1. Introduction
One of the actual tasks of modern chemistry is the search of new pharmacologically active substances. This is due to the need to create drugs against new diseases, reduce toxicity and side effects, and improve the effectiveness of existing drugs. Additional problem, which leads to the need to search for new antibacterial agents, is the developing bacterial resistance to widely used substances. A known direction for the obtaining of new biologically active compounds is the functionalization of known biologically active molecules.
Azomethines, commonly known as Shiff bases, have firmly established their importance in modern chemical research due to their remarkable versatility and wide-ranging applications, particularly in the fields of medical chemistry [
1,
2,
3,
4] and advanced materials science. These compounds, characterized by the imine group >C=N-, serve as pivotal ligands in coordination chemistry, forming complexes with diverse metal ions that exhibit biological activity [
5,
6], magnetic behavior and photoluminescent properties.
Numerous studies have documented that azomethines exhibit significant antibacterial properties [
7,
8], demonstrating effectiveness against pathogens such as
Staphylococcus aureus and
Escherichia coli, with some compounds showing activity comparable to standard antibiotics like Ciprofloxacin [
8]. Beyond their antibacterial potential, this class of compounds is also recognized for possessing antifungal [
9], anticancer [
10], antioxidant [
7,
11] and analgesic [
12] activities, highlighting their broad pharmacological value. Aromatic azomethines also show tyrosinase inhibition activity [
13].
The enduring scientific interest in the azomethines is driven by their structural adaptability, which allows for the strategic design of derivatives with tailored functionalities for specific technological and biomedical needs. The incorporation of long-chain substituents into azomethine frameworks presents a particularly promising research direction, as it enables precise modulation of their physicochemical characteristics, such as solubility, lipophilicity and etc., influencing practical efficacy. Azomethines with long alkyl chains and diethylamine substituent were synthesized [
14] and biological activity measurements shown high efficacy against Gram positive and Gram negative bacterial strains and fungi. Adding 3d-metal complexes, the biocidal activity was increased remarkably, which was attributed to increased lipophilicity due to complexation with azomethines and enhanced penetration of metal ions through the lipid membrane, resulting in improved interaction with ferments [
14].
The simplest way to obtain azomethines is reaction between aldehyde and amine [
15]. Through the varying structures of both reagents a lot of combinations are possible. Such variability is important to obtain structure – properties correlations, which with relatively simple synthetic procedures for a lot of azomethines makes them attractive for robotic systems and autonomous (self-driving) labs [
16].
Including another pharmacophore group into azomethine structure is also good way to obtain biologically active compounds. Thus, introduction triazole fragment gives substances with antifungal activity [
17]. Azomethines with thiazole ring show anti-inflammatory and antioxidant activities [
18]. A novel strategy for one-pot solvothermal synthesis were developed [
19] and used to obtain phenylpyrazole Shiff bases, which works well as insecticides. Interestingly that this activity is attributed to the blocking of ionic channels due calcium chloride binding to C=N bond.
Nowadays a lot of new chemical substances are synthesized and biological activity measurements for all of them are hardly possible. So initial “filtration” with the real testing of the best revealed candidates seems more realistic scenario. Widely used method for biological activity predictions is docking, which is relatively fast for one molecule and one “target” (ferment molecule, DNA, RNA and etc.). However, in a living organism there are a lot of biomolecules for possible interactions and in combination with the necessity to predict activity for many molecules such investigations became very expensive in terms of computational time. Alternative way for predictions is quantitative structure-activity relationships (QSAR) modelling [
20], which became powerful tool for high-throughput screening with the development of machine learning (ML) and artificial intelligence (AI).
Lipophilicity is very important property in drug discovery and design [
21,
22,
23] and it is also necessary for good prediction of molecule bioactivity by machine learning. Usually lipophilicity is quantified as octanol-water partition coefficient (log
P) and can be measured directly through the concentrations of substance in two phases by the shake-flask [
24], slow-stirring [
25] and generator column techniques [
26]. Another possibility to determine lipophilicity is undirect methods, for example high-performance liquid chromatography [
27], when log
P is determined through the retention time, but such method works well only with the presence of reference substance with the known value of log
P.
Experimental determination of lipophilicity can be unapplicable or give unreliable results for the poorly soluble substances. Similar problem with the highly hydrophilic or highly lipophilic compounds. In such cases computational approaches (for example, quantum-chemical calculations) or machine learning methods [
28] can be the way for lipophilicity determination. Quantum-chemical determination of log
P value is based on calculation of the Gibbs energy change upon transfer between solvents [
29].
The main aim of the work is the search of compounds with antibacterial properties and simple synthetic pathways, especially in combination with properties modelling technics, to use, in perspective, such systems for the development of autonomous labs [
16], when predictions of artificial intelligence are tested by experiments on robotic installations and feedback is received on the results. The synthesized azomethines showed antibacterial activity at the level of some commercial antiseptics. Lipophilicity of the synthesized and isomeric substances was predicted using three different approaches: quantum chemical calculations, machine learning and fragment-based description. Machine learning-based methods (GraphormerLogP) demonstrate acceptable accuracy, sensitivity to isomerism, and orders of magnitude higher throughput, making them an optimal tool for high-throughput screening. Quantum-chemical calculations can take into account any spatial arrangements, but much more computational time it is necessary for it.
2. Materials and Methods
Commercially available reagents and solvents were used for synthesis.
The 1H and 13C{1H} NMR spectra were recorded on Bruker Avance III 400 MHz spectrometer. Fourier Transform infrared spectroscopy (FT-IR) were measured using a FT-IR Spectrometer Spectrum two PerkinElmer with UATR (Single Reflection Diamond). Elemental analysis was made using PerkinElmer® 2400 Series II CHNS/O Elemental Analyzer (2400 Series II).
The antifungal and antibacterial activity of the chemical compounds was studied using test cultures of opportunistic microflora. The following strains were used: Staphylococcus aureus (ATCC 29213), Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 27853), Bacillus cereus, and Candida albicans (ATCC 885-653). All compounds are extremely poorly soluble in water, so 1% solutions in ethanol were used in the study.
24-hour cultures of microorganisms were washed from meat-peptone agar slants with saline and standardized to a turbidity standard of 0.5 McFarland (1.5·108 CFU/ml). The culture media were inoculated with a swab soaked in the standardized culture. Wells were then cut in the contaminated nutrient agar and added with the test drugs and comparison drugs, Miramistin, Chlorhexidine and Kodan. Sabouraud's medium for Candida yeasts and Mueller-Hinton medium for pathogenic and opportunistic microflora of humans and animals were used as nutrient media. The dishes were incubated at 35 °C for 24-48 hours, after which the growth inhibition zone size was assessed, measuring it to an accuracy of 0.1 mm.
To complement the experimental investigation, the lipophilicity parameter (log
P, the octanol-water distribution coefficient) of the synthesized and isomeric compounds was estimated using quantum-chemical calculations, which were made using Orca program (version 6) [
30,
31]. r
2SCAN-3c composite method [
32] was chosen as good cost-benefit-ratio method for structural optimizations and energy calculations [
33]. SMD model [
34] were used for accounting solvent effects; octanol and water were taken as solvents, structures were optimized at both solvents. Initial structures for calculations were made using Avogadro program [
35]. Chemcraft [
36] was used for calculations results viewing and visualization. Gibbs free energies were taken from calculations and log
P was calculated using the formula:
where ∆
Gwater – Gibbs free energy in water, ∆
Goctanol – Gibbs free energy in octanol,
R – universal gas constant,
T – temperature (298 K).
Additionally, the lipophilicity parameter was estimated using a graph-based deep learning approach. For this purpose, we employed the GraphormerLogP model described in a recent publication [
37], which demonstrated superior predictive accuracy compared to traditional descriptor-based and machine-learning methods. The GraphormerLogP model is based on a fine-tuned GraphormerMapper encoder architecture [
38] that directly processes molecular graphs derived from SMILES strings. The molecular structures of the synthesized compounds were converted from SMILES representations into molecular graphs using the Chython library [
39]. These graphs were subsequently processed by the pre-trained GraphormerLogP model with the original weights.
As an additional computational approach, lipophilicity (log
P) values for the investigated compounds were estimated using the Crippen atom contribution model [
40], as implemented in the Chem.Crippen module of the RDKit library [
41]. The molecular structures of the investigated compounds were standardized and converted from SMILES representations into RDKit molecule objects. The lipophilicity was then computed using the MolLogP function, which reproduces the Wildman-Crippen model coefficients without additional parameter optimization. Due to its computational efficiency and reproducibility, this approach serves as a reliable baseline reference for comparing quantum-chemical log
P estimates with machine learning-based predictions such as those obtained from the GraphormerLogP model.
3. Results
3.1. Compounds Synthesis and Characterization
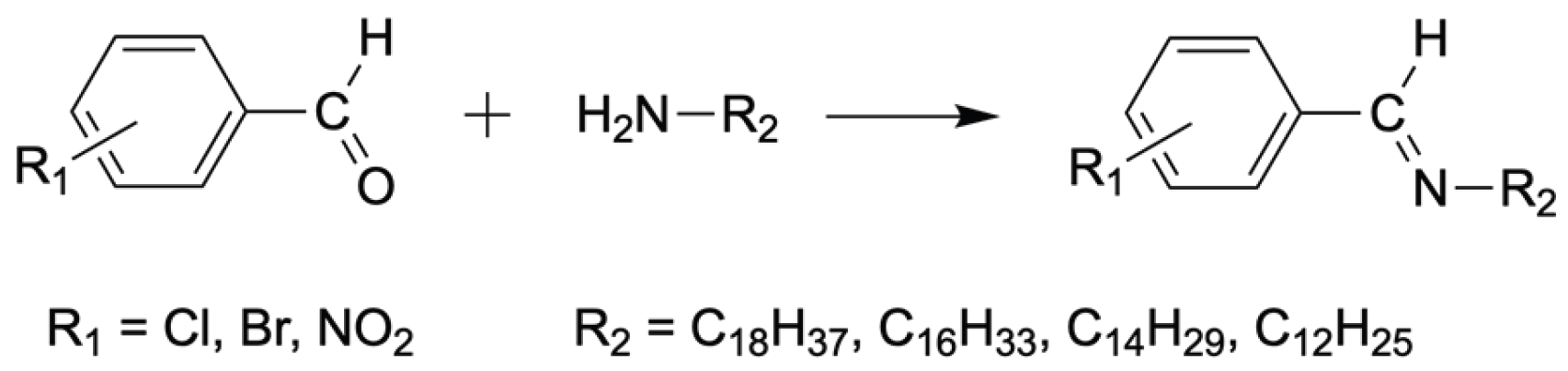
Azomethines were synthesized according to the previously reported procedure [
42] by the reaction between aromatic aldehyde with long-chain amine. Reaction in shown in
Scheme 1; synthetic details are given in ESI1.
The conformity of the obtained product with the expected structure was established on the basis of a set of data from elemental analysis, NMR and IR spectra (for details see ESI1). The yields are bigger than 90% (exact values can be found within synthetic details).
3.2. Rotation of Imine Fragment
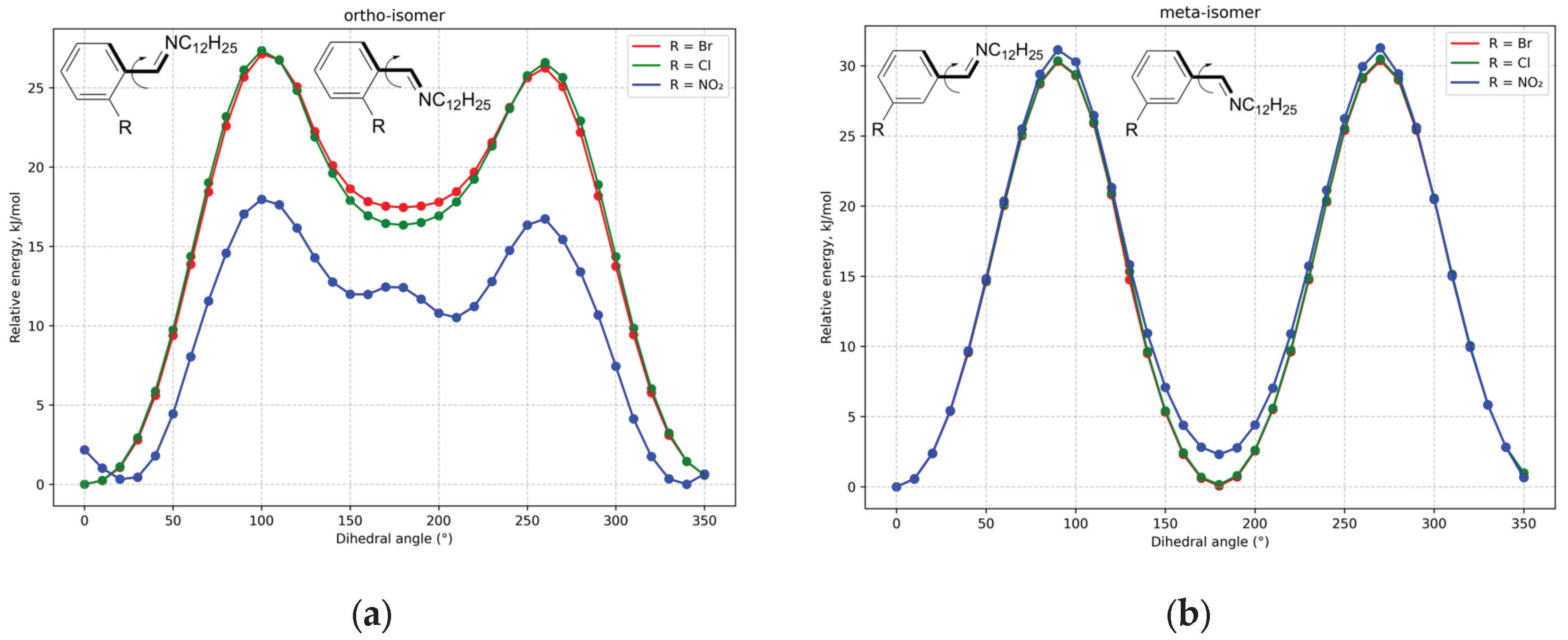
Theoretically, the C=N double bond of imine fragment is in conjugation with phenyl ring and rotation around C(phenyl)-C(imine) bond may be difficult, so two ortho and two meta structures can be possible. That’s why relaxed surface scans for the corresponding dihedral angle were made, results are shown in
Figure 1 for the substances with C12 alkyl chain.
As can be seen from the
Figure 1, two minima are presented: in the first dihedral angle is 0° and nitrogen atom is in the opposite side of C(phenyl)-C(imine) bond relative to phenyl ring substituent; in the second angle is 180° and nitrogen is in the same side with heteroatomic group.
For the meta-isomer two minima have practically the same energies, whereas for ortho-isomer the difference is more than 15 kJ/mol for bromine and chlorine derivatives (for the nitro-substituted compound the difference is smaller – near 10 kJ/mol). Such situation can be explained by the steric hindrance between phenyl ring substituent and nitrogen atom of imine bond in the structures of ortho-isomers with the dihedral angle near 180°, when the atoms of mentioned fragments are very close to each other. In the meta-isomer substituent is in a more distant position from imine fragment, steric hindrance between those two groups is absent and energies of two minima are practically the same.
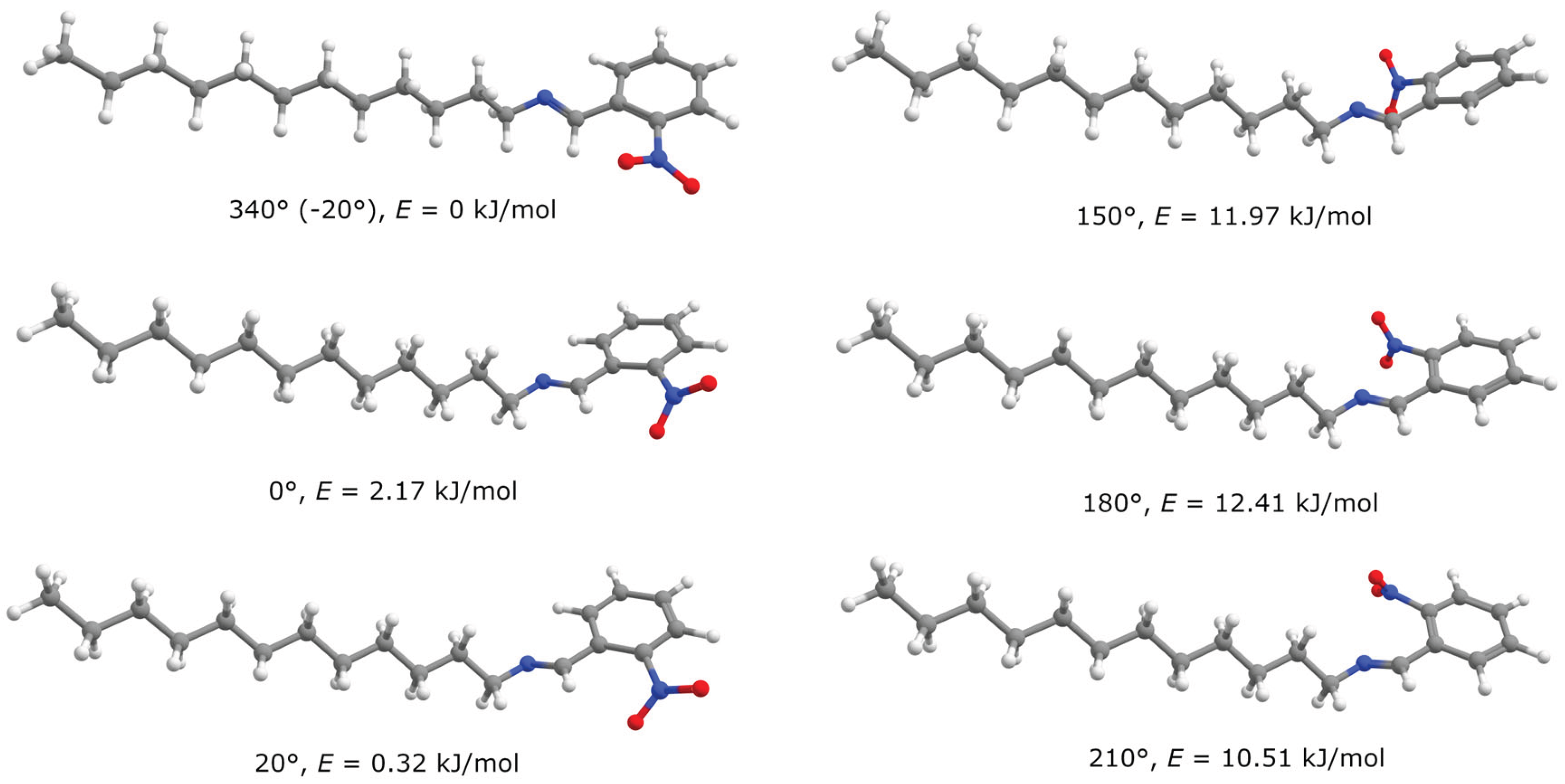
In the ortho-isomer of nitro-substituted compound minima are “twinned” – there are two small maxima at 0° and 180° with two minima at the both side of each. It should be mentioned that such “twinning” is absent in meta-isomer. Such situation is also the result of steric hindrance – at the “ideal” angles 0° and 180° nitro-group of ortho-isomer can’t be located in the plane of phenyl ring, that’s why conjugation between them is violated and energy is increased at 0° and 180° dihedral angles. Minima are shifted by some value (near 20-30°) to obtain optimal conjugation between the phenyl ring and both groups – nitro substituent and imine fragment. Structures, clarifying this situation, are given in
Figure 2.
As can be seen from the
Figure 1, energy barriers for the rotation are relatively low (the maximal value is near 30 kJ/mol), that’s why the isomerization is highly possible at room temperature and structures for both local minima can be presented in solution. However, taking into account the energy difference between two minima for ortho-isomer, mole fraction of the structure with close arrangement of phenyl ring substituent and nitrogen atom of imine group is smaller than 1%. Similar equilibration between two structures of azomethine was observed for more complicated situation with cyclization [
43].
3.3. Antibacterial Activity
Antibacterial activity testing results are given in
Table 1.
As can be seen from the given data in
Table 1, nine investigated compounds show the activity on the same level as antiseptic “Kodan” and works better for several test cultures than Miramistin and Chlorhexidine.
3.4. Lipophilicity Modelling
Lipophilicity modeling results obtained by three different methods (quantum chemical calculations (QC), GraphormerLogP model (GLP) and RDKit library (RDK)) are shown in
Table 2 (optimized structures, xyz-matrices and energies for all isomers in
Table 2 for both solvents (water and octanol) are given in ESI2).
To evaluate predictive accuracy, quantum-chemically computed logP values of azomethines were used as reference data. The GraphormerLogP deep learning model and the Wildman-Crippen atom contribution model were compared on this dataset. The GraphormerLogP model achieved higher overall accuracy (RMSE = 1.054 for all compounds, MAE = 0.943) than the RDKit predictions (RMSE = 1.407, MAE = 1.331).
As can be seen from the
Table 2, logP modelling on the base of quantum-chemical calculations gives different results for all five structures (ortho, ortho’, meta, meta’ and para) as expected from different atom arrangements in 3d-space. GraphormerLogP approach doesn’t distinguish ortho/ortho’ and meta/meta’ pairs, which is not in controversary with relatively easy isomerization due to rotation (see section 3.2). Additionally, GLP method gives slightly different values for ortho-, meta- and para-isomers, the biggest difference is obtained for nitro derivative with C12 alkyl chain (0.27 log units, see
Table 2). However, the relationships between values for ortho-, meta- and para-isomers in two methods (QC and GLP) are not the same for several substances in the
Table 2. At the same time, RDKit method in such case is the worst of the used because doesn’t distinguish the position of substituent in phenyl ring (see
Table 2).
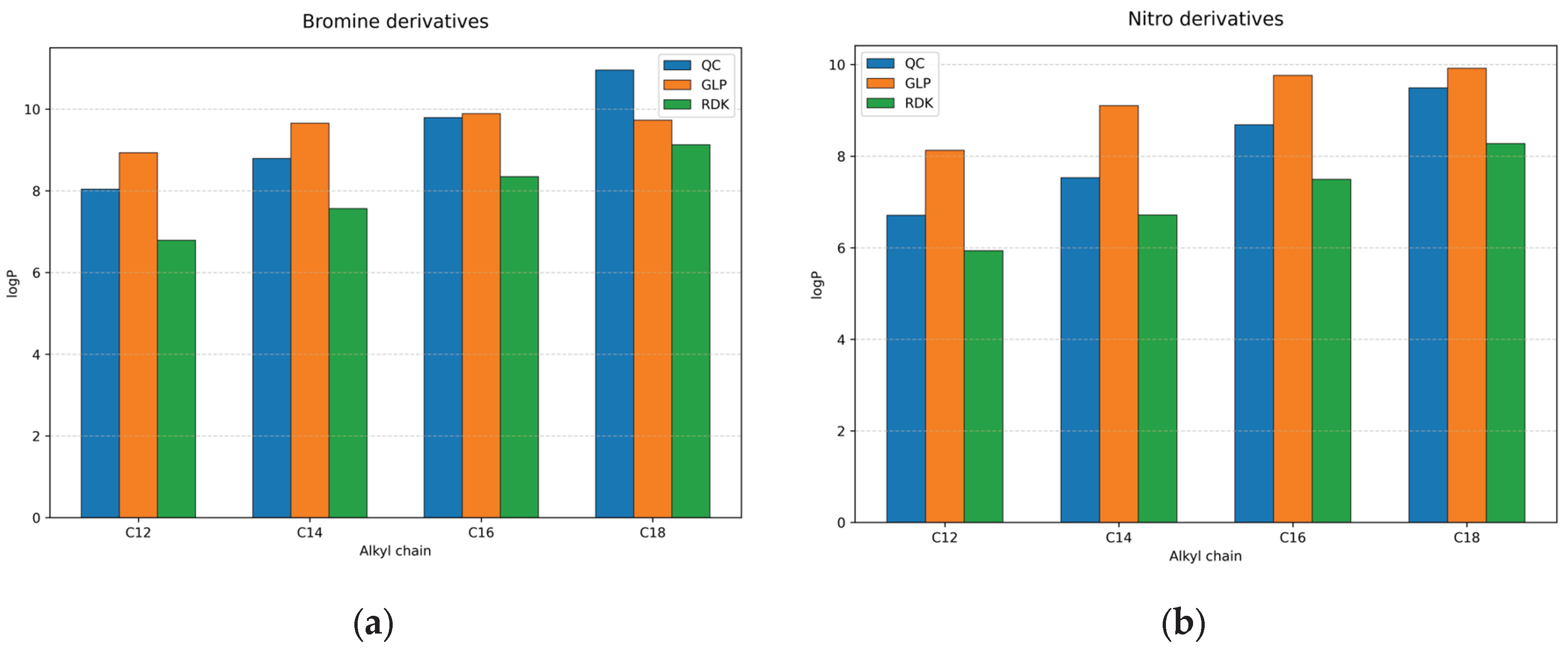
Dependencies of modelled logP values from alkyl chain length are given in the
Figure 3 for bromine and nitro derivatives (such dependency for chlorine derivatives is not shown, because it is very similar for bromine, see
Table 2). The increasing of alkyl chain length leads to the increasing of logP value by QC method (adding two carbon atoms to the chain increase logP by the value near 1 log units).
For bromine derivatives (
Figure 3a) with C12 and C14 alkyl chains, GraphormerLogP slightly overestimated lipophilicity, while for C16 compounds it showed the best agreement with QC values. Predictions for C18 compounds approached the upper limit of the model’s training range (logP = 9.98), causing mild saturation and downward deviation relative to QC values. For nitro derivatives (
Figure 3b) the best agreement between two methods (QC and GLP) is observed for C18 alkyl chain.
In contrast to this, the fragment-based model (RDK) consistently underestimates logP for all homologs, with MAE = 1.049, 1.111, 1.467, and 1.699 (mean values for bromine, chlorine and nitro derivatives together) for C12–C18, respectively.
It should be mentioned, that the three approaches differed markedly in computational efficiency. The quantum-chemical logP estimation required hundreds of hours for all 60 molecules, while GraphormerLogP completed predictions in several tens of seconds, and the Wildman–Crippen (RDKit) method finished in a few seconds, including environment initialization.
4. Discussion
Synthesized azomethines can be considered as perspective antibacterial agents due to their activity (see
Table 1) and relatively simple synthesis (see experimental details in ESI1). However, very poor solubility in water and relatively low solubility in ethanol reduce these prospects. Despite this azomethines continue to be a good platform for the search for new pharmacological drugs, because changing substituents opens easy way to vary the properties of compounds.
Combination of three substituents (bromine, chlorine and nitro-group) at three positions (ortho-, meta- and para-) with four alkyl chains (C12, C14, C16 and C18) gives 36 possible compounds for the described platform with phenyl ring. Experimental synthesis and properties investigation of all substances “by hand” even at this relatively simple case looks impossible (if the developing field of the laboratory robotic systems and self-driving labs is not taken into account), so the initial “filtration” before real experiments is needed.
One of the important properties for the biological activity predictions is lipophilicity. Experimental measurements are very difficult for substances with very low solubility in water, so computational approaches come to the fore. In the present work three different methods were used: quantum-chemical calculations (QC), machine learning (GLP) and fragment-based approach (RDK).
On the base of quantum-chemical calculations logP can be determined for any structure, which corresponds to local (or global) minima, so the influence of conformational change, isomerization and tautomerization can be taken into account. However, this method is very time-consuming. Fragment based approach is very fast, but it is insensitive for the isomers with the same fragments (as in the investigated case with ortho-, meta- and para-isomers) and can give quite different results (underestimated for the studied azomethines). Machine-learning approach, such as used GraphormerLogP, is also fast, but is sensitive for isomers and can give better results than RDK.
Additionally, the initial 3d-structures are necessary for quantum-chemical calculations and generation of it is also the time-consuming step, whereas machine learning and fragment-based approaches need just text representation of molecule (for example, SMILES), generation of which can be easily automated for the similar compounds.
Thus, while quantum-chemical methods remain the reference standard for precision, deep learning and fragment-based approaches provide several orders of magnitude faster alternatives suitable for large-scale screening. Overall, GraphormerLogP predictions can be considered reliable within the explored lipophilicity range, offering a meaningful balance between accuracy and computational efficiency, with quantitative agreement to the quantum-chemical estimates.
5. Conclusions
Synthesized azometines represent promising antibacterial agents due to their pronounced activity and relatively simple synthesis. However, their potential is limited by their extremely low aqueous solubility. Despite this, this class of compounds remains a convenient platform for discovering new pharmacological substances, as varying substituents allows for targeted modification of the molecule's properties. Due to the need for preliminary virtual screening to select target structures from a multitude of possible derivatives, assessing lipophilicity (logP), which is difficult to measure experimentally, is crucial. Three computational approaches are compared: quantum chemical calculation (QC), machine learning (GLP), and fragment-based description (RDK). Despite the benchmark QC accuracy, which requires significant computational resources and three-dimensional structures, machine learning-based methods (GraphormerLogP) demonstrate acceptable accuracy, sensitivity to isomerism, and orders of magnitude higher throughput, making them an optimal tool for high-throughput screening.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figures S1-1 – S1-13: NMR spectra; Figures S1-14 – S1-25: IR spectra; Figures S2-1 – S2-120: optimized structures of all compounds in water and octanol; Tables S2-1 – S2-120: cartesian coordinates for optimized structures; Tables S2-121 – S2-126: Gibbs free energies for optimized structures.
Author Contributions
Conceptualization, N.Yu.S. and Kh.R.Kh.; methodology, I.V.G. and T.R.G.; software, V.A.G. and T.R.G.; formal analysis, N.Yu.S., Kh.R.Kh., I.V.G. and T.R.G.; investigation, Kh.R.Kh., M.P.Sh. and V.A.G.; resources, N.Yu.S., I.V.G. and T.R.G.; data curation, Kh.R.Kh., M.P.Sh. and V.A.G.; writing—original draft preparation, N.Yu.S. and Kh.R.Kh.; writing—review and editing, N.Yu.S., Kh.R.Kh., V.A.G. and T.R.G.; visualization, N.Yu.S. and Kh.R.Kh.; supervision, N.Yu.S., I.V.G. and T.R.G.; project administration, N.Yu.S. and T.R.G.; funding acquisition, N.Yu.S. All authors have read and agreed to the published version of the manuscript.
Funding
The work was funded by financial support from the government assignment for FRC Kazan Scientific Center of RAS.
Data Availability Statement
The synthesis details, experimental spectra, optimized structures and calculated Gibbs energies are included in the supplementary materials.
Acknowledgments
The authors express sincere appreciation to Kazan Scientific Center of Russian Academy of Science for providing the research infrastructure and intellectual environment that enabled the quantum-chemical calculations and machine learning.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| AI |
artificial intelligence |
| CFU |
colony forming units |
| GLP |
GraphormerLogP model |
| FT-IR |
fourier transform infrared spectroscopy |
| MAE |
mean absolute error |
| ML |
machine learning |
| RDK |
RDKit library |
| RMSE |
root mean squared error |
| SMD |
solvation model based on density |
| SMILES |
simplified molecular input line entry system |
| QC |
quantum chemistry |
| QSAR |
quantitative structure-activity relationships |
References
- Hameed, A.; al-Rashida, M.; Uroos, M.; Abid Ali, S.; Khan, K.M. Schiff Bases in Medicinal Chemistry: A Patent Review (2010-2015). Expert Opin. Ther. Pat. 2017, 27, 63–79. [Google Scholar] [CrossRef]
- Kajal, A.; Bala, S.; Kamboj, S.; Sharma, N.; Saini, V. Schiff Bases: A Versatile Pharmacophore. J. Catal. 2013, 2013, 893512. [Google Scholar] [CrossRef]
- Ceramella, J.; Iacopetta, D.; Catalano, A.; Cirillo, F.; Lappano, R.; Sinicropi, M.S. A Review on the Antimicrobial Activity of Schiff Bases: Data Collection and Recent Studies. Antibiotics 2022, 11, 191. [Google Scholar] [CrossRef]
- Pencheva, I.; Peikova, L.; Tsvetkova, D. Pharmacological Applications of Azomethine Derivatives in the Therapy of Different Diseases. Pharmacia 2025, 72, 1–17. [Google Scholar] [CrossRef]
- Shekhar, S.; Khan, A.M.; Sharma, S.; Sharma, B.; Sarkar, A. Schiff Base Metallodrugs in Antimicrobial and Anticancer Chemotherapy Applications: A Comprehensive Review. Emergent Mater. 2022, 5, 279–293. [Google Scholar] [CrossRef]
- Pervaiz, M.; Sadiq, A.; Sadiq, S.; Saeed, Z.; Imran, M.; Younas, U.; Majid Bukhari, S.; Rashad Mahmood Khan, R.; Rashid, A.; Adnan, A. Design and Synthesis of Schiff Base Homobimetallic-Complexes as Promising Antimicrobial Agents. Inorg. Chem. Commun. 2022, 137, 109206. [Google Scholar] [CrossRef]
- Shanty, A.A.; Philip, J.E.; Sneha, E.J.; Prathapachandra Kurup, M.R.; Balachandran, S.; Mohanan, P.V. Synthesis, Characterization and Biological Studies of Schiff Bases Derived from Heterocyclic Moiety. Bioorganic Chem. 2017, 70, 67–73. [Google Scholar] [CrossRef]
- Iqbal, A.; Siddiqui, H.L.; Ashraf, C.M.; Ahmad, M.; Weaver, G.W. Synthesis, Characterization and AntibacterialActivity of Azomethine Derivatives Derived from 2-Formylphenoxyacetic Acid. Molecules 2007, 12, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.A.; Prabhakara, C.T.; Halasangi, B.M.; Toragalmath, S.S.; Badami, P.S. DNA Cleavage, Antibacterial, Antifungal and Anthelmintic Studies of Co(II), Ni(II) and Cu(II) Complexes of Coumarin Schiff Bases: Synthesis and Spectral Approach. Spectrochim. Acta. A. Mol. Biomol. Spectrosc. 2015, 137, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Sztanke, K.; Maziarka, A.; Osinka, A.; Sztanke, M. An Insight into Synthetic Schiff Bases Revealing Antiproliferative Activities in Vitro. Bioorg. Med. Chem. 2013, 21, 3648–3666. [Google Scholar] [CrossRef]
- Zubair, M.; Sirajuddin, M.; Ullah, K.; Haider, A.; Perveen, F.; Hussain, I.; Ali, S.; Tahir, M.N. Synthesis, Structural Peculiarities, Theoretical Study and Biological Evaluation of Newly Designed O-Vanillin Based Azomethines. J. Mol. Struct. 2020, 1205, 127574. [Google Scholar] [CrossRef]
- Chinnasamy, R.; Sundararajan, R.; Govindaraj, S. Synthesis, Characterization, and Analgesic Activity of Novel Schiff Base of Isatin Derivatives. J. Adv. Pharm. Technol. Res. 2010, 1, 342. [Google Scholar] [CrossRef]
- Bae, S.J.; Ha, Y.M.; Park, Y.J.; Park, J.Y.; Song, Y.M.; Ha, T.K.; Chun, P.; Moon, H.R.; Chung, H.Y. Design, Synthesis, and Evaluation of (E)-N-Substituted Benzylidene–Aniline Derivatives as Tyrosinase Inhibitors. Eur. J. Med. Chem. 2012, 57, 383–390. [Google Scholar] [CrossRef]
- Negm, N.A.; Zaki, M.F.; Salem, M.A.I. Cationic Schiff Base Amphiphiles and Their Metal Complexes: Surface and Biocidal Activities against Bacteria and Fungi. Colloids Surf. B Biointerfaces 2010, 77, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Sprung, M.A. A Summary of the Reactions of Aldehydes with Amines. Chem. Rev. 1940, 26, 297–338. [Google Scholar] [CrossRef]
- Sanchez-Lengeling, B.; Aspuru-Guzik, A. Inverse Molecular Design Using Machine Learning: Generative Models for Matter Engineering. Science 2018, 361, 360–365. [Google Scholar] [CrossRef]
- Sun, X.; Bai, Y.; Liu, Y.; Chen, B.; Jia, Y.; Zeng, Z. Synthesis and Biological Activity of 4,5-Dihydro-1,2,4-Triazole-5-Thione Schiff Base. Chin. J. Chem. 2009, 27, 1397–1400. [Google Scholar] [CrossRef]
- Geronikaki, A.V. Paola; Incerti, Matteo; Hadjipavlou-Litina, Dimitra Thiazolyl and Isothiazolyl Azomethine Derivatives with Anti-Inflammatory and Antioxidant Activities. Arzneimittelforschung 2004, 54, 530–537. [Google Scholar] [CrossRef]
- Chen, L.; Wu, Z.; Du, Y.; Huang, Y.; Jin, S. Solvothermal Synthesis of Novel Phenylpyrazole Schiff Base Fluorescent Insecticides Fused Extended Conjugate Units for Enhancing Bioactivities, Photophysical and Electrochemical Properties. J. Mol. Struct. 2019, 1196, 555–566. [Google Scholar] [CrossRef]
- Muratov, E.N.; Bajorath, J.; Sheridan, R.P.; Tetko, I.V.; Filimonov, D.; Poroikov, V.; Oprea, T.I.; Baskin, I.I.; Varnek, A.; Roitberg, A.; et al. QSAR without Borders. Chem. Soc. Rev. 2020, 49, 3525–3564. [Google Scholar] [CrossRef]
- Waring, M.J. Lipophilicity in Drug Discovery. Expert Opin. Drug Discov. 2010, 5, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Arnott, J.A.; Planey, S.L. The Influence of Lipophilicity in Drug Discovery and Design. Expert Opin. Drug Discov. 2012, 7, 863–875. [Google Scholar] [CrossRef] [PubMed]
- Tsopelas, F.; Giaginis, C.; Tsantili-Kakoulidou, A. Lipophilicity and Biomimetic Properties to Support Drug Discovery. Expert Opin. Drug Discov. 2017, 12, 885–896. [Google Scholar] [CrossRef]
- Ma, Y.; Chen, X.; Javeria, H.; Du, Z. High-Throughput Screening of LogD by Using a Sample Pooling Approach Based on the Traditional Shake Flask Method. J. Chromatogr. B 2023, 1227, 123804. [Google Scholar] [CrossRef] [PubMed]
- De Bruijn, J.; Busser, F.; Seinen, W.; Hermens, J. Determination of Octanol/Water Partition Coefficients for Hydrophobic Organic Chemicals with the “Slow-Stirring” Method. Environ. Toxicol. Chem. 1989, 8, 499–512. [Google Scholar] [CrossRef]
- Woodburn, K.B.; Doucette, W.J.; Andren, A.W. Generator Column Determination of Octanol/Water Partition Coefficients for Selected Polychlorinated Biphenyl Congeners. Environ. Sci. Technol. 1984, 18, 457–459. [Google Scholar] [CrossRef]
- Terada, H. Determination of Log Poct by High-Performance Liquid Chromatography, and Its Application in the Study of Quantitative Structure-Activity Relationships. Quant. Struct.-Act. Relatsh. 1986, 5, 81–88. [Google Scholar] [CrossRef]
- Win, Z.-M.; Cheong, A.M.Y.; Hopkins, W.S. Using Machine Learning To Predict Partition Coefficient (Log P) and Distribution Coefficient (Log D) with Molecular Descriptors and Liquid Chromatography Retention Time. J. Chem. Inf. Model. 2023, 63, 1906–1913. [Google Scholar] [CrossRef]
- Roy, D.; Patel, C. Revisiting the Use of Quantum Chemical Calculations in LogPoctanol-Water Prediction. Molecules 2023, 28, 801. [Google Scholar] [CrossRef]
- Neese, F. The ORCA Program System. WIREs Comput. Mol. Sci. 2012, 2, 73–78. [Google Scholar] [CrossRef]
- Neese, F. Software Update: The ORCA Program System—Version 6.0. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2025, 15, e70019. [Google Scholar] [CrossRef]
- Grimme, S.; Hansen, A.; Ehlert, S.; Mewes, J.-M. r2SCAN-3c: A “Swiss Army Knife” Composite Electronic-Structure Method. J. Chem. Phys. 2021, 154, 064103. [Google Scholar] [CrossRef]
- Bursch, M.; Mewes, J.-M.; Hansen, A.; Grimme, S. Best-Practice DFT Protocols for Basic Molecular Computational Chemistry**. Angew. Chem. Int. Ed. 2022, 61, e202205735. [Google Scholar] [CrossRef]
- Marenich, A.V.; Cramer, C.J.; Truhlar, D.G. Universal Solvation Model Based on Solute Electron Density and on a Continuum Model of the Solvent Defined by the Bulk Dielectric Constant and Atomic Surface Tensions. J. Phys. Chem. B 2009, 113, 6378–6396. [Google Scholar] [CrossRef] [PubMed]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An Advanced Semantic Chemical Editor, Visualization, and Analysis Platform. J. Cheminformatics 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Chemcraft - Graphical Software for Visualization of Quantum Chemistry Computations. Version 1.8, Build 780. Https. Available online: Www.Chemcraftprog.Com.
- Grigorev, V.; Serov, N.; Gimadiev, T.; Poyezzhayeva, A.; Sidorov, P. Graph-based transformer to predict the octanol-water partition coefficient. J. Chemoinformatics. In press.
- Nugmanov, R.; Dyubankova, N.; Gedich, A.; Wegner, J.K. Bidirectional Graphormer for Reactivity Understanding: Neural Network Trained to Reaction Atom-to-Atom Mapping Task. J. Chem. Inf. Model. 2022, 62, 3307–3315. [Google Scholar] [CrossRef]
- Fallani, A.; Nugmanov, R.; Arjona-Medina, J.; Wegner, J.K.; Tkatchenko, A.; Chernichenko, K. Pretraining Graph Transformers with Atom-in-a-Molecule Quantum Properties for Improved ADMET Modeling. J. Cheminformatics 2025, 17, 25. [Google Scholar] [CrossRef] [PubMed]
- Wildman, S.A.; Crippen, G.M. Prediction of Physicochemical Parameters by Atomic Contributions. J. Chem. Inf. Comput. Sci. 1999, 39, 868–873. [Google Scholar] [CrossRef]
- RDKit. Available online: http://www.rdkit.org/ (accessed on 14 October 2025).
- Galkina, I.V.; Kiyamova, E.R.; Gaynullin, A.Z.; Bakhtiyarov, D.I.; Shulaeva, M.P.; Pozdeev, O.K.; Egorova, S.N.; Ivshin, K.A.; Kataeva, O.N.; Bakhtiyarova, Y.V.; et al. Synthesis and Structure of Novel Phosphorylated Azomethines. Phosphorus Sulfur Silicon Relat. Elem. 2016, 191, 1679–1681. [Google Scholar] [CrossRef]
- Salih, K.S.M. Solvent Influence on Absorption Spectra and Tautomeric Equilibria of Symmetric Azomethine-Functionalized Derivatives: Structural Elucidation and Computational Studies. ChemistryOpen 2022, 11, e202100237. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).