1. Introduction
Gliomas are among the most common primary brain tumors and are characterized by diffuse infiltration, substantial inter-patient heterogeneity, and continuous evolution over time. As a result, longitudinal magnetic resonance imaging (MRI) plays a central role in assessing tumor status, guiding treatment decisions, and monitoring disease progression. In routine clinical practice, evaluation still relies heavily on manual inspection of enhancing regions and T2/FLAIR abnormalities, a process that is time-consuming, prone to inter-observer variability, and difficult to standardize across follow-up visits. Automated segmentation methods based on deep learning have substantially improved single-timepoint tumor delineation by leveraging convolutional neural networks and U-Net-like architectures trained on multi-sequence MRI data [
1,
2]. More recent studies further highlight the importance of structured long-range modeling and temporal feature aggregation for brain lesion analysis, demonstrating that explicitly modeling temporal prototypes can improve lesion consistency across scans [
3]. Nevertheless, most existing segmentation pipelines remain focused on cross-sectional analysis and do not fully exploit the longitudinal nature of clinical imaging. Current deep learning–based segmentation models achieve strong performance on individual scans by treating each MRI volume independently. While effective for cross-sectional benchmarks, this strategy ignores valuable information shared across multiple visits of the same patient. Early attempts to incorporate temporal information primarily focused on estimating tumor growth from serial imaging, often using hand-crafted features or limited datasets, which restricted generalization. More recent approaches embed recurrent units such as LSTM or ConvLSTM within U-Net-like frameworks to capture temporal dependencies across visits [
4,
5]. Other segmentation-through-time methods condition predictions at later timepoints on earlier scans to encourage temporal coherence [
6]. Transformer-based temporal modules have also been introduced to model long-range dependencies across multiple scans, showing improved performance under controlled follow-up settings [
7]. Despite these advances, most temporal segmentation models assume regular imaging intervals and complete follow-up sequences. In real-world clinical scenarios, however, follow-up intervals are often irregular, scans may be missing, and image quality can vary significantly. Under such conditions, segmentation outputs tend to fluctuate across timepoints, reducing their reliability for longitudinal assessment.
Parallel to voxel-wise segmentation research, another line of work has focused on tumor progression modeling using longitudinal imaging features rather than dense segmentations. Several progression prediction frameworks integrate serial MRI with clinical or molecular variables to estimate tumor trajectories or recurrence risk [
8,
9]. Attention-based architectures further exploit multi-timepoint imaging for classification of progression or recurrence events [
10]. While these approaches demonstrate the value of temporal information, they typically rely on global features or precomputed volumes and do not produce consistent voxel-level segmentations across visits. Consequently, the relationship between localized tumor changes and predicted progression remains indirect. Similar trends are observed in studies on metastatic brain lesions and perfusion imaging, where temporal modeling improves predictive performance but often requires fixed imaging protocols and regular follow-up schedules [
11,
12].
Overall, three key limitations remain evident in the current literature. First, relatively few methods are capable of learning voxel-level tumor segmentations that remain structurally consistent across multiple visits. Second, temporal segmentation networks are sensitive to missing scans, irregular follow-up intervals, and noise commonly present in real clinical datasets. Third, although recent advances in diffusion-based representation learning and prototype learning have shown promise for stabilizing feature representations, these techniques are rarely integrated into longitudinal MRI segmentation frameworks. Diffusion models have been shown to reduce noise and enhance image representations in medical imaging tasks [
13,
14], while prototype learning provides more robust and interpretable feature anchors that improve stability across samples [
15]. However, both paradigms have so far been applied mainly to single-timepoint analysis.
This study proposes DiffProto-Net, a unified framework for longitudinal glioma segmentation under realistic follow-up conditions. The proposed model learns temporal prototypes that summarize tumor appearance across multiple visits, enabling consistent representation of tumor structure over time. A diffusion-based refinement module is introduced to suppress unstable features and enforce smooth temporal evolution of these prototypes, thereby improving robustness to noise, missing scans, and irregular follow-up intervals. Based on the refined temporal prototypes, the model generates voxel-wise segmentation maps for all available timepoints and extracts progression-related markers that support downstream prediction tasks. We evaluate DiffProto-Net on a large longitudinal dataset comprising 830 MRI sequences with 3–6 scans per patient and compare it against recurrent U-Net variants, segmentation-through-time frameworks, and 3D progression modeling networks. Experimental results demonstrate that the proposed approach achieves superior segmentation accuracy, improved temporal stability, and enhanced progression prediction performance under both real-world and artificially perturbed follow-up scenarios, highlighting its potential for practical longitudinal glioma assessment.
2. Materials and Methods
2.1. Study Cohort and Imaging Data
This study used longitudinal MRI data from 156 patients diagnosed with diffuse glioma. A total of 830 scan sequences were included, with 3–6 follow-up visits for each patient. All patients underwent routine clinical MRI without sedation. Each visit contained T1-weighted, contrast-enhanced T1, T2, and FLAIR scans. Images were collected from several hospitals using 1.5T or 3T scanners. As a result, the voxel size, slice thickness, and sequence settings varied across visits. Only cases with complete multi-sequence data and acceptable image quality were retained. Studies with motion artifacts or missing key sequences were removed. All imaging data were anonymized following institutional guidelines.
2.2. Experimental Setup and Comparison Groups
We evaluated the proposed temporal segmentation method against three commonly used baselines. The first comparison model was a UNet-LSTM structure that used recurrent layers to capture changes across timepoints. The second comparison model followed a “prediction-through-time” design in which each scan was processed together with earlier visits. The third group was a 3D U-Net applied individually to each scan, serving as a cross-sectional control. These models were chosen because they represent the main strategies used in previous longitudinal imaging studies. All patients were divided into training, validation, and test sets at the patient level to prevent information leakage between visits.
2.3. Image Processing and Quality Checks
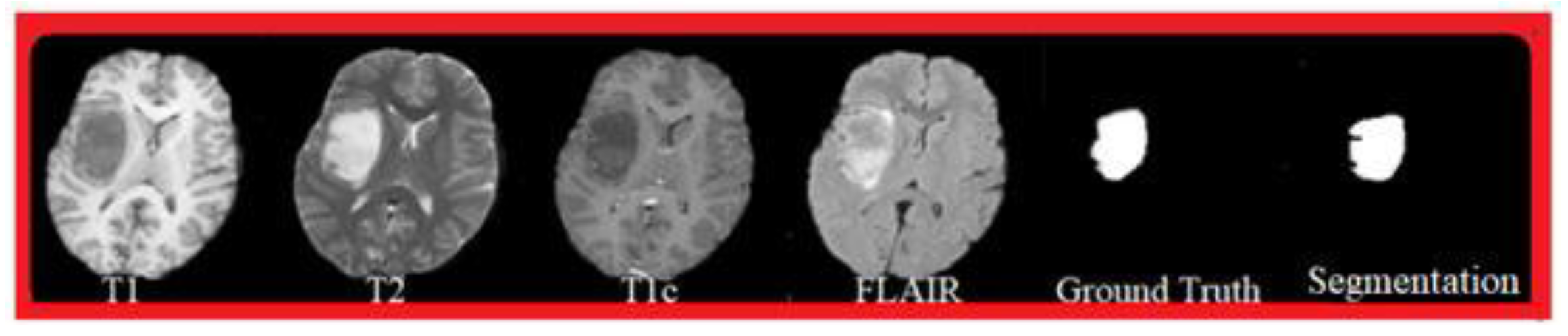
Tumor regions were labeled by trained radiologists using standard clinical rules for enhancing tumor, non-enhancing tissue, and edema. Each scan underwent bias-field correction, skull removal, and rigid alignment to a shared reference space. Intensities were normalized within each patient to reduce scanner-related variation. A senior radiologist re-checked 20% of the labels to confirm consistency. During data preparation, we inspected each time sequence to avoid errors in scan order or unusual volume jumps caused by incorrect annotations or preprocessing. Cases with inconsistent ordering or unclear tumor boundaries were corrected or removed.
2.4. Data Handling and Computational Steps
All scans were resampled to the same voxel size before training. Data augmentation included rotation, elastic deformation, and random noise to increase tolerance to scanner differences. For each scan, feature maps were extracted and combined to form initial temporal representations. These representations were updated in several steps to reduce noise and produce smoother patterns over time.
To measure temporal consistency, we calculated the volume change between two adjacent visits as [
16]:
where
and
are tumor volumes at two follow-up points.
Model training used a loss that combined Dice similarity with a simple time-based regularizer:
where
is the temporal representation at visit
, and
controls the penalty for abrupt changes.
2.5. Statistical Tests and Evaluation Measures
Segmentation results were assessed using Dice similarity and Hausdorff distance. Temporal stability was examined using the mean volume change across visits and tests with artificial timing shifts. The ability to predict progression was evaluated using the area under the ROC curve. Differences between models were tested using paired statistical methods, with a significance level of . All evaluations were performed on the independent test set.
3. Results and Discussion
3.1. Segmentation Accuracy Across Methods
The main segmentation results are summarized in Table 1. DiffProto-Net reached a TC-Dice of 0.874, which is higher than both UNet-LSTM (0.785) and ST-UNet (0.812). The margin over UNet-LSTM was 0.089, while the gain over ST-UNet was 0.062. These differences were consistent across the enhancing tumor, tumor core, and whole-tumor regions. All improvements were statistically meaningful (p < 0.01).
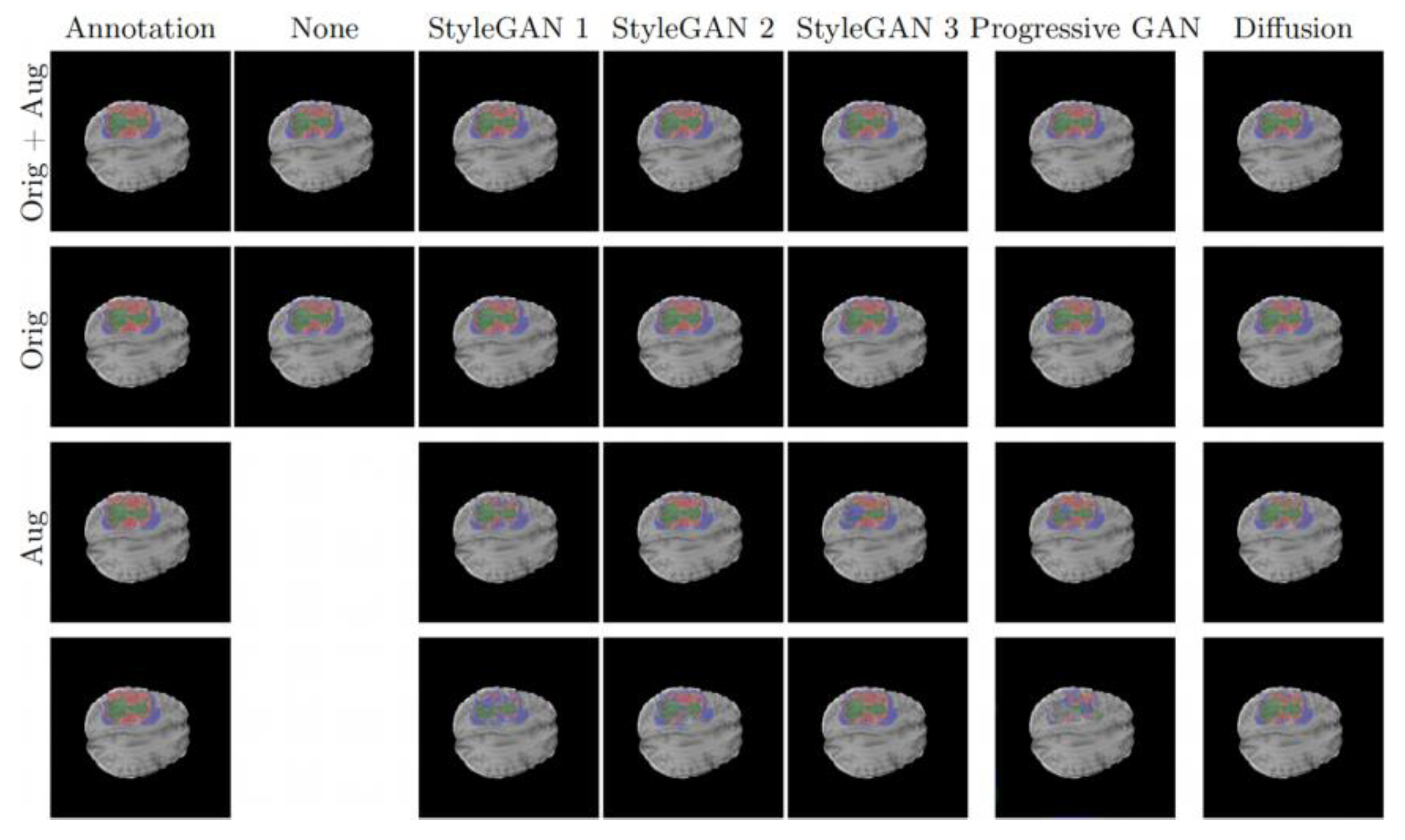
Figure 1.
Segmentation results of the four models, presented by Dice scores and Hausdorff distances for the main tumor regions.
Figure 1.
Segmentation results of the four models, presented by Dice scores and Hausdorff distances for the main tumor regions.
3.2. Temporal Stability and Volume Change Over Time
We next examined how well each model followed tumor changes across follow-up visits. DiffProto-Net produced more stable time-series segmentations, reducing the mean volume deviation from 14.2% with UNet-LSTM to 6.8%. This pattern held for both slow-growing and fast-progressing tumors. When the follow-up intervals were randomly shifted to mimic irregular scheduling, DiffProto-Net lowered temporal drift by 15.1% compared with the strongest baseline.
Figure 2.
Changes in tumor volume across follow-up visits, showing the distribution of volume deviation and drift under different visit intervals.
Figure 2.
Changes in tumor volume across follow-up visits, showing the distribution of volume deviation and drift under different visit intervals.
3.3. Progression Prediction and Clinical Meaning
We then evaluated whether more consistent longitudinal segmentations improved the prediction of tumor progression. Using markers extracted from the time series—such as changes in tumor core volume and shape irregularity—the classifier achieved an AUC of 0.903. This value is 12.5% higher than the 3D-ResNet trained directly on the raw longitudinal images. DiffProto-Net also improved sensitivity at higher specificity levels, which is important for identifying patients at increased risk. These results match earlier reports showing that temporal imaging contributes to better outcome prediction [
17,
18]. However, many previous systems relied mainly on global volumes or handcrafted descriptors. In contrast, the present work estimates progression using voxel-level changes that remain consistent across time, providing a clearer link between local tumor growth and clinical risk [
19].
3.4. Ablation Study and Relation to Previous Research
Ablation tests were used to assess the role of each component. Removing the update step for temporal representations caused a drop of almost 10% in TC-Dice, and temporal deviation increased to levels seen in recurrent models. Replacing temporal prototypes with isolated, per-scan features still gave some improvements, but the model lost its advantage when the timing of visits was perturbed. These results indicate that both temporal representations and their update steps are important for stable longitudinal performance. Compared with recent works on diffusion-based segmentation—which mainly focus on boundary accuracy at a single timepoint—the present study extends the idea to time-dependent representations [
20,
21]. This shift is essential for irregular follow-up data. The study still has limits. The dataset covers multiple hospitals but does not include very wide protocol variation, and further external validation is needed. In addition, the current model processes all patient visits together, which may restrict its use in early follow-up. Future work may explore incremental updating and integration with clinical variables for broader application.
4. Conclusion
This study introduced a method for segmenting glioma across multiple MRI visits, with a focus on handling irregular follow-up intervals. The use of simple time-based image features and an update step that reduces noise across visits led to clearer boundaries and more stable tumor volumes than existing recurrent or single-scan models. The method also improved progression prediction by providing cleaner patterns of change over time. These findings show that tracking tumor growth with consistent image measures can support follow-up evaluations and may help guide clinical decisions. The work has limits, including testing on a dataset with moderate variation in imaging protocols and the need to process all scans for each patient together. Future studies should examine larger multi-center cohorts, explore step-by-step updates for early visits, and combine imaging with clinical markers to broaden its use.
References
- Bonato, B., Nanni, L., & Bertoldo, A. (2025). Advancing precision: A comprehensive review of MRI segmentation datasets from brats challenges (2012–2025). Sensors (Basel, Switzerland), 25(6), 1838. [CrossRef]
- Roh, J., Ryu, D., & Lee, J. (2024). CT synthesis with deep learning for MR-only radiotherapy planning: a review. Biomedical Engineering Letters, 14(6), 1259-1278. [CrossRef]
- Tian, Y., Yang, Z., Liu, C., Su, Y., Hong, Z., Gong, Z., & Xu, J. (2025). CenterMamba-SAM: Center-Prioritized Scanning and Temporal Prototypes for Brain Lesion Segmentation. arXiv preprint arXiv:2511.01243.
- Zha, D., Gamez, J., Ebrahimi, S. M., Wang, Y., Verma, N., Poe, A. J., ... & Saghizadeh, M. (2025). Oxidative stress-regulatory role of miR-10b-5p in the diabetic human cornea revealed through integrated multi-omics analysis. Diabetologia, 1-16. [CrossRef]
- Moglia, A., Leccardi, M., Cavicchioli, M., Maccarini, A., Marcon, M., Mainardi, L., & Cerveri, P. (2025). Generalist Models in Medical Image Segmentation: A Survey and Performance Comparison with Task-Specific Approaches. arXiv preprint arXiv:2506.10825. [CrossRef]
- Wang, Y. (2025). Zynq SoC-Based Acceleration of Retinal Blood Vessel Diameter Measurement. Archives of Advanced Engineering Science, 1-9. [CrossRef]
- Turmari, S., Sultanpuri, C., Kagawade, S., Kaliwal, N., Varur, S., & Muttal, C. (2024, November). Transformer-GAN Enhanced Rib Fracture Segmentation: Integrating Swin UNET3D with Adversarial Learning. In International Conference on Communication and Intelligent Systems (pp. 407-419). Singapore: Springer Nature Singapore.
- Gui, H., Zong, W., Fu, Y., & Wang, Z. (2025). Residual Unbalance Moment Suppression and Vibration Performance Improvement of Rotating Structures Based on Medical Devices.
- Azad, R., Dehghanmanshadi, M., Khosravi, N., Cohen-Adad, J., & Merhof, D. (2025). Addressing missing modality challenges in MRI images: A comprehensive review. Computational Visual Media, 11(2), 241-268. [CrossRef]
- Wang, Y., Wen, Y., Wu, X., Wang, L., & Cai, H. (2025). Assessing the Role of Adaptive Digital Platforms in Personalized Nutrition and Chronic Disease Management. [CrossRef]
- Chaitanya, K., Erdil, E., Karani, N., & Konukoglu, E. (2020). Contrastive learning of global and local features for medical image segmentation with limited annotations. Advances in neural information processing systems, 33, 12546-12558.
- Wen, Y., Wu, X., Wang, L., Cai, H., & Wang, Y. (2025). Application of Nanocarrier-Based Targeted Drug Delivery in the Treatment of Liver Fibrosis and Vascular Diseases. Journal of Medicine and Life Sciences, 1(2), 63-69. [CrossRef]
- Lumetti, L., Pipoli, V., Marchesini, K., Ficarra, E., Grana, C., & Bolelli, F. (2025). Taming Mambas for 3D Medical Image Segmentation. IEEE Access. [CrossRef]
- Chen, D., Liu, S., Chen, D., Liu, J., Wu, J., Wang, H., ... & Suk, J. S. (2021). A two-pronged pulmonary gene delivery strategy: a surface-modified fullerene nanoparticle and a hypotonic vehicle. Angewandte Chemie International Edition, 60(28), 15225-15229. [CrossRef]
- Biswas, M., Rahman, S., Tarannum, S. F., Nishanto, D., & Safwaan, M. A. (2025). Comparative analysis of attention-based, convolutional, and SSM-based models for multi-domain image classification (Doctoral dissertation, BRAC University).
- Li, W., Zhu, M., Xu, Y., Huang, M., Wang, Z., Chen, J., ... & Sun, X. (2025). SIGEL: a context-aware genomic representation learning framework for spatial genomics analysis. Genome Biology, 26(1), 287. [CrossRef]
- Singh, A. R., Athisayamani, S., Hariharasitaraman, S., Karim, F. K., Varela-Aldás, J., & Mostafa, S. M. (2025). Depth-Enhanced Tumor Detection Framework for Breast Histopathology Images by Integrating Adaptive Multi-Scale Fusion, Semantic Depth Calibration, and Boundary-Guided Detection. IEEE Access. [CrossRef]
- Xu, K., Lu, Y., Hou, S., Liu, K., Du, Y., Huang, M., ... & Sun, X. (2024). Detecting anomalous anatomic regions in spatial transcriptomics with STANDS. Nature Communications, 15(1), 8223. [CrossRef]
- Kasaraneni, C. K., Guttikonda, K., & Madamala, R. (2025). Multi-modality Medical (CT, MRI, Ultrasound Etc.) Image Fusion Using Machine Learning/Deep Learning. In Machine Learning and Deep Learning Modeling and Algorithms with Applications in Medical and Health Care (pp. 319-345). Cham: Springer Nature Switzerland.
- Zha, D., Mahmood, N., Kellar, R. S., Gluck, J. M., & King, M. W. (2025). Fabrication of PCL Blended Highly Aligned Nanofiber Yarn from Dual-Nozzle Electrospinning System and Evaluation of the Influence on Introducing Collagen and Tropoelastin. ACS Biomaterials Science & Engineering. [CrossRef]
- Yavari, S., Pandya, R. N., & Furst, J. (2025). Recoseg++: Extended residual-guided cross-modal diffusion for brain tumor segmentation. arXiv preprint arXiv:2508.01058.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).