1. Introduction
An abnormal cell proliferation in the brain is referred to as a brain tumor. The human brain has a complex structure, with different areas devoted to different nervous system functions. Any part of the brain, involving the protective membranes, the base of the brain, the brain stem, the sinuses, the nasal cavity, and many other places, can develop tumors [
1].
In the United States, brain tumors are detected in roughly 30 out of 100,000 people. Because they have the potential to penetrate or put strain on healthy brain tissue, these tumors present serious hazards. It is possible for some brain tumors to be malignant or to eventually turn malignant. Because they eliminate the flow of cerebral fluid, they can cause problems by raising the pressure inside the brain. Furthermore, certain tumor types may spread to distant brain regions through cerebrospinal fluid. [
2].
Gliomas are classified as primary tumors since they are derived from glial cells, which assist neurons in the brain. Astrocytomas, oligodendrogliomas, and ependymomas are among the various kinds of meningiomas that are commonly seen in the adult population. Usually benign, meningiomas grow slowly and develop from the protective coverings that envelop the brain[
3].
In order to detect the problem, diagnostic methods include MRI or CT scans followed by biopsies. Potential treatment options could include chemotherapy, radiation therapy, particular treatments, surgery, or a combination of these treatments. There are sorts of tumors: malignant and benign. The word "cancer" refers only to malignant tumors [
4]. Although all types of cancer present as tumors, it is crucial to remember that not all tumors are malignant. Compared to other cancer forms, brain tumors in particular are linked to a more severe survival rate. Brain tumors’ uneven forms, varying morphology, various locations, and indistinct borders make early identification extremely difficult [
5]. For medical specialists to make accurate treatment decisions, accurate tumor identification at this early stage is essential. Primary and secondary tumors are types of tumors. While secondary cancer, or metastases, develop from cells that start in other body areas and then move to the brain, primary tumors cause the tumor cells to begin with inside the brain so as to spread to different parts of the body[
6].
Either manual, semi-automatic, or fully automated systems requiring user input handle magnetic resonance (MR) images. For medical image processing, precise segmentation and classification are critical, but they often require manual intervention by physicians, which can take a lot of time [
7].A precise diagnosis allows patients to start with the right treatments, which may result in longer lifespans. As a result, the development and application of novel frameworks targeted at lessening the workload of radiologists in detecting and classifying tumors is of vital importance in the field of artificial intelligence [
8].
Assessing the grade and type of a tumor is crucial, especially at the onset of a treatment strategy. The precise identification of abnormal is vital for diagnosing accurate [
9],which necessity is underscored existence of effective methods which utilize classifying, statementing, or a combing qualitatively analyze the brain.
The precision and reliability challenges that affect traditional imaging techniques frequently require the implementation of complex computational methods in order to improve analysis. Deep learning has become well-known in this domain due to the fact of its excellent ability to extract hierarchical feature from data with no any processing functions, which significantly improves image classification tasks in medical diagnostics [
10].
CNN appears to have ended up being a usual approach in clinical image classifications. With its great classification performance, CNN is advanced popularity strategies primarily relied on classical function extractions, mainly on big datasets. [
11].
VGG was designed by the Visual Geometry Research Group at Oxford University. Its primary role is to improve network depth. VGG16 and VGG19 are sorts of VGG structures [
12]. VGG16 is extra popular ,and VGG19 is a deep network, that invloves 16 convolutional layers and 3 fully connected layer, which could extract reliable feature. So, VGG16 was selected in classifying MRI brain tumor images [
13].
Although high performance of a conventional neural network, its restricted nearby receptive usual overall performance, while the vision transformer network extracts global data that is rely on self attention. Every transformers module composed of a multi head attention layer.[
14] Although transformer vision networks lack the conventional neural network, results would be aggressive. Unlike conventional fine-tuning techniques, in which they are most effective, the FTVT structure involves a custom classifier head block that includes BN, ReLU, and dropout layers, which has an alternate structure to analyze particular representations immediately from the data set[
15].
An automated system developed particularly for the classification and segmentation of brain tumors is presented in this paper. This advanced technology has the potential to greatly improve specialists’ and other medical professionals’ diagnostic abilities, especially when it comes to analyzing cancers found in brain for prompt and precise diagnosis. Facilitating effective and accessible communication is one of the main goals of that study. The technology seeks to narrow the knowledge gap between medical experts by simplifying the way magnetic resonance imaging (MRI) results are presented.[
16]
We introduce a hybrid of VGG-16 with fine-tuned vision transformer models to enhance brain tumors classification. Using MR images. The fusion framework demonstrated outstanding performance, suggesting a promising direction for automated brain tumors detection.
The structure of this paper is as follows:
Section 2 introduces related research .
Section 3 illustrates material and method.
Section 4 discussed the experimental results. discussion is presented in
Section 5. Finally, conclusions presented in
Section 6.
2. Related Work
The classification of brain tumors through MRI has witnessed vast improvements via the combination of deep learning techniques, but chronic demanding situations in feature representation, generalizability, and computational performance necessitate ongoing innovation. This section synthesizes seminal and contemporary works, delineating their contributions and barriers to contextualize the proposed hybrid framework. A lot of papers presented are selected for detecting brain tumors, and discussions are covered in the last five years in this section.
The capacity of Convolutional Neural Networks (CNNs) to autonomously obtain structural attributes makes them an essential component of medical image analysis. Ozkaraca et al. (2023) [
19]used VGG16 and DenseNet to obtain 97% accuracy, while Sharma et al. (2022)[
20] used VGG-19 with considerable data augmentation to achieve 98% accuracy. For tumors with diffuse boundaries (like gliomas), CNNs are excellent at capturing local information but have trouble with global contextual linkages. VGG-19 and other deeper networks demand a lot of resources without corresponding increases in accuracy [
19].Data Dependency: Without extensive, annotated datasets, performance significantly declines [
20].Eman et.al (2024) [
33] proposed framework of using CNN and EfficientNetV2B3’s flattened outputs before feeding them into the KNN classifier.
In the study of ResNet50 and DenseNet (97.32% accuracy), Sachdeva et al. (2024) [
21] observed that skip connections enhanced gradient flow but added redundancy to feature maps. The necessity for architectures that strike a balance between discriminative capability and depth was brought to light by their work.Rahman et al. (2023) [
18] proposed PDCNN (98.12% accuracy) to fuse multi-scale features. While effective, the model’s complexity increased training time by 40% compared to standalone CNNs.Ullah et al. (2022) [
22] and Alyami et al. (2024) [
26] paired CNNs with SVMs (98.91% and 99.1% accuracy, respectively).Global Context at a Cost ViTs, introduced by Dosovitskiy et al. (2020) [
15], revolutionized image analysis with self-attention mechanisms.Tummala et al. (2022) [
16] achieved 98.75% accuracy but noted ViTs’ dependency on large-scale datasets (>100K images). Training from scratch on smaller medical datasets led to overfitting [24].Reddy et al. (2024) [
27] fine-tuned ViTs (98.8% accuracy) using transfer learning, mitigating data scarcity. However, computational costs remained high (30% longer inference times than CNNs) . Amin et al. (2022) [
28] merged Inception-v3 with a Quantum Variational Classifier (99.2% accuracy), showcasing quantum computing’s potential. However,Quantum implementations require specialized infrastructure, limiting clinical deployment . CNNs and ViTs excel at local and global features, respectively, but no framework optimally combined both [
15,
16,
19,
20,
21,
27]. Most models operated as "black boxes," failing to meet clinical transparency needs [
22,
26] .Hybrid models often sacrificed speed for accuracy [
18,
27].The VGG-16 and FTVT-b16 hybrid model innovates by using VGG-16 captures local textures (e.g., tumor margins), while FTVT-b16 models global context (e.g., anatomical relationships).
The proposed hybrid VGG-16 and FTVT-b16 framework addresses these gaps by: Harmonizing CNN-derived local features with ViT-driven global attention maps. Using transfer learning to optimize data efficiency. Generation of interpretable attention maps for clinical transparency. This work builds on foundational studies while introducing a novel fusion strategy that outperforms existing benchmarks (99.46% accuracy), as detailed in
Section 4.
3. The Proposed Framework and Methods
We suggest a hybrid of VGG-16 with fine tuned vision transformers model to enhance brain tumors classification using MR images. The system includes choosing 27 capabilities from layer order 3 of VGG-16, which incorporates 13,696 capabilities, to reap the first-class category performance.
3.1. Image Preprocessing
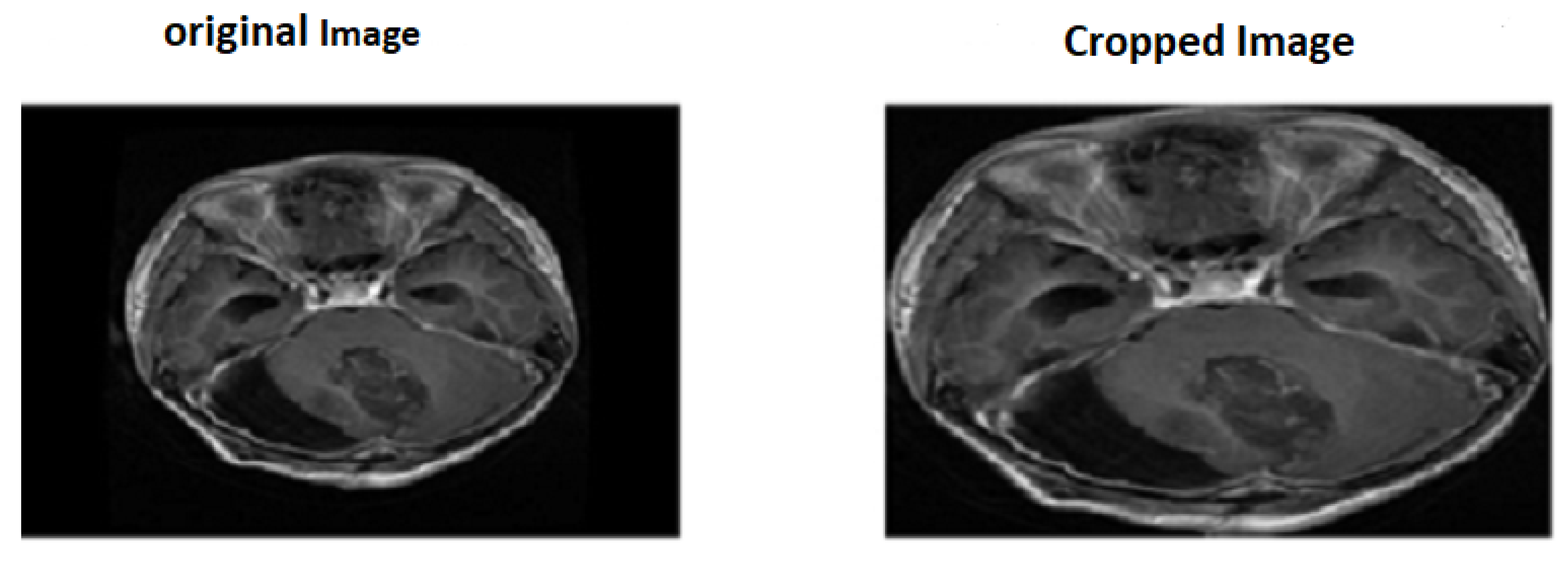
There is an amount of noise in MRI pictures that may be because of the external effects of instrumentality. So the primary step is to eliminate noise from MR images. There are two methods utilized for noise elimination: linear or nonlinear. In linear methods, the pixel value is replaced by a neighborhood-weighted average, which affects image quality [
28] .But, in nonlinear methods, sides are preserved, but the fine structures are degraded. Here, we used a median filter to remove noise from the images[
29].So we choose the nonlinear median filter technique, followed by cropping input images as seen in
Figure 1.
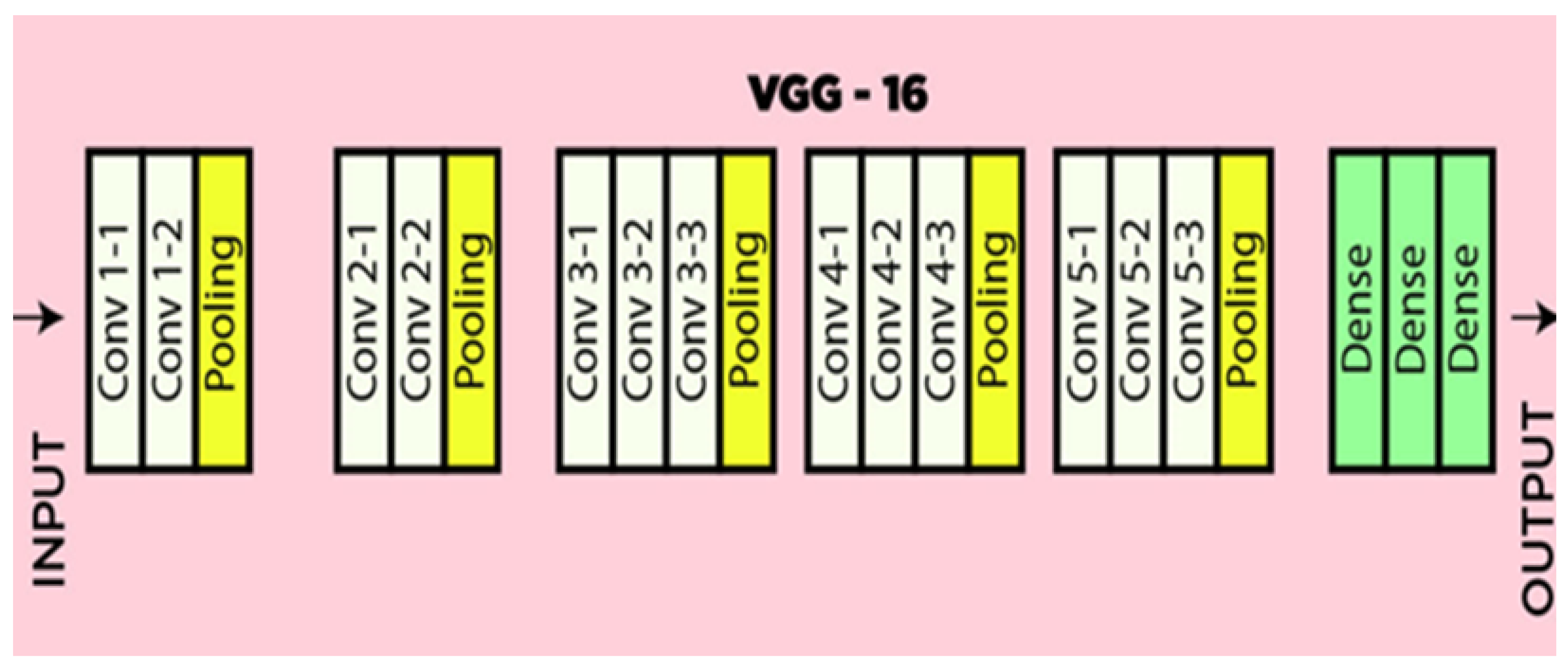
3.2. VGG-16
The VGG16 structure is a deep convolutional neural networks (CNN) utilized for classifying images. VGG-16 is characterized as having a simple structure, smooth recognization and implementation. The VGG16 parameters commonly includes 16 layers, along with 13 convolutional layers and 3 fully connected layers, as shown in
Figure 2.The layers were prepared into blocks, as every block composed of more than one convolutional layer observed via means of a max-pooling layer for downsampling [
30].
3.3. FTVT-B16
The structure of the vision transformers models, as ViT with a “B” version and sixteen layers,includes 12 transformer blocks[
23]. Vit/B16 models was pre trained on the Image-Net datasets using a 1e-4 Adam optimizer and loss entropy feature within 10 epochs with an input image of 224 × 224 pixels. The FTVT/B16 model consists of the ViT/B16 model with a custom classifier head that contains BN, ReLU, and dropout layers [
31]. These layers are enhancing the capacity model to discover critical functions applicable to the direct class. The parameters are being initialized randomly with prevailing VIT/b16 model parameters [
24]. The collection of introduced layers starts off evolved with BN, linear transformer layers, ReLU , and dropout layers. At last, the function vector is converted to the output area through linear layers, figuring out the range of output lessons considering brain tumor types[
32].
Figure 3 shows FTVT architecture.
3.3.1. VGG16- FTVTB16
The typical structure of VGG-16 with fine-tuned Vision Transformers models is:
The input image is split into several patches.
The images were flattened and furnished with class labels.
The outcomes of transformers community were dispatched to the multilayer perception modules.
Constructing the Fused Model Constructing the model fusion of VGG-16 and FTVT-b16 includes many steps:
Feature Extraction: Initial function extraction takes the area via the VGG-16 backbone, which analyzes the MRI images and extracts deep features.
Attention Mechanism: function input to the FTVT-b16 model, which makes use of its self-attention layers to create awareness of salient features and contextual cues that signify distinctive tumor types.
Integration Layer: An integration layer merges the outputs of each model at the same time as leveraging strategies, inclusive of concatenation or weighted averaging to create a unified feature representation.
Classification: The final features from the mixing layer are being fed into fully connected layers for the final types of tumor types, inclusive of gliomas, meningiomas, and pituitary tumors.
3.4. Dataset
The Kaggle database website (
https://www.kaggle.com/datasets/masoudnickparvar/brain-tumor-mri-dataset) is the open-access dataset used in this study[
27]. Gliomas, meningiomas, pituitary, and no tumor are four distinct groups into which the 7,023 images captured by MRI of the human brain are classified. This dataset’s main goal is to help researchers create a highly accurate model for detect and classify brain tumors. The dataset, that includes all four classes, has been divided into 80% for train stage and 20% for test stage.
4. Experimental Results
The overall accuracy of the proposed framework may be computed using preferred metrics, which include accuracy,and precision. Experiments impled that the hybrid structure surpasses the overall performance of both standalone versions, specifically in dealing with many tumor shows and noise in MRI images.
Training and execution of the three models using Google Co-laboratory (Co-lab), which is based totally on Jupyter Notebooks,virtual GPU powered with the aid of a Nvidia Tesla K80 with 16 GB of RAM,Keras library ,and tensor Flow.
Table 1 and
Table 2 summarize the experimental results of the three models run (VGG-16, FTVT-B16, and VGG-16-FTVT-B16).
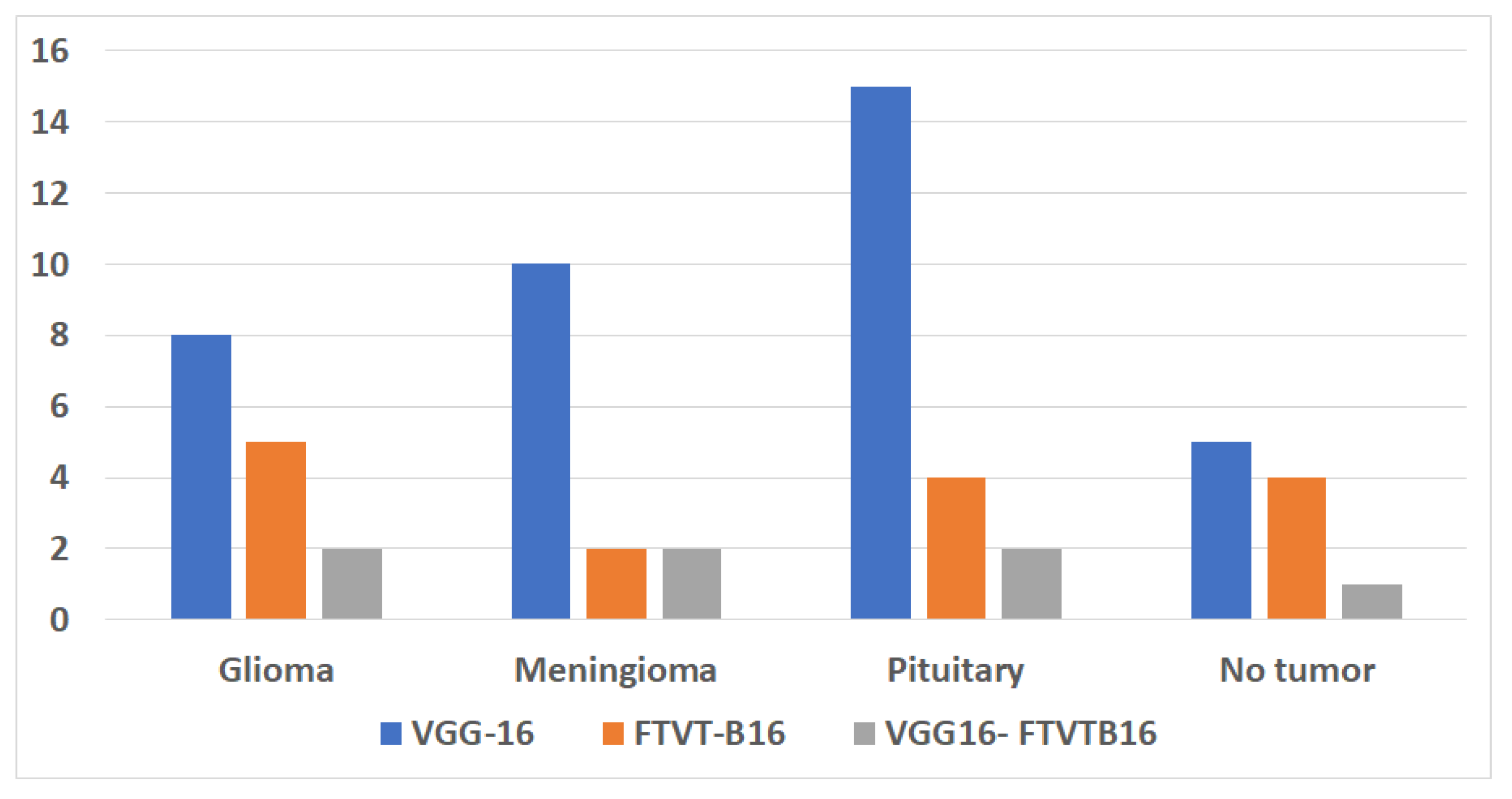
Figure 4 shows the accuracy according to each class of the data set.
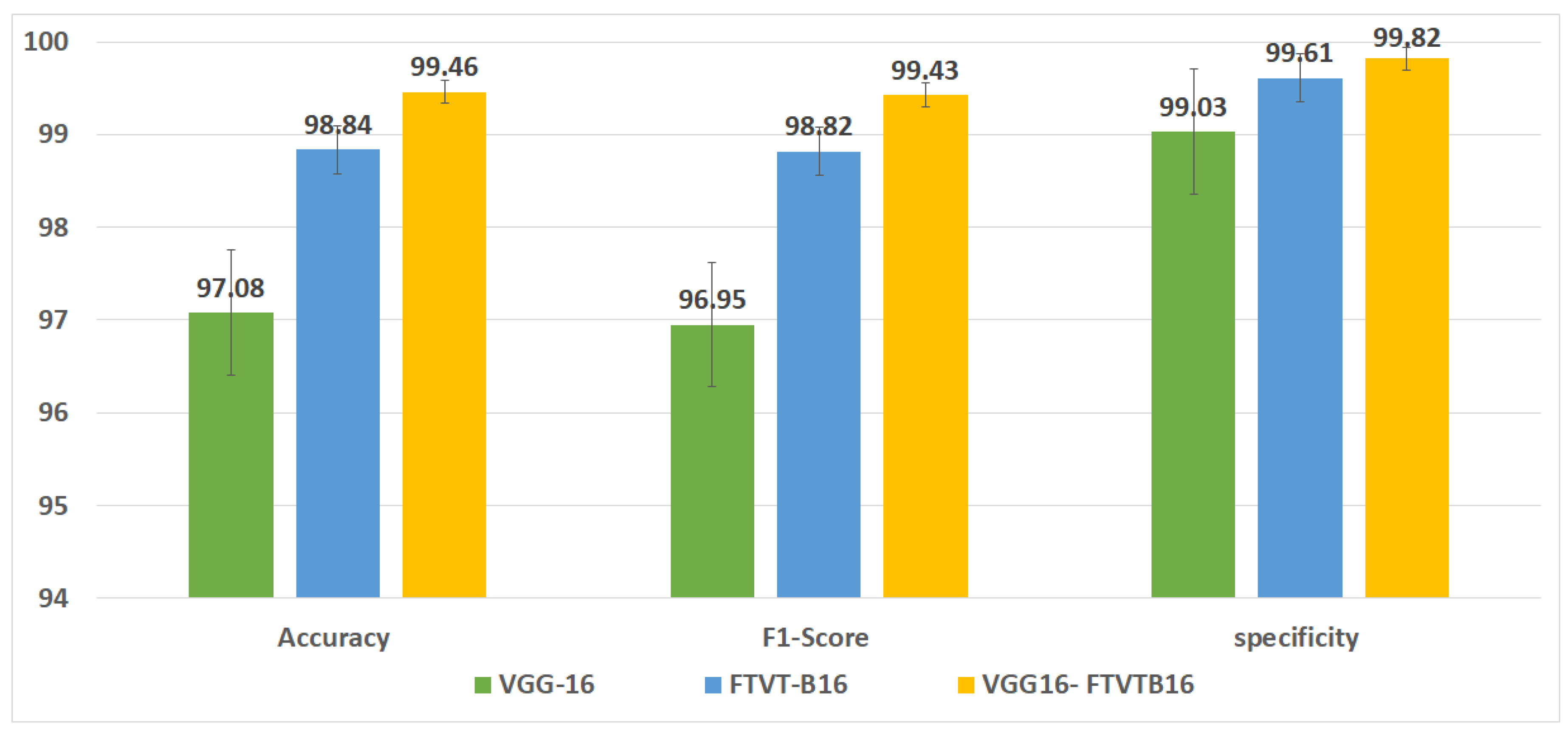
Figure 5 shows the overall accuracy of the suggested models.
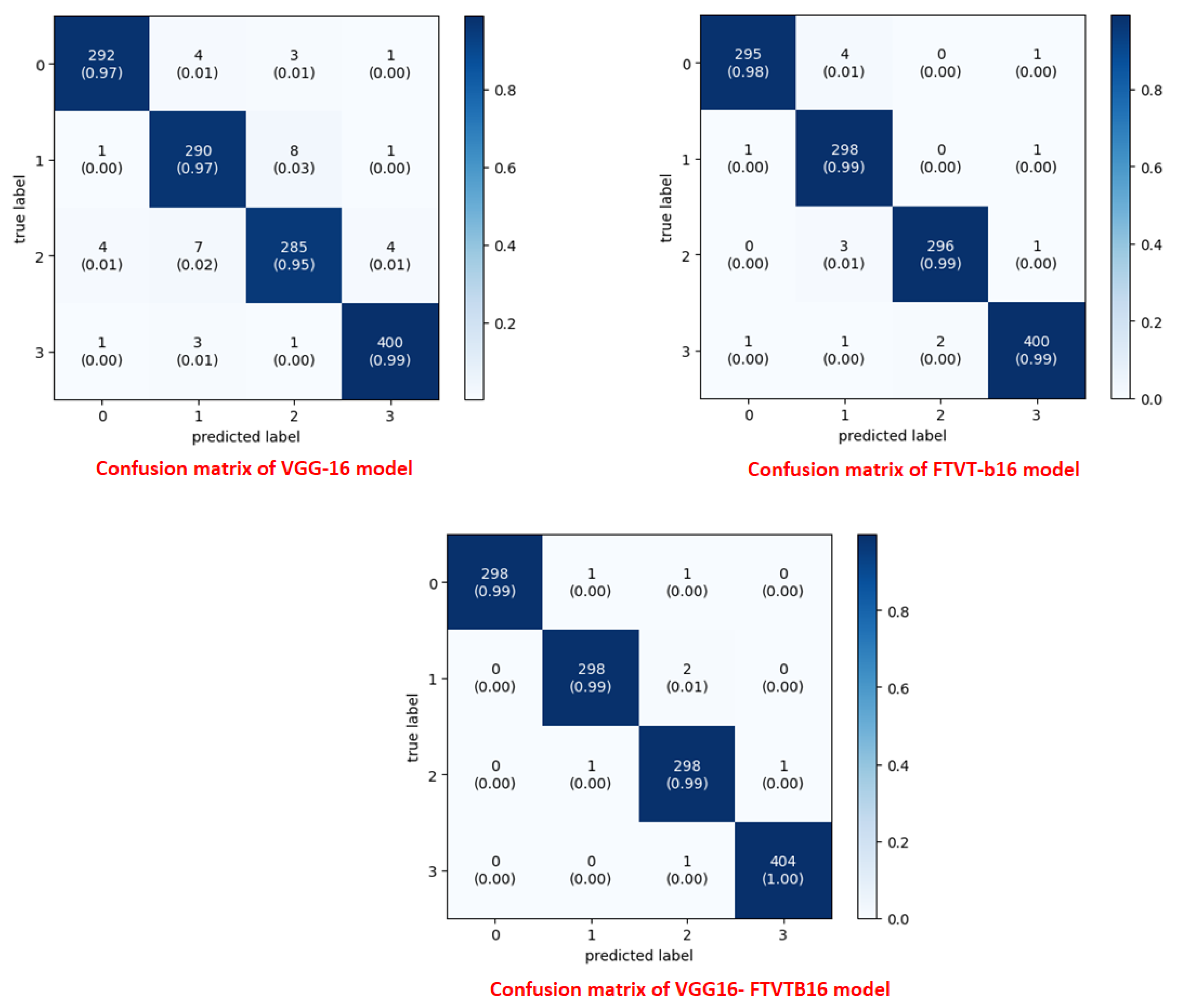
A confusion matrix was normalized by means of dividing every element’s cost in each class, improving the visible illustration of misclassifications in every class,which shows that G, P, M, and no-tumor, or 0, 1, 2, and 3, confer with glioma, pituitary, meningioma, and no-tumor, respectively. distinct values may be used from the confusion matrix to illustrate overall classification accuracy, Recall for every class in dataset.
Figure 6 illustrates confusion matrix for each model.
5. Results Discussion
This study concerned with classifying brain tumors type via pre-processing stage,and using hybrid approach of VGG-16 with fine-tuning vision transformer models, and comparing overall performance metrics such as the accuracy, precision, and recall. The fusion model demonstrates 99.46% for classification accuracy,surpassing the performance of standalone VGG-16 (97.08%) and FTVT-b16 (98.84%).In this paper, We introduce a hybrid of VGG-16 with fine-tuned Vision Transformer models to improve brain tumors Classification Using MR images. The suggested framework demonstrated outstanding performance, suggesting a promising direction of automated brain tumors detection. Sensitivity, precision, and recall.
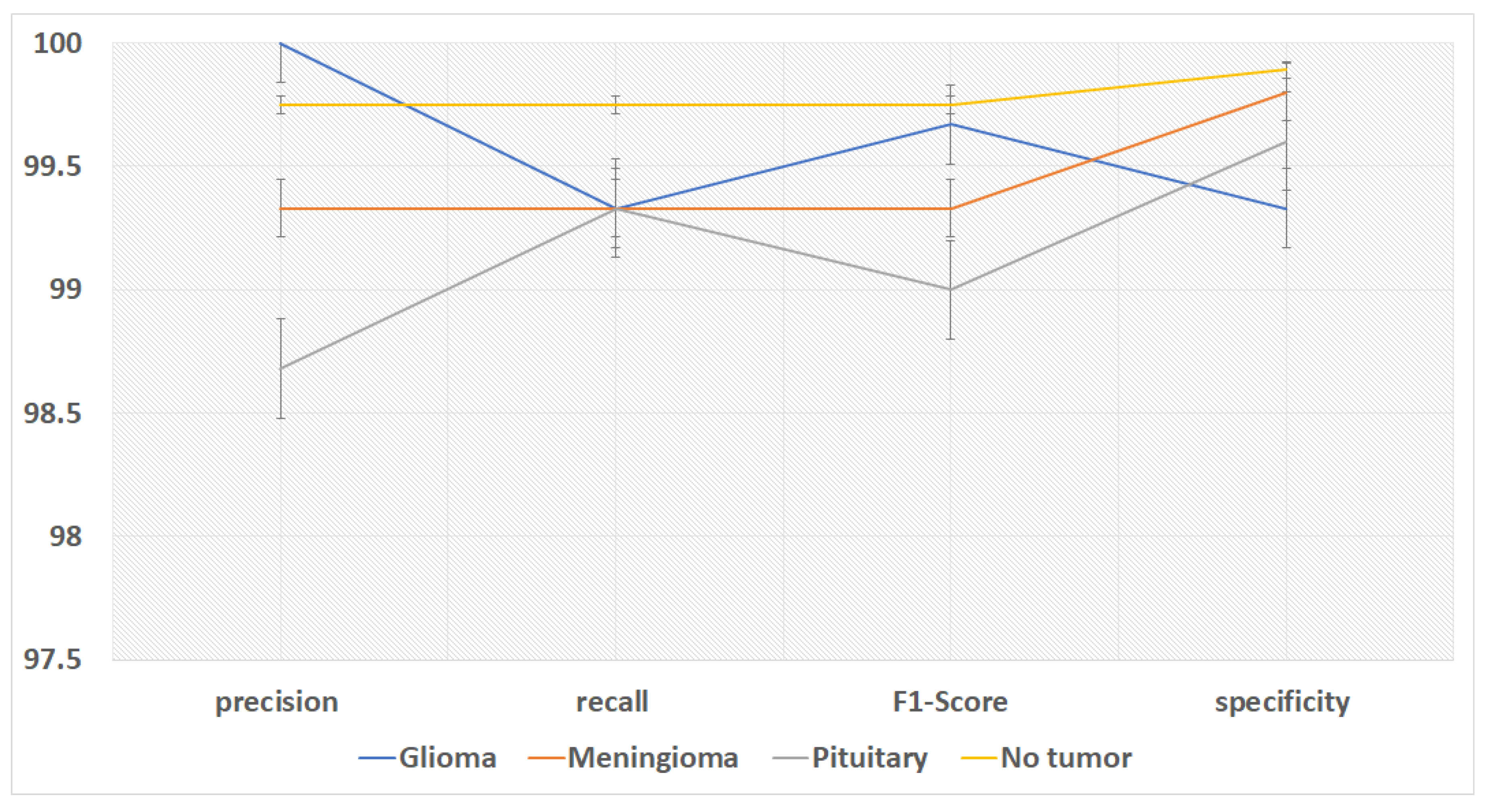
Figure 7 shows the performance metrics (precision, recall, f1-score,specificity) on the fusion of VGG-16 and FTVT-b16 models. We compare the accuracy of recent papers tested on the same dataset in
Table 3.
The hybrid model performs better than current benchmarks when compared to modern approaches. For example, it outperformed the parallel deep convolutional neural network (PDCNN, 98.12%; Rahman et al., 2023) and the Inception-QVR framework (99.2%; Amin et al., 2022), both of which depend on homogenous designs, in terms of accuracy. While pure ViTs require large amounts of training data (Khan et al., 2022) and conventional CNNs like VGG-19 are limited by their localized receptive fields (Mascarenhas & Agarwal, 2021), the suggested hybrid makes use of FTVT-b16’s ability to resolve long-range pixel interactions and VGG-16 effectiveness in hierarchical feature extraction. Confusion matrix analysis (
Figure 6) also showed that 15 pituitary cases were incorrectly classified by the solo VGG-16, most likely as a result of its incapacity to contextualize anatomical relationships outside of local regions. To illustrate the transforming function of transformer-driven attention in disambiguating heterogeneous tumor appearances, the hybrid model, on the other hand, decreased misclassifications to two cases per class (Tummala et al., 2022).
The model’s balanced class-wise measures and excellent specificity (99.82%) offer significant promise for improving diagnostic workflows from a clinical standpoint. The methodology tackles a widespread issue in neuro-oncology, where incorrect diagnosis can result in unnecessary surgical procedures, by reducing false positives (van den Bent et al., 2023). For instance, the model’s incorporation of both global and local variables improves the accuracy of distinguishing pituitary adenomas from non-neoplastic cysts, a task that has historically been subjective. Additionally, the FTVT-b16 component produces attention maps that show the locations of tumors, providing radiologists with visually understandable explanations (
Figure 3).This is in line with the increasing calls for explainable AI in medical imaging and contrasts with opaque "black-box" systems (Jiang et al., 2022). However, claims of generalization are limited by the study’s dependence on a single-source dataset that lacks metadata on demographic diversity or scanner variability. To evaluate robustness against population-specific biases and varied imaging procedures, future validation across multi-institutional cohorts is essential (Rajiah et al., 2023).
6. Conclusions
Brain tumors classification in MRI considered a critical issues in neuro-oncology, necessitating advanced computational frameworks to enhance diagnostic performance. This paper proposed a hybrid deep learning framework which extracting features capabilities using VGG-16 with the global contextual modeling of FTVT-b16, a fine tuned Vision Transformer (ViT), to address the limitations of standalone convolutional neural networks (CNN) and transformer based architectures. The proposed framework tested on dataset of 7,023 MRI images spanning four classes glioma, meningioma, pituitary, and no-tumor,and achieved a classification accuracy of 99.46%, surpassing both VGG-16 (97.08%) and FTVT-b16 (98.84%) . Key performance metrics, including precision (99.43%), recall (99.46%), and specificity (99.82%), underscored the model’s robustness, particularly in reducing misclassifications of anatomically complex cases such as pituitary tumors.
The "black-box" constraints of traditional deep learning systems were addressed by combining CNN-derived local features with ViT-driven global attention maps, which improved interpret-ability through displayed interest regions and increased diagnostic precision. This dual feature gives radiologists both excellent accuracy and useful insights into model decisions, which is in line with clinical processes. A comparison with cutting-edge techniques like PDCNN (98.12%) and Inception-QVR (99.2%) confirmed the hybrid model’s superiority in striking a balance between diagnostic granularity and computational costs.
Despite these developments, generalization is limited by the study’s reliance on a single-source dataset because demographic variety and scanner variability were not taken into consideration. Multi-institutional validation should be given top priority in future studies in order to evaluate robustness across diverse demographics and imaging procedures. Furthermore, investigating advanced explainability tools (e.g., Grad-CAM, SHAP) and multi modal data integration (e.g., diffusion-weighted imaging, PET) could improve diagnostic value and clinical acceptance. To advance AI-driven precision in neuro-oncology, real-world application through collaborations with healthcare organizations and benchmarking against advanced technology like Transformers will be essential.
In conclusion, this paper lays a foundational paradigm for hybrid deep learning in medical image analysis, by showing that the fusion of CNN and ViT can bridge the gap between computational innovation and clinical utility. By enhancing both accuracy and interpret ability, the proposed framework has great potential to improve patient outcomes through earlier, more accurate diagnosis by increasing both accuracy and interpret ability.
Funding
This work was supported and funded by the Deanship of Scientific Research at Imam Mohammad Ibn Saud Islamic University (IMSIU) (grant number IMSIU-DDRSP2502)..
Conflicts of Interest
The authors declare no conflicts of interest.
References
- van den Bent, M. J., Geurts, M., French, P. J., Smits, M., Capper, D., Bromberg, J. E., & Chang, S. M. (2023). Primary brain tumours in adults. The Lancet, 402(10412), 1564-1579.
- Solanki, S., Singh, U. P., Chouhan, S. S., & Jain, S. (2023). Brain tumor detection and classification using intelligence techniques: an overview. IEEE Access, 11, 12870-12886.
- Patel, A. (2020). Benign vs malignant tumors. JAMA oncology, 6(9), 1488-1488.
- Jiang, Y., Zhang, Y., Lin, X., Dong, J., Cheng, T., & Liang, J. (2022). SwinBTS: A method for 3D multimodal brain tumor segmentation using swin transformer. Brain sciences, 12(6), 797.
- Ranjbarzadeh, R., Caputo, A., Tirkolaee, E. B., Ghoushchi, S. J., & Bendechache, M. (2023). Brain tumor segmentation of MRI images: A comprehensive review on the application of artificial intelligence tools. Computers in biology and medicine, 152, 106405.
- Raghavendra, U., Gudigar, A., Paul, A., Goutham, T. S., Inamdar, M. A., Hegde, A., ... & Acharya, U. R. (2023). Brain tumor detection and screening using artificial intelligence techniques: Current trends and future perspectives. Computers in Biology and Medicine, 163, 107063.
- Ranjbarzadeh, R., Caputo, A., Tirkolaee, E. B., Ghoushchi, S. J., & Bendechache, M. (2023). Brain tumor segmentation of MRI images: A comprehensive review on the application of artificial intelligence tools. Computers in biology and medicine, 152, 106405.
- Ottom, M. A., Rahman, H. A., & Dinov, I. D. (2022). Znet: deep learning approach for 2D MRI brain tumor segmentation. IEEE Journal of Translational Engineering in Health and Medicine, 10, 1-8.
- Rajiah, P. S., François, C. J., & Leiner, T. (2023). Cardiac MRI: state of the art. Radiology, 307(3), e223008.
- Nazir, M., Shakil, S., & Khurshid, K. (2021). Role of deep learning in brain tumor detection and classification (2015 to 2020): A review. Computerized medical imaging and graphics, 91, 101940.
- Younis, E.; Mahmoud, M.N.; Ibrahim, I.A. Python Libraries Implementation for Brain tumour Detection Using MR Images Using Machine Learning Models. In Advanced Interdisciplinary Applications of Machine Learning Python Libraries for Data Science; Biju, S., Mishra, A., Kumar, M., Eds.; IGI Global: 2023; pp. 243–262. [CrossRef]
- Mascarenhas, S., & Agarwal, M. (2021, November). A comparison between VGG16, VGG19 and ResNet50 architecture frameworks for Image Classification. In 2021 International conference on disruptive technologies for multi-disciplinary research and applications (CENTCON) (Vol. 1, pp. 96-99). IEEE.
- Kaur, T., & Gandhi, T. K. (2019, December). Automated brain image classification based on VGG-16 and transfer learning. In 2019 international conference on information technology (ICIT) (pp. 94-98). IEEE.
- Khan, S., Naseer, M., Hayat, M., Zamir, S. W., Khan, F. S., & Shah, M. (2022). Transformers in vision: A survey. ACM computing surveys (CSUR), 54(10s), 1-41.
- More, R. B., & Bhisikar, S. A. (2021, May). Brain tumor detection using deep neural network. In Techno-Societal 2020: Proceedings of the 3rd International Conference on Advanced Technologies for Societal Applications—Volume 1 (pp. 85-94). Cham: Springer International Publishing.
- Saleh, A., Sukaik, R., & Abu-Naser, S. S. (2020, August). Brain tumor classification using deep learning. In 2020 International Conference on Assistive and Rehabilitation Technologies (iCareTech) (pp. 131-136). IEEE.
- Sachdeva, J., Sharma, D., & Ahuja, C. K. (2024). Comparative Analysis of Different Deep Convolutional Neural Network Architectures for Classification of Brain Tumor on Magnetic Resonance Images. Archives of Computational Methods in Engineering, 1-20.
- Sharma, S., Gupta, S., Gupta, D., Juneja, A., Khatter, H., Malik, S., & Bitsue, Z. K. (2022). [Retracted] Deep Learning Model for Automatic Classification and Prediction of Brain Tumor. Journal of Sensors, 2022(1), 3065656.
- Özkaraca, O., Bağrıaçık, O. İ., Gürüler, H., Khan, F., Hussain, J., Khan, J., & Laila, U. E. (2023). Multiple brain tumor classification with dense CNN architecture using brain MRI images. Life, 13(2), 349.
- Rahman, T., & Islam, M. S. (2023). MRI brain tumor detection and classification using parallel deep convolutional neural networks. Measurement: Sensors, 26, 100694.
- Alyami, J., Rehman, A., Almutairi, F., Fayyaz, A. M., Roy, S., Saba, T., & Alkhurim, A. (2024). Tumor localization and classification from MRI of brain using deep convolution neural network and Salp swarm algorithm. Cognitive Computation, 16(4), 2036-2046.
- Ullah, N., Khan, J. A., Khan, M. S., Khan, W., Hassan, I., Obayya, M., ... & Salama, A. S. (2022). An effective approach to detect and identify brain tumors using transfer learning. Applied Sciences, 12(11), 5645.
- Dosovitskiy, A. (2020). An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929.
- Tummala, S., Kadry, S., Bukhari, S. A. C., & Rauf, H. T. (2022). Classification of brain tumor from magnetic resonance imaging using vision transformers ensembling. Current Oncology, 29(10), 7498-7511.
- Reyes, D., & Sánchez, J. (2024). Performance of convolutional neural networks for the classification of brain tumors using magnetic resonance imaging. Heliyon, 10(3).
- Amin, J., Anjum, M. A., Sharif, M., Jabeen, S., Kadry, S., & Moreno Ger, P. (2022). A new model for brain tumor detection using ensemble transfer learning and quantum variational classifier. Computational intelligence and neuroscience, 2022(1), 3236305.
- Msoud Nickparvar. (2021). Brain Tumor MRI Dataset [Data set]. Kaggle. [CrossRef]
- Kovásznay, L. S., & Joseph, H. M. (1955). Image processing. Proceedings of the IRE, 43(5), 560-570.
- Maharana, K., Mondal, S., & Nemade, B. (2022). A review: Data pre-processing and data augmentation techniques. Global Transitions Proceedings, 3(1), 91-99.
- Guan, Q., Wang, Y., Ping, B., Li, D., Du, J., Qin, Y., ... & Xiang, J. (2019). Deep convolutional neural network VGG-16 model for differential diagnosing of papillary thyroid carcinomas in cytological images: a pilot study. Journal of Cancer, 10(20), 4876.
- Dai, Y., Gao, Y., & Liu, F. (2021). Transmed: Transformers advance multi-modal medical image classification. Diagnostics, 11(8), 1384.
- Reddy CKK, Reddy PA, Janapati H, Assiri B, Shuaib M, Alam S, Sheneamer A. A fine-tuned vision transformer based enhanced multi-class brain tumor classification using MRI scan imagery. Front Oncol. 2024 Jul 18;14:1400341. PMID: 39091923; PMCID: PMC11291226. [CrossRef]
- Younis, E. M., Mahmoud, M. N., Albarrak, A. M., & Ibrahim, I. A. (2024). A Hybrid Deep Learning Model with Data Augmentation to Improve Tumor Classification Using MRI Images. Diagnostics, 14(23), 2710.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).