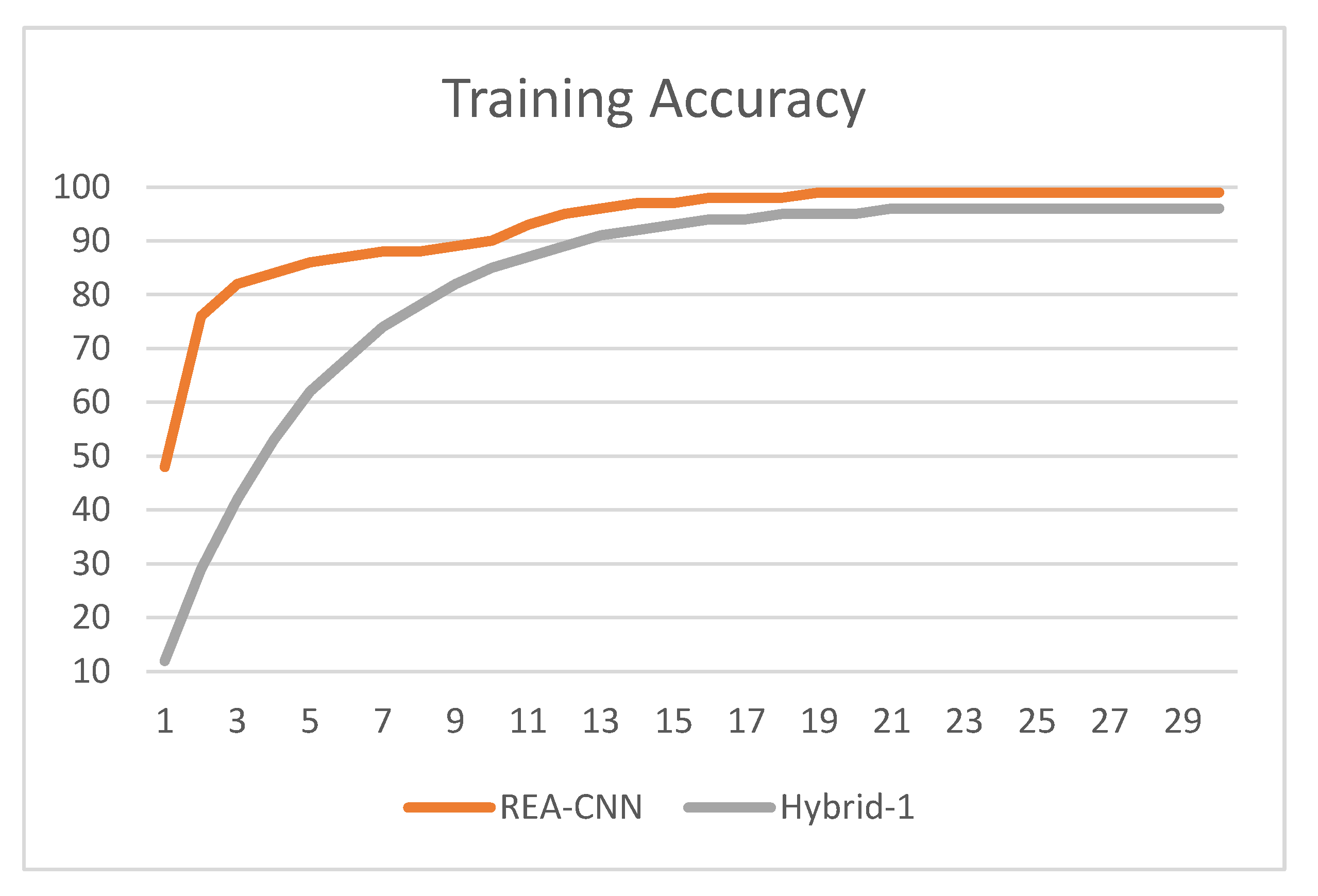
- acc: training accuracy,
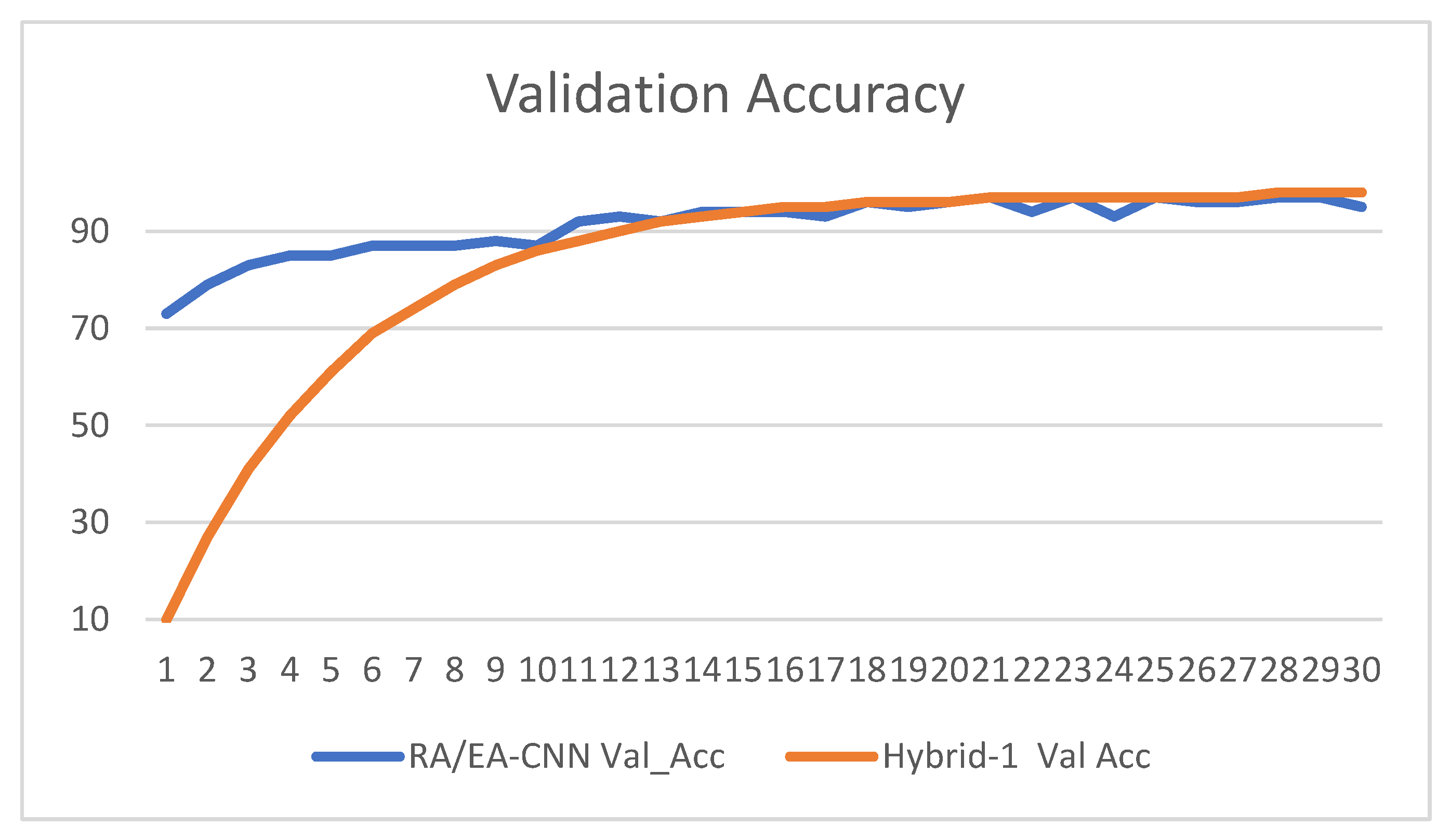
- val_acc: validation accuracy,
- test_acc: final test accuracy, representing the true generalization performance.
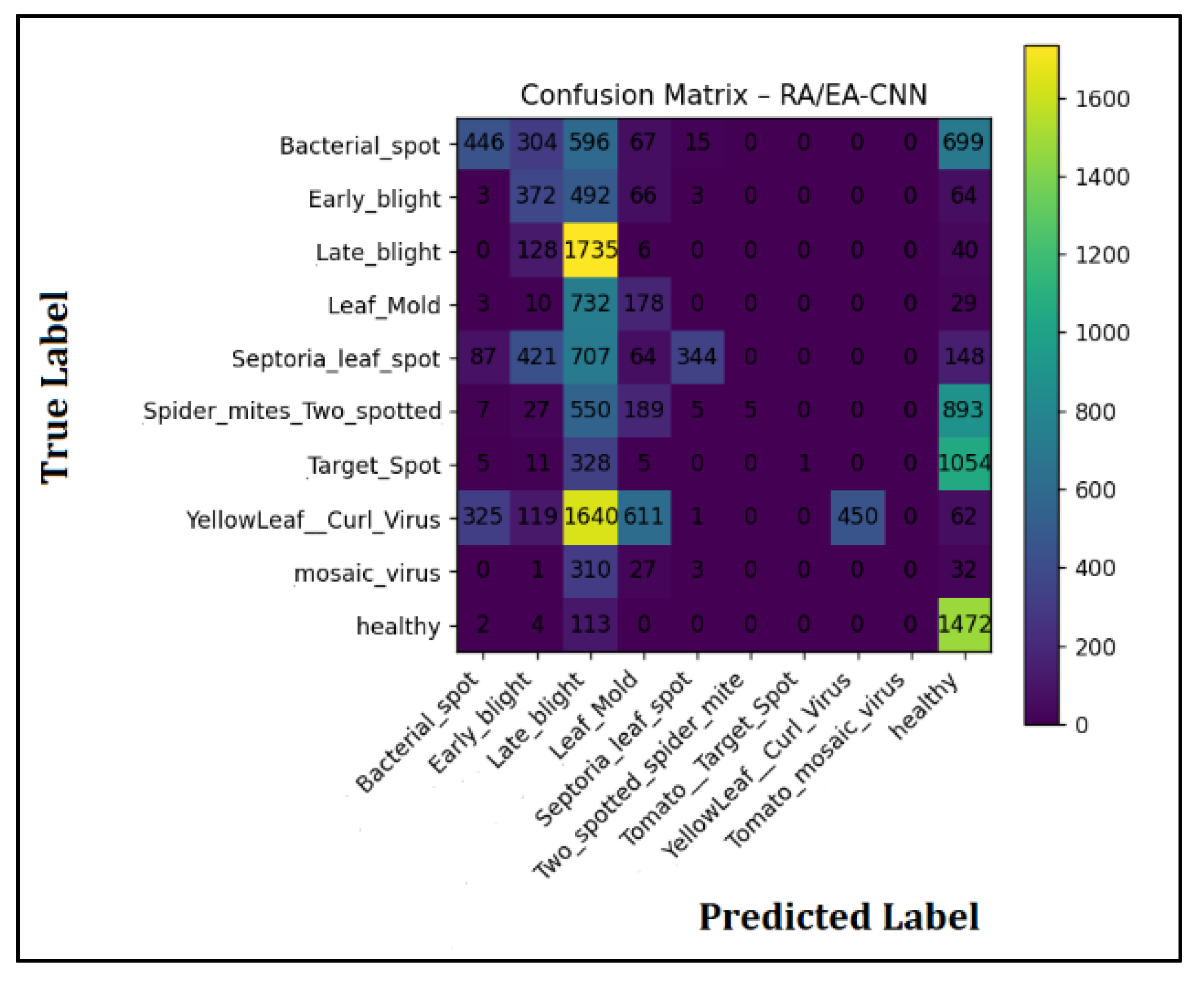
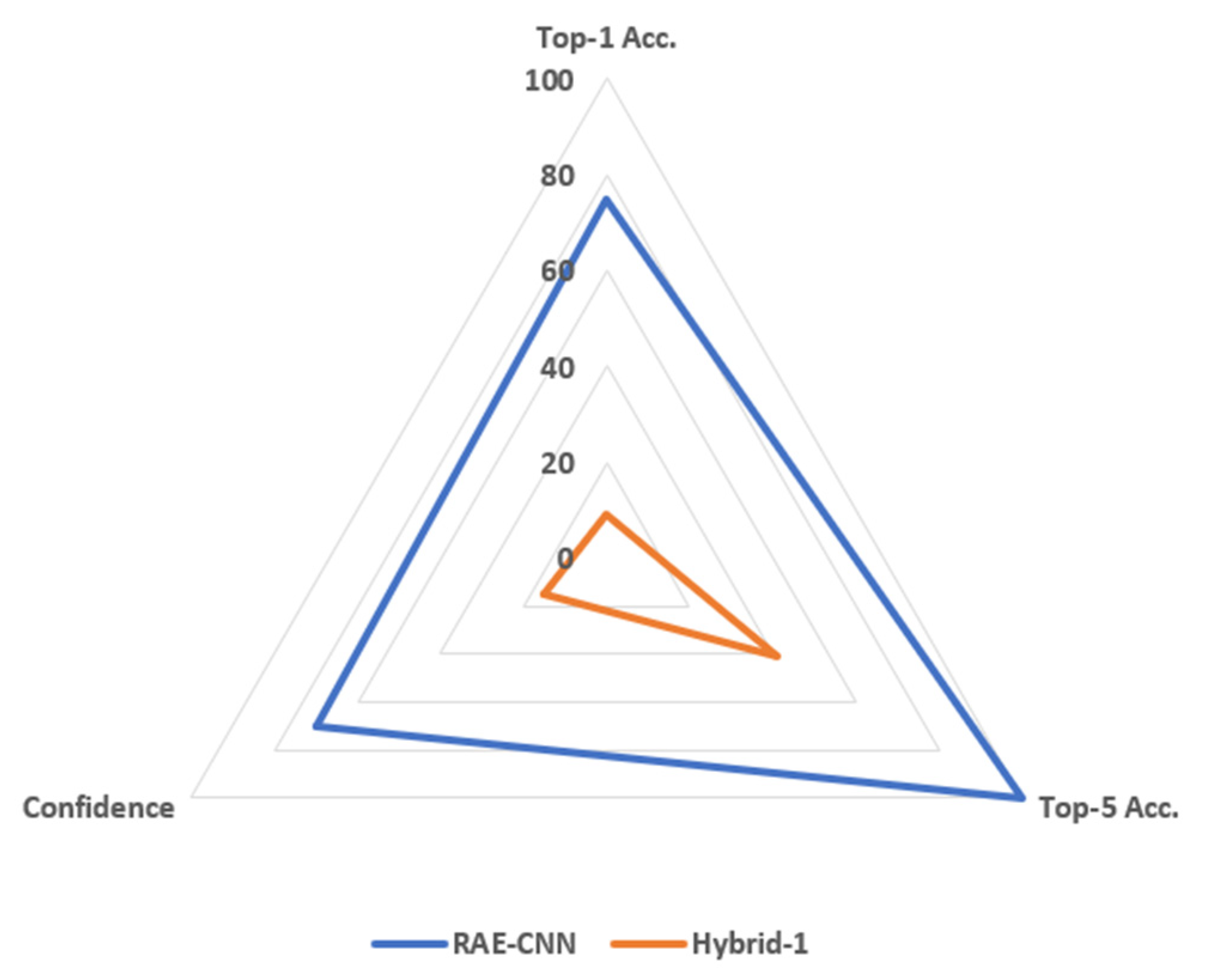
3.2. Result Analysis
| Metric | REA-CNN | Hybrid-1 |
|---|---|---|
| Top-1 Correct (out of 12) | 9 (75%) | 1 (8.5%) |
| Top-5 Correct (out of 12) | 12 (100 %) | 5 (41%) |
| Average Confidence (%) | 70+ | 1.25 |
4. Conclusion
References
- Yağ, I.; Altan, A. Artificial Intelligence-Based Robust Hybrid Algorithm Design and Implementation for Real-Time Detection of Plant Diseases in Agricultural Environments. Biology (Basel) 2022, 11, 1732. [Google Scholar] [CrossRef] [PubMed]
- Kamencay, P.; Benco, M.; Mizdos, T.; Radil, R. A New Method for Face Recognition Using Convolutional Neural Network. Advances in Electrical and Electronic Engineering 2017, 15. [Google Scholar] [CrossRef]
- Bedi, P.; Gole, P. Plant disease detection using hybrid model based on convolutional autoencoder and convolutional neural network. Artificial Intelligence in Agriculture 2021, 5, 90–101. [Google Scholar] [CrossRef]
- Hashmi, N.; Haroon, M. A Hybrid AI-Based Approach for Early and Accurate Rice Disease Detection. Fusion: Practice and Applications 2026, 21. [Google Scholar] [CrossRef]
- Kabir, M.F.; Rahat, I.S.; Beverley, C.; Uddin, R.; Kant, S. Tealeafnet-gwo: an intelligent CNN-Transformer hybrid framework for tea leaf disease detection using gray wolf optimization. Discover Artificial Intelligence 2025, 5, 377. [Google Scholar] [CrossRef]
- Khalid, M.; Talukder, M.D.A. A Hybrid Deep Multistacking Integrated Model for Plant Disease Detection. IEEE Access 2025, 13, 116037–116053. [Google Scholar] [CrossRef]
- Aboelenin, S.; Elbasheer, F.A.; Eltoukhy, M.M.; El-Hady, W.M.; Hosny, K.M. A hybrid Framework for plant leaf disease detection and classification using convolutional neural networks and vision transformer. Complex & Intelligent Systems 2025, 11, 142. [Google Scholar] [CrossRef]
- Huang, X.; Chen, A.; Zhou, G.; Zhang, X.; Wang, J.; Peng, N.; Yan, N.; Jiang, C. Tomato Leaf Disease Detection System Based on FC-SNDPN. Multimed Tools Appl 2023, 82, 2121–2144. [Google Scholar] [CrossRef]
- Baser, P.; Saini, J.R.; Kotecha, K. TomConv: An Improved CNN Model for Diagnosis of Diseases in Tomato Plant Leaves. Procedia Computer Science 2023, 218, 1825–1833. [Google Scholar] [CrossRef]
- Arafath, M.; Nithya, A.A.; Gijwani, S. Tomato Leaf Disease Detection Using Deep Convolution Neural Network. Procedia Computer Science 2023, 236–245. [Google Scholar] [CrossRef]
- Indira, K.; Mallika, H. Classification of Plant Leaf Disease Using Deep Learning. Journal of The Institution of Engineers (India): Series B 2024, 105, 609–620. [Google Scholar] [CrossRef]
- Roy, K.; et al. Detection of Tomato Leaf Diseases for Agro-Based Industries Using Novel PCA DeepNet. IEEE Access 2023, 11, 14983–15001. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, G.; Lu, C.; Chen, A.; Wang, Y.; Li, L.; Cai, W. MMDGAN: A fusion data augmentation method for tomato-leaf disease identification. Applied Soft Computing 2022, 123, 108969. [Google Scholar] [CrossRef]
- Sunil, C.K.; Jaidhar, C.D.; Patil, N. Tomato plant disease classification using Multilevel Feature Fusion with adaptive channel spatial and pixel attention mechanism. Expert Syst Appl 2023, 228, 120381. [Google Scholar] [CrossRef]
- Arshad, F.; Mateen, M.; Hayat, S.; Wardah, M.; Al-Huda, Z.; Gu, Y.H.; Al-antari, M.A. PLDPNet: End-to-end hybrid deep learning framework for potato leaf disease prediction. Alexandria Engineering Journal 2023, 78, 406–418. [Google Scholar] [CrossRef]
- Rashid, J.; Khan, I.; Ali, G.; Almotiri, S.H.; AlGhamdi, M.A.; Masood, K. Multi-Level Deep Learning Model for Potato Leaf Disease Recognition. Electronics (Basel) 2021, 10, 2064. [Google Scholar] [CrossRef]
- Khobragade, P.; Shriwas, A.; Shinde, S.; Mane, A.; Padole, A. Potato Leaf Disease Detection Using CNN. International IEEE Conference on Smart Generation Computing, Communication and Networking (SMART GENCON) 2022, 1–5. [Google Scholar] [CrossRef]
- Mahum, R.; Munir, H.; Mughal, Z.; Awais, M.; Khan, F.S.; Saqlain, M.; Mahmad, S.; Tlili, I. A novel framework for potato leaf disease detection using an efficient deep learning model. Human and Ecological Risk Assessment: An International Journal 2023, 29, 303–326. [Google Scholar] [CrossRef]
- Goyal, B.; Pandey, A.K.; Kumar, R.; Gupta, M. Disease Detection in Potato Leaves Using an Efficient Deep Learning Model. International IEEE Conference on Data Science and Network Security (ICDSNS) 2023, 01–05. [Google Scholar] [CrossRef]
- Das, P.K. Leaf Disease Classification in Bell Pepper Plant using VGGNet. Journal of Innovative Image Processing 2023, 5, 36–46. [Google Scholar] [CrossRef]
- Kapoor, K.; Singh, S.; Singh, N.P.; Priyanka. Bell-Pepper Leaf Bacterial Spot Detection Using AlexNet and VGG-16; 2023; pp. 507–519. [Google Scholar] [CrossRef]
- Mahesh, T.Y.; Mathew, M.P. Detection of Bacterial Spot Disease in Bell Pepper Plant Using YOLOv3. IETE J Res 2024, 70, 2583–2590. [Google Scholar] [CrossRef]
- Begum, S. S. A.; Syed, H. GSAtt-CMNetV3: Pepper Leaf Disease Classification Using Osprey Optimization. IEEE Access 2024, 12, 32493–32506. [Google Scholar] [CrossRef]
- Fatima, S.; Kaur, R.; Doegar, A.; Srinivasa, K.G. CNN Based Apple Leaf Disease Detection Using Pre-trained GoogleNet Model; 2023; pp. 575–586. [Google Scholar] [CrossRef]
- Hasan, S.; Jahan, S.; Islam, M.I. Disease detection of apple leaf with combination of color segmentation and modified DWT. Journal of King Saud University - Computer and Information Sciences 2022, 34, 7212–7224. [Google Scholar] [CrossRef]
- Rehman, Z.U.; Khan, M.A.; Ahmed, F.; Damasevicius, R.; Naqvi, S.R.; Nisat, W.; Javed, K. Recognizing apple leaf diseases using a novel parallel real-time processing framework based on MASK RCNN and transfer learning: An application for smart agriculture. IET Image Process 2021, 15, 2157–2168. [Google Scholar] [CrossRef]
- Kaur, N.; Devendran, V. A novel framework for semi-automated system for grape leaf disease detection. Multimed Tools Appl 2023, 83, 50733–50755. [Google Scholar] [CrossRef]
- URao, S.; Swathi, R.; Sanjana, V.; Arpitha, L.; Chandrasekhar, K.; Chimayi; Naik, P.K. Deep Learning Precision Farming: Grapes and Mango Leaf Disease Detection by Transfer Learning. Global Transitions Proceedings 2021, 2, 535–544. [Google Scholar] [CrossRef]
- Altalak, M.; Uddin, M.A.; Alajmi, A.; Rizg, A. A Hybrid Approach for the Detection and Classification of Tomato Leaf Diseases. Applied Sciences 2022, 12, 8182. [Google Scholar] [CrossRef]


| Aspect | [1] | [2] | [3] | [4] | [5] | [6] | [7] | RA/EA-CNN |
|---|---|---|---|---|---|---|---|---|
| Publication venue | Biology MDPI | AEEE Journal | Elsevier SciDirect |
Springer | Springer | IEEE Access | Springer | — |
| Application domain | Plant diseases | Face Recognition | Plant diseases | Rice diseases | Tea leaf diseases | Plant diseases | Plant leaf diseases | Plant diseases |
| Input type | Leaf images | Face images | Leaf images | Leaf images | Leaf images | Leaf image sequences | Leaf images | Leaf images |
| Background removal | No | No | No | segmentation | Partial | No | No | Explicit |
| Region awareness | No | No | No | Yes | Partial | No | Partial (ViT attention) | Yes |
| Image enhancement | No | No | No | Yes | CLAHE | No | Indirect | Explicit |
| Feature extraction | Hand-crafted (2D-DWT) | PCA / LBP / CNN | CAE latent space | Hybrid fusion | CNN + Transformer | VGG16 CNN | CNN ensemble + ViT | Region-guided CNN |
| Temporal modeling | No | No | No | No | No | MBi-LSTM | No | No |
| Optimization strategy | Flower Pollination Algorithm | Manual | Implicit | African Vultures Optimization | Gray Wolf Optimization | None | None | Not required |
| Classifier / backbone | SVM + CNN | CNN / KNN | CNN | Depth-Separable NN | CNN-Transformer | CNN + Bi-LSTM | CNN + Vision Transformer | CNN |
| End-to-end learning | Partial | Mixed |
Semi |
Semi | Semi | Semi | Semi | Full |
| Model complexity | High | Medium | Low | Moderate–High | Moderate–High | High | High | Moderate |
| Explainability | Weak |
Weak |
Weak | Moderate | Moderate | Moderate | Moderate | Strong |
| Real-time suitability | Possible | Limited | Yes | Limited | Moderate | Limited | Limited | Yes |
| Generalizability | oderate | Low | oderate | Moderate | Moderate | Moderate | Moderate | High |
| Robust to real-world images | No | No | Limited | Limited | Limited | Limited | Limited | High |
| Title | Crop Studied | AI Model Used | Accuracy (%) | Relevance with respect to REA-CNN |
|---|---|---|---|---|
| Tomato Leaf Disease Detection System Based on FC-SNDPN [8] | Tomato | FC-SNDPN (FCN + Dual-Path + Switchable Norm) | 97.59 | Partial (segmentation, no explicit attention) |
| An Improved CNN Model for Diagnosis of Diseases in Tomato Crop [9] | Tomato | Hybrid CNN (VGG + Inception) | 99.17 | No |
| Tomato Leaf Disease Detection Using Deep CNN[10] | Tomato | Deep CNN (Batch Norm + Dropout) | 98.00 | No |
| Classification of Plant Leaf Disease Using Deep Learning [11] | Multi (incl. Tomato) |
CNN / AlexNet / MobileNet | 84.24 / 91.19 / 97.33 | No |
| Detection of Tomato Leaf Diseases… PCA DeepNet [12] | Tomato | PCA DeepNet + GAN + Faster R-CNN | 99.60 | Partial (region detection, no attention) |
| MMDGAN : Fusion Augmentation + B-ARNet [13] | Tomato | GAN-based + B-ARNet | 97.12 | No |
| Tomato Plant Disease Classification… Adaptive Attention [14] | Tomato | Multilevel fusion + attention | ≈99.83 | No |
| PLDPNet: Hybrid CNN Framework [15] | Potato / Tomato | Hybrid CNN | 94.25 (Tomato) | No |
| Multi-Level Deep Learning Model [16] | Potato / Tomato | Multi-level CNN | 96.71 (Tomato) | No |
| Potato Leaf Disease Detection Using CNN [17] | Potato | CNN | 98.07 | No |
| Efficient DenseNet-Based Potato Leaf Disease Detection) [18] | Potato | Efficient DenseNet-201 | 97.20 | No |
| Disease Detection in Potato Leaves Using DL + SVM [19] | Potato | Enhanced DL + SVM | 99.42 | No |
| Leaf Diseases Classification in Bell Pepper Using VGGNet [20] | Bell Pepper | VGG16 / VGG19 | 97 / 96 | No |
| Bell-Pepper Leaf Bacterial Spot Detection [21] | Bell Pepper | AlexNet / VGG-16 | 97.87 | No |
| Detection of Bacterial Spot Disease Using YOLOv3 [22] | Bell Pepper | YOLOv3 (detection) | 90 | Partial (region detection only) |
| Optimized GSAtt-CMNetV3 for Pepper Leaf Disease [23] | Pepper | GSAtt-CMNetV3 + Os-OA | 97.87 | Partial (attention, no RA pre-processing) |
| CNN Based Apple Leaf Disease Detection [24] | Apple | GoogleNet (pretrained) | 99.79 | No |
| Apple Leaf Disease Detection using Color + DWT + RF [25] | Apple | Color + DWT + Random Forest | 98.63 | No |
| Recognizing Apple Leaf Diseases using Mask R-CNN [26] | Apple | Mask R-CNN + Transfer Learning | 96.6 | Partial (segmentation, no attention) |
| Efficient Deep Learning Approach for Apple Leaf Diseases | Apple | CNN (transfer learning) | 97.82 | No |
| A Novel Framework for Grape Leaf Disease Detection [27] | Grape | Hybrid segmentation + ensemble classifier | 98.7 | Partial |
| Deep Learning Precision Farming: Grapes & Mango Leaf Diseases [28] | Grapes / Mango | AlexNet (pretrained) | 99 (Grapes) 89 (Mango) |
No |
| AI System Type | Level-1: Input & Preparation | Level-2: Feature Extraction | Level-3: Feature Refinement / Intelligence | Level-4: Decision / Classification |
|---|---|---|---|---|
| 2-Level AI System | Preprocessing / Normalization / Augmentation | CNN / Feature Extractor | — | — |
| 3-Level AI System | Preprocessing / Augmentation / Normalization | CNN / Transfer Learning | Attention / Optimization / Feature Selection | — |
| 4-Level AI System (Advanced / Hybrid) | Preprocessing + Augmentation + Normalization | CNN Backbone | CBAM / Attention / Hybrid Intelligence | Dense / Softmax / SVM |
| Level | Possible Techniques |
|---|---|
| Level-1 (Input & Preparation) | Pre-processing, normalization, data augmentation, CLAHE, background removal, segmentation |
| Level-2 (Feature Extraction) | CNN, ResNet, VGG, DenseNet, EfficientNet, Autoencoder, Transfer Learning |
| Level-3 (Refinement / Intelligence) | CBAM, SE-block, Attention, ViT, PCA, GWO, PSO, LSTM |
| Level-4 (Decision Layer) | Dense + Softmax, SVM, KNN, Random Forest, XGBoost |
| CNN Model | Hybrid-1 [29] | Proposed Hybrid |
|---|---|---|
| Input Leaf Image ↓ Image Pre-processing Resize, normalization, augmentation ↓ CNN Backbone ResNet50 pretrained ↓ Global Feature Vector Global Average Pooling ↓ Fully Connected Layer ↓ Softmax ↓ Disease / Healthy Label |
Input Leaf Image ↓ Image Pre-processing Resize, normalize , augmentation ↓ CNN Backbone ResNet50 pretrained ↓ CBAM (Channel + Spatial Attention) ↓ Deep Feature Vector ↓ SVM Classifier ↓ Disease / Healthy Label |
Input Leaf Image ↓ Region-Aware Processing Leaf-focused, BG Suppression GrabCut-based leaf extraction ↓ CNN Backbone ResNet50 pretrained ↓ CBAM Attention Module ↓ Global Average Pooling ↓ Fully Connected Layer ↓ Softmax ↓ Disease / Healthy Label |
| Stage 1: CNN + CBAM trained using Softmax loss Stage 2: CNN frozen and SVM trained on extracted deep features |
Single-stage, end-to-end training | |
| Joint optimization of feature extraction and classification | Extract strong features, then classify robustly. | Focus on the right regions, attend to disease patterns, and learn everything end-to-end. |
|
✓ End-to-end training ✓Single loss function × No explicit attention ×No region isolation Best for: Clean benchmark datasets (e.g., PlantVillage) Weakness: Sensitive to background and dataset bias |
× Not end-to-end ✓Attention focuses on disease regions ✓ SVM improves margin-based generalization Best for: Small datasets, controlled environments Weakness: More complex training, harder deployment |
✓End-to-end ✓Attention-enhanced ✓Region-focused ✓Robust to background & noise ✓Simple deployment Best for: Real-world images and practical deployment Weakness: Slightly more complex than CNN-only |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





