1. Introduction
Rotator cuff tears (RCTs) represent one of the most prevalent and disabling musculoskeletal disorders, particularly in aging populations. Epidemiological studies indicate that the prevalence of RCTs increases significantly with age, affecting up to 50% of individuals over the age of 60 [
1,
2,
3]. Among the rotator cuff tendons, the supraspinatus is the most frequently injured due to its unique anatomical position beneath the coracoacromial arch and its susceptibility to both intrinsic degeneration and extrinsic impingement [
4,
5,
6]. Clinically, accurate differentiation among partial-thickness tears, full-thickness tears, and tendinosis is critical, as it dictates the therapeutic pathway—ranging from conservative management and physical therapy to surgical repair [
3,
7,
8]. Furthermore, associated features such as tendon retraction and fatty infiltration of the muscle belly are key prognostic factors for repairability, postoperative healing and functional outcomes [
1,
9,
10].
Magnetic Resonance Imaging (MRI) is currently the non-invasive gold standard for diagnosing RCTs, offering superior soft-tissue contrast for evaluating tendon integrity and muscle quality [
8,
11,
12,
13]. However, shoulder MRI interpretation is complex and subject to significant interobserver variability, particularly when distinguishing high-grade partial tears from small full-thickness tears [
2,
11,
14,
15]. While magnetic resonance arthrography (MRA) can increase diagnostic sensitivity, nevertheless it is invasive and not routinely performed for all patients [
8,
16]. Consequently, there is growing clinical demand for automated, objective, and reproducible diagnostic support tools to assist radiologists in accurately grading the tendon pathology [
8,
11,
13,
15].
In recent years, Artificial Intelligence (AI), particularly deep learning (DL), has revolutionized orthopedic imaging [
15,
17,
18]. Early computational approaches relied on machine-learning classifiers (e.g., Support Vector Machines) using handcrafted texture features such as Gray-Level Co-occurrence Matrix (GLCM) or Local Binary Pattern (LBP) [
11,
19]. However, these methods often lacked generalizability across different MRI scanners. The advent of Convolutional Neural Networks (CNNs) marked a paradigm shift, achieving expert-level performance in tear detection [
2,
4,
15]. Most existing studies have employed 2D CNN architectures (e.g., ResNet, VGG, Xception) that analyze MRI data on a slice-by-slice basis [
13,
19,
20,
21,
22]. While effective, these 2D slice-centric models have a fundamental limitation: they often fail to capture the 3D volumetric context and the continuity of the tendon across adjacent slices [
22,
23]. Although some studies have explored 3D CNNs to address this limitation, such models are computationally expensive and often struggle with the “black-box” nature of deep learning, offering limited explainability [
1,
2,
22].
A current debate in the field concerns whether assessing the shoulder as a sequence of images is superior to analyzing 3D volumes directly or treating 2D slices independently [
2,
22]. This has motivated the exploration of Vision Transformers (ViTs), a novel architecture that uses self-attention mechanisms to model long-range dependencies [
4,
11]. Unlike CNNs, which are constrained by local receptive fields, Transformers can attend to global anatomical relationships, more closely reflecting how radiologists scroll through MRI stacks to form a patient-level diagnosis [
4,
22]. Recent hybrid models combining CNNs for feature extraction and Transformers for sequence modeling have shown promise in other medical imaging domains but remain underexplored for rotator cuff grading [
4].
The primary aim of this study is to develop and validate a patient-level, explainable Vision Transformer framework for automated classification of supraspinatus tendon pathology (Normal, Partial-Thickness Tear, Full-Thickness Tear). We hypothesize that a Transformer-based architecture that treats the MRI stack as a coherent anatomical sequence will outperform traditional slice-based CNNs in diagnostic accuracy. Furthermore, by leveraging the attention mechanism inherent to Transformers, we aim to generate high-resolution, slice-specific attention maps. These visualizations help bridge the explainability gap by enabling clinicians to verify that the model focuses on relevant anatomical structures—such as the tendon footprint—rather than imaging artifacts. Overall, this study emphasizes clinical integration by moving beyond binary detection to provide a graded, explainable, and patient-centric diagnosis.
2. Materials and Methods
The study was conducted in accordance with the Declaration of Helsinki. The research protocol, entitled “Multimodal Diagnosis and Virtual Assistant Use in Magnetic Resonance Imaging for Rotator Cuff Tear Diagnosis,” was reviewed and approved by the Non-Interventional Clinical Research Ethics Committee of Bilecik Şeyh Edebali University (Approval Date: October 30, 2025; Meeting Number: 2025/10; Decision Number: 9).
In this section, we comprehensively delineate the study population, the proposed computational framework—the Patient-Aware Vision Transformer (Pa-ViT)—and the experimental protocols designed for automated, patient-level diagnosis of rotator cuff tears (RCTs) using volumetric magnetic resonance imaging (MRI). The methodology is organized into four principal components: dataset characteristics, mathematical problem formulation, architectural innovations, and the optimization landscape.
2.1. Dataset and Study Population
To ensure the clinical relevance and robustness of our evaluation, we utilized a large-scale retrospective dataset comprising 2,447 shoulder MRI examinations, as introduced by Kim et al. [
24]. The dataset consists exclusively of T2-weighted images, with a particular emphasis on coronal slices, which are widely regarded as the gold standard for evaluating the integrity of the rotator cuff tendons due to the high contrast between hypointense (dark) tendon fibers and hyperintense (bright) fluid signals associated with tears.
Imaging studies were included if they contained complete T2-weighted sequences acquired in oblique coronal, sagittal, and axial planes, which together constitute the standard shoulder MRI protocol for comprehensive rotator cuff assessment. Examinations that did not meet ideal shoulder MRI acquisition criteria—including incomplete imaging planes, missing T2-weighted series, severe motion artifacts, or technically inadequate image quality—were excluded from the study.
2.1.1. Data Characteristics and Class Distribution
As summarized in
Table 1, the dataset exhibits a pronounced long-tail distribution that closely reflects real-world clinical prevalence. Normal examinations constitute the majority of cases (66.51%), followed by full-thickness tears (27.06%), while partial-thickness tears represent a smaller but clinically significant minority (6.43%).
2.1.2. Data Partitioning Strategy
To ensure a robust and unbiased evaluation, a strict patient-level split was employed, allocating 1,959 examinations for training and 488 for validation/testing. This strategy ensures that imaging slices from the same patient do not appear in both the training and evaluation sets, thereby preventing data leakage and enabling a reliable assessment of the model’s generalization capability.
2.2. Data Preprocessing and Augmentation
A standard clinical MRI examination of the shoulder consists of a sequence of T2-weighted coronal slices that spatially traverse the glenohumeral joint from the anterior to the posterior aspect. Within this volumetric stack, the number of slices () varies stochastically across the cohort depending on the patient’s physical dimensions and the specific acquisition protocols of the MRI scanner.
To standardize the input for the deep learning model, the spatial dimensions ( and ) of each slice were normalized to pixels via bicubic interpolation. The channel dimension was constructed by replicating the grayscale intensity to accommodate the pre-trained weights of the encoder architecture.
To improve the generalization capability of the model and prevent overfitting given the class imbalance, we devised a comprehensive data augmentation pipeline applied strictly during the training phase using the torchvision library. The specific transformations included:
Geometric Transformations: Random rotations (), random affine transformations (scaling range 0.9–1.1, translation 10%), and random horizontal flips (
).
Photometric Augmentations: Color Jittering (brightness and contrast adjusted by a factor of 0.1) was employed to simulate the variability in signal-to-noise ratios observed across different MRI scanner manufacturers.
Normalization: All images were normalized using the standard ImageNet mean ([0.485, 0.456, 0.406]) and standard deviation ([0.229, 0.224, 0.225]).
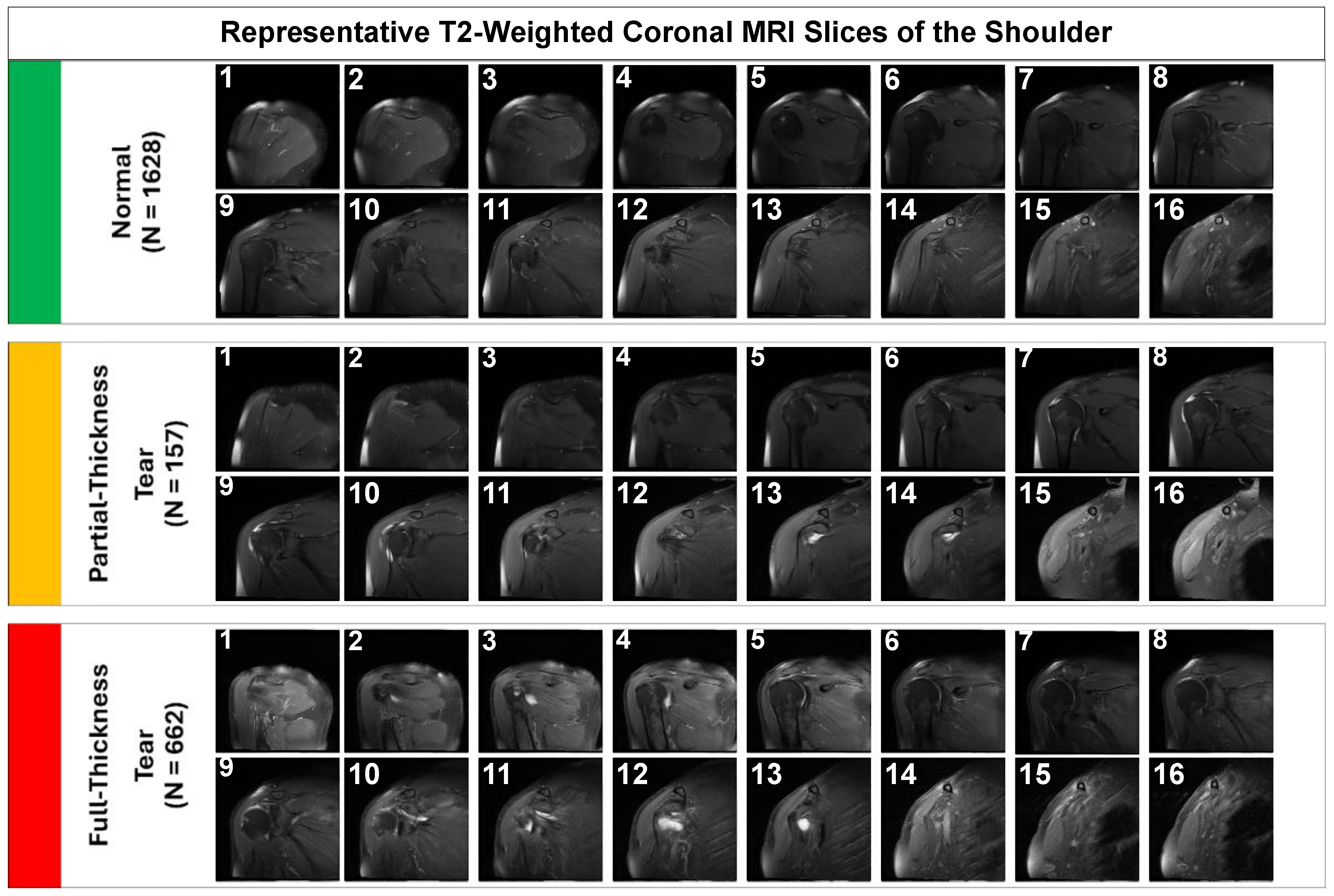
Representative samples illustrating the visual complexity of the diagnostic task are provided in
Figure 1. Note the subtle focal hyperintensity of the partial-thickness tear compared to the gross disruption seen in the full-thickness tear.
2.3. Problem Formulation: Volumetric Multiple Instance Learning
The diagnosis of RCTs presents a unique set of challenges distinct from standard computer vision tasks: the input is a variable-length sequence of 2D slices rather than a static image; the pathological signal is often subtle and localized to a specific anatomical sub-region (the supraspinatus footprint); and the remaining slices may display healthy anatomy or irrelevant structures. Traditional Deep Learning approaches that rely on slice-level classification treat each image as an independent sample, a formulation that erroneously discards the critical inter-slice context and necessitates labor-intensive slice-level annotations.
To address these challenges, we rigorously formulate the diagnostic task as a Weakly-Supervised Multiple Instance Learning (MIL) problem. Let
represent the high-dimensional input space of MRI examinations. We define a single patient sample
not as a static tensor, but as a “bag” of instances composed of a variable sequence of
slices:
The objective of our framework is to learn a non-linear mapping function that predicts the patient-level diagnostic label , essentially treating the patient label as a “weak label” for the entire bag. This formulation forces the algorithm to implicitly learn a relevance function for each instance within the bag, aggregating local evidences to form a global prediction.
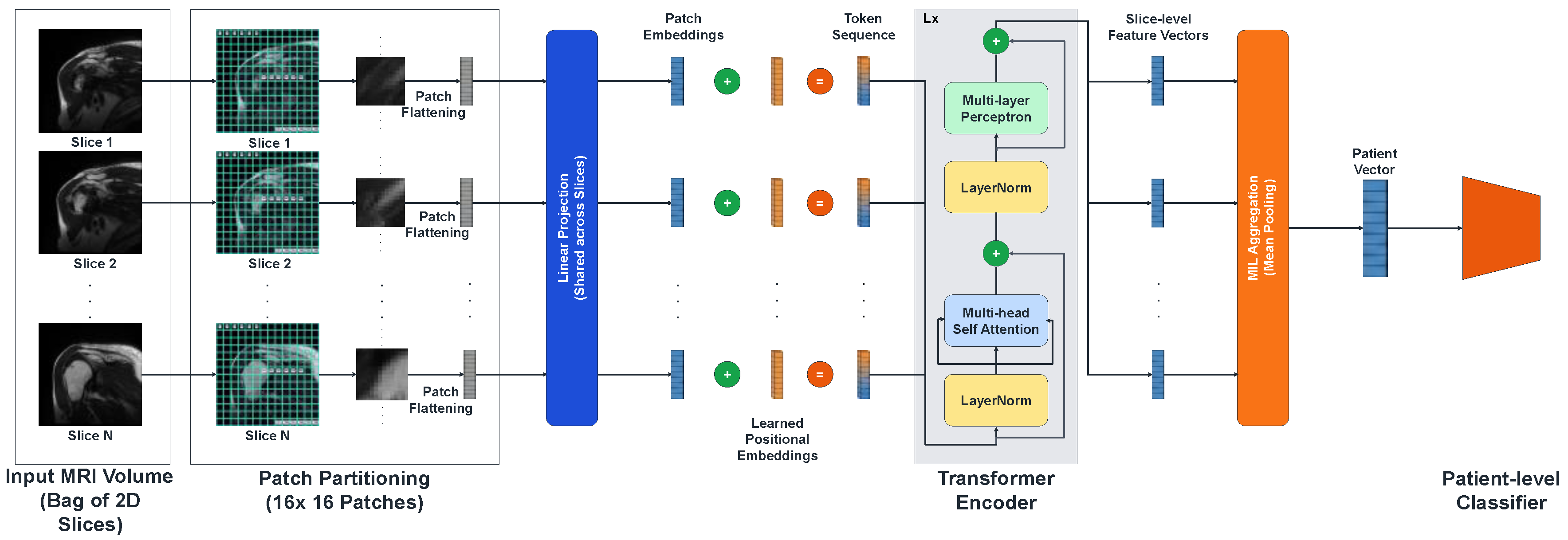
2.4. Proposed Method: The Patient-Aware Vision Transformer (Pa-ViT) Architecture
Here, the term “patient-aware” refers to a patient-level learning paradigm in which information from all MRI slices belonging to a single patient is aggregated to generate a unified diagnosis; no demographic or clinical metadata were used. While Convolutional Neural Networks (CNNs) have been explored for RCT diagnosis, they process images via local convolution operations, inherently limiting their effective receptive field. However, the radiological signs of a rotator cuff tear often involve subtle semantic relationships between spatially distant anatomical structures. To capture these long-range global dependencies, we propose the Patient-Aware Vision Transformer (Pa-ViT) framework.
Figure 2.
The proposed Patient-Aware Vision Transformer (Pa-ViT) architecture.
Figure 2.
The proposed Patient-Aware Vision Transformer (Pa-ViT) architecture.
The pipeline begins with the ingestion of a volumetric MRI sequence treated as a bag of 2D slices. Each slice is independently tokenized into patches and processed by a shared Vision Transformer encoder to extract high-dimensional semantic embeddings. These embeddings are subsequently aggregated via a permutation-invariant mean pooling mechanism to form a global patient descriptor.
2.4.1. Slice Encoding and Self-Attention Mechanism
The core feature extraction engine of our framework is a Vision Transformer (ViT-Base) encoder, implemented via the timm library (vit_base_patch16_224). The process begins with Patch Partitioning, where each 2D slice is tessellated into a grid of non-overlapping patches of resolution , where . This operation effectively flattens the 2D image into a sequence of vectors, which are then linearly projected into a latent embedding dimension . Crucially, to allow the model to learn anatomical context—distinguishing the superior aspect of the tendon from the inferior glenoid labrum—we utilize pre-trained 1D positional embeddings.
The sequence of patch embeddings, denoted as
, is processed through a stack
Transformer layers. The critical computational component within these layers is the Multi-Head Self-Attention (MSA) mechanism. For a given attention head, the input sequence is projected into Query (
), Key (
), and Value (
) matrices. The attention weights are computed via the scaled dot-product interaction:
Where is a scaling factor dependent on the embedding dimension ( per head) to ensure numerical stability during gradient backpropagation. This global attention mechanism enables the model to dynamically focus on hyperintense regions indicative of fluid and tears while simultaneously suppressing irrelevant background noise from the deltoid muscle or subcutaneous fat, resulting in a robust slice-level feature vector .
2.4.2. Permutation-Invariant Patient Aggregation
Following the extraction of slice-level embeddings
, the MIL framework necessitates a differentiable aggregation strategy to synthesize a unified patient-level representation
. We employ a Global Mean Pooling strategy. This choice is theoretically grounded in the requirement for permutation invariance; the diagnostic output of the system should not be dependent on the specific ordering of slices. The aggregation operation is mathematically defined as:
Unlike Max Pooling, which extracts the maximum activation across the volume and risks latching onto a single slice containing a noisy artifact, Mean Pooling aggregates information across the entire volume. This aligns with the radiological reality that true tears are volumetric defects spanning multiple contiguous slices.
2.4.3. Classification Head and Regularization
The aggregated patient vector serves as the input to the final classification head. Given the high dimensionality of the feature space relative to the number of unique patients, overfitting is a significant concern. To address this, we designed a custom Multi-Layer Perceptron (MLP) head that incorporates extensive regularization:
Layer Normalization: Stabilizes the input distribution.
Linear Projection 1: Projects to a hidden dimension of 512.
GELU Activation: Utilizing the Gaussian Error Linear Unit for probabilistic non-linearity.
Dropout (p=0.5): Prevents the co-adaptation of neurons.
Linear Projection 2: Projects to a dimension of 256, followed by GELU and a second Dropout (
Final Projection: Maps features to the
2.5. Optimization Landscape and Implementation Details
This section delineates the technical configuration and optimization strategies employed to ensure the robust performance and reproducibility of the proposed framework.
2.5.1. Loss Function and Class Imbalance Mitigation
A pervasive challenge in medical image analysis is the inherent class imbalance. To ameliorate this, we implemented a Weighted Cross-Entropy Loss combined with Label Smoothing. The class weights
are strictly calculated using the inverse square root of the class frequency to provide a balanced penalty:
The final loss function
incorporates these weights along with label smoothing (
). Label smoothing replaces the “hard” one-hot target vectors with “soft” targets, preventing the model from becoming over-confident:
2.5.2. Training Protocol and Evaluation Metrics
For optimization, we utilized the AdamW algorithm to decouple weight decay () from gradient updates. We employed a Cosine Annealing learning rate scheduler and a differential learning rate strategy: the pre-trained backbone was fine-tuned with a conservative learning rate of , while the classification head was trained at . To efficiently manage GPU memory with volumetric 3D data, we utilized Automatic Mixed Precision (AMP) via torch.amp.GradScaler and implemented Gradient Accumulation (accumulation steps = 16) to simulate an effective batch size of 16 patients, ensuring stable optimization and convergence.
To comprehensively evaluate the model’s performance, we employed standard classification metrics, including Accuracy, F1-Score (Macro-averaged), Sensitivity, and Specificity. Furthermore, confusion matrices were generated to analyze class-wise performance, particularly for the challenging partial-thickness tear class.
2.5.3. Software, Hardware, and Reproducibility
The entire computational framework was implemented using Python (version 3.8) and the PyTorch deep learning library (version 2.1.0). We leveraged the PyTorch Image Models (timm) library for the Vision Transformer backbone and scikit-learn for metric evaluation. All experiments were conducted on a Google Colab L4 GPU environment with 24GB of VRAM. A fixed random seed (seed=42) was set for all random number generators (numpy, torch, python) to ensure the strict reproducibility of the results.
3. Results
In this section, we present a comprehensive experimental evaluation of the proposed Pa-ViT framework. The evaluation protocol assesses the model’s performance from three distinct perspectives: quantitative benchmarking against established deep learning architectures and traditional machine learning classifiers, granular error analysis to evaluate diagnostic capabilities on clinically challenging classes, and visual validation through Explainable AI (XAI) to ensure the model’s decision-making process aligns with radiological pathology.
3.1. Quantitative Benchmarking and Comparative Analysis
To establish the diagnostic efficacy of the Pa-ViT model, we conducted a rigorous comparative analysis. The proposed model was benchmarked against a diverse suite of traditional machine learning classifiers—including Random Forest, XGBoost, and Support Vector Machines—as well as a strong prior deep learning baseline for this domain proposed by Kim et al. [
24], which utilizes a VGG-16 backbone with Weighted Linear Combination.
The quantitative results, detailed in
Table 2, demonstrate that our Vision Transformer-based approach demonstrates improved performance on this dataset. While traditional machine learning methods struggled to capture the complex features of MRI data, generally yielding accuracies between 52% and 74%, deep learning approaches showed significantly superior performance.
As presented in
Table 2, the Pa-ViT model achieved an Overall Accuracy of 91%, representing a notable improvement over the 87% accuracy reported by the CNN baseline. More importantly, the model achieved a macro-averaged F1-Score of 0.91. This metric is particularly salient in medical imaging, as it confirms that the high accuracy is not merely an artifact of class imbalance (i.e., predicting the majority “Normal” class). The balanced Precision (0.92) and Recall (0.91) indicate that the model effectively minimizes both False Positives (over-diagnosis) and False Negatives (missed diagnosis). This balance is critical for a clinical screening tool, ensuring that healthy patients are not subjected to unnecessary procedures while ensuring pathological cases are not overlooked.
3.2. Diagnostic Accuracy and Error Analysis
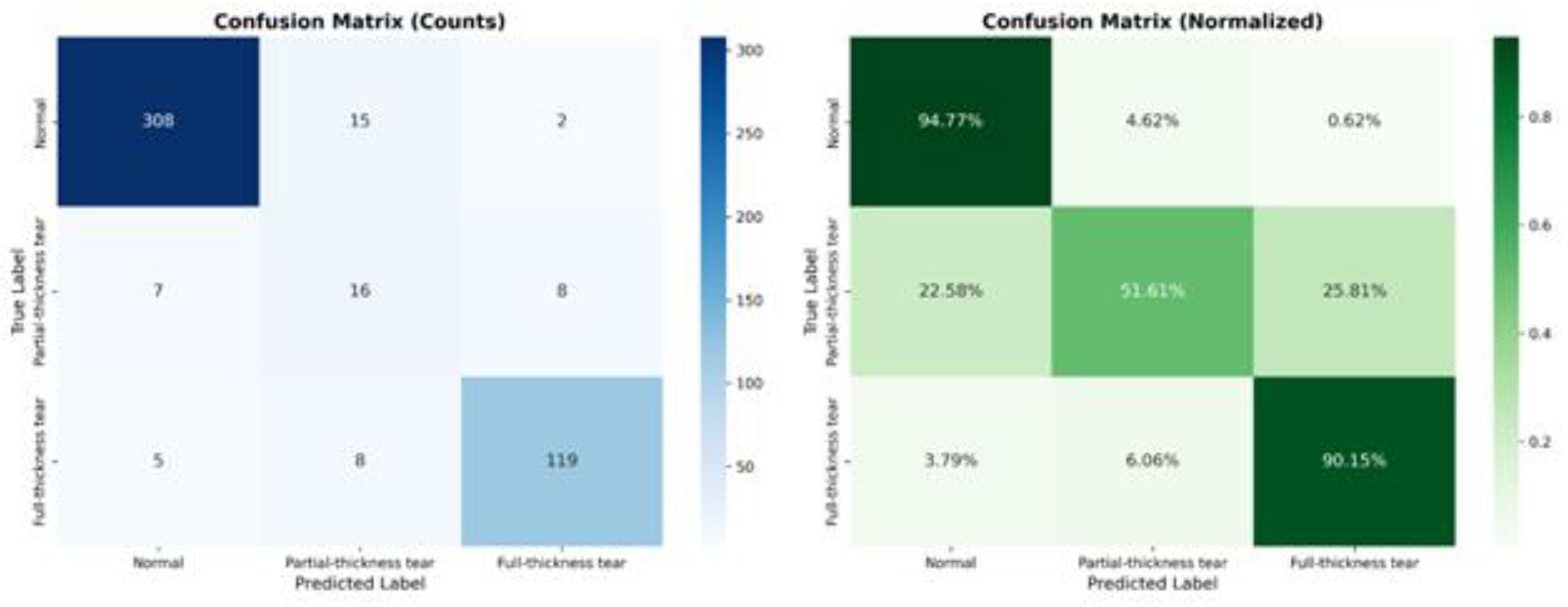
Beyond global metrics, it is crucial to understand the model’s behavior at a granular level, particularly for classes that are historically difficult to diagnose. To provide insight into the model’s decision-making process, specifically regarding the “Partial-Thickness Tear” class, we examined the Confusion Matrix (
Figure 3). This visualization allows us to pinpoint exactly where the model succeeds and where it struggles relative to the ground truth labels.
As illustrated in
Figure 3, the model exhibits exceptional specificity for “Normal” examinations, correctly classifying 94.77% of healthy patients. Similarly, for “Full-Thickness Tears,” the sensitivity is remarkably high at 90.15%, indicating the model’s robustness in detecting severe pathology. The most critical insight is derived from the “Partial-Thickness Tear” class. Previous works have struggled significantly with this class due to its subtle morphological features, often reporting accuracies as low as 38%. Our model advances this frontier, correctly identifying 51.61% of partial tears. While misclassifications exist, they are clinically interpretable: 25.81% of partial tears were classified as “Full-Thickness Tears,” likely representing high-grade partial tears that are morphologically similar to full tears. Conversely, only 22.58% were missed as “Normal,” suggesting that the model is sensitive to the presence of pathology even if the severity grading is occasionally overestimated.
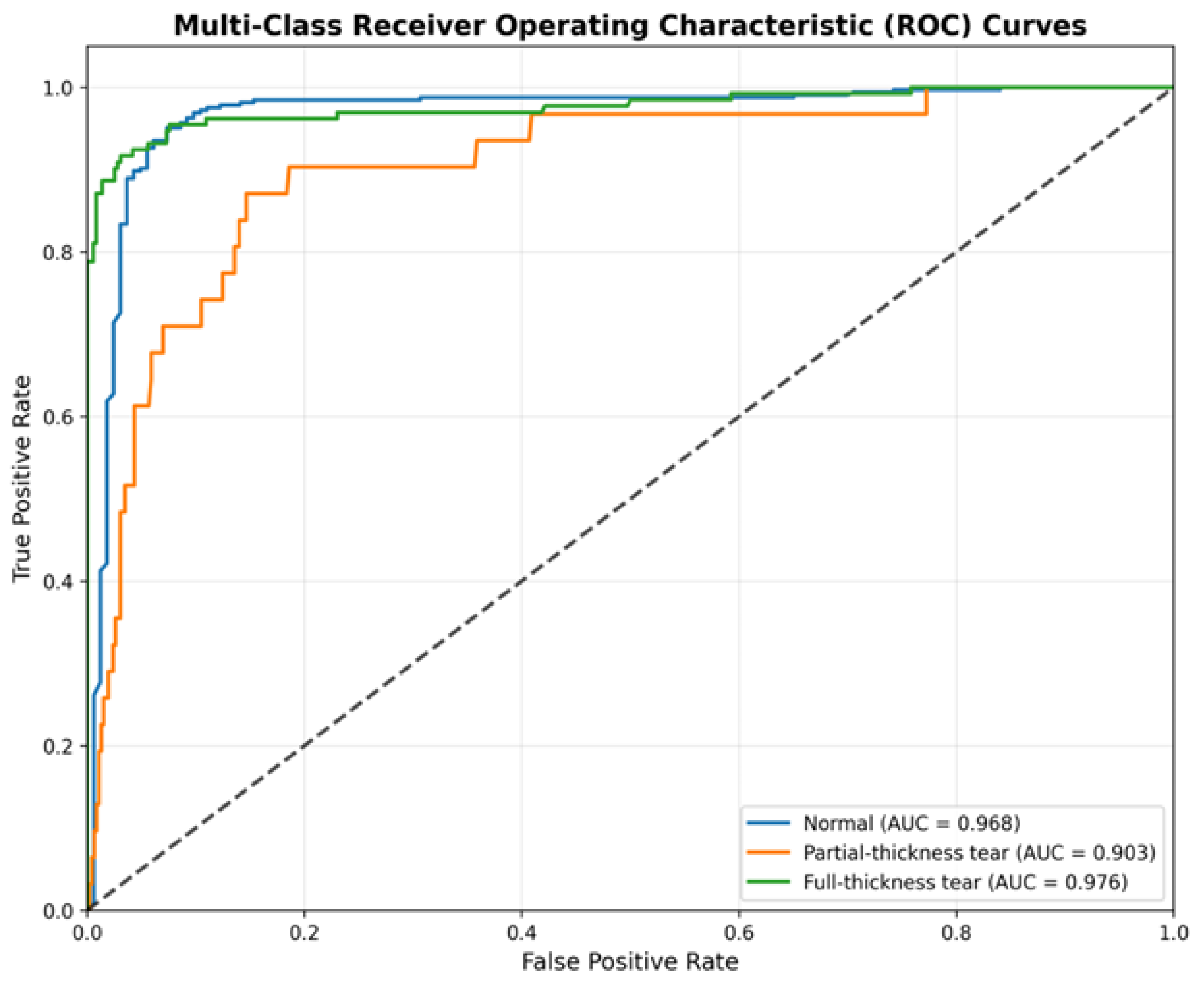
We further quantified the discriminative power of the model using One-vs-Rest Receiver Operating Characteristic (ROC) curves (
Figure 4). This analysis assesses the model’s performance independent of specific decision thresholds.
The ROC analysis corroborates the robustness of the classifier. The model achieves an AUC of 0.976 for the “Full-Thickness Tear” class and 0.968 for the “Normal” class, indicating near-perfect separability. Most notably, for the minority “Partial-Thickness Tear” class, the model achieves an AUC of 0.903. This high AUC value, despite the moderate accuracy observed in the confusion matrix, implies that the model consistently assigns higher probability scores to partial tears compared to negatives. This suggests that the sensitivity for partial tears could be further optimized in a clinical setting by calibrating the decision threshold specifically for this class.
3.3. Explainable AI for Clinical Validation
A critical barrier to the clinical adoption of deep learning systems is their perceived “black box” nature. To validate that the Pa-ViT model relies on genuine anatomical features rather than spurious correlations or artifacts, we utilized Attention Rollout to visualize the self-attention maps generated by the final Transformer block. These heatmaps represent the regions of the MRI slice that most influenced the model’s decision.
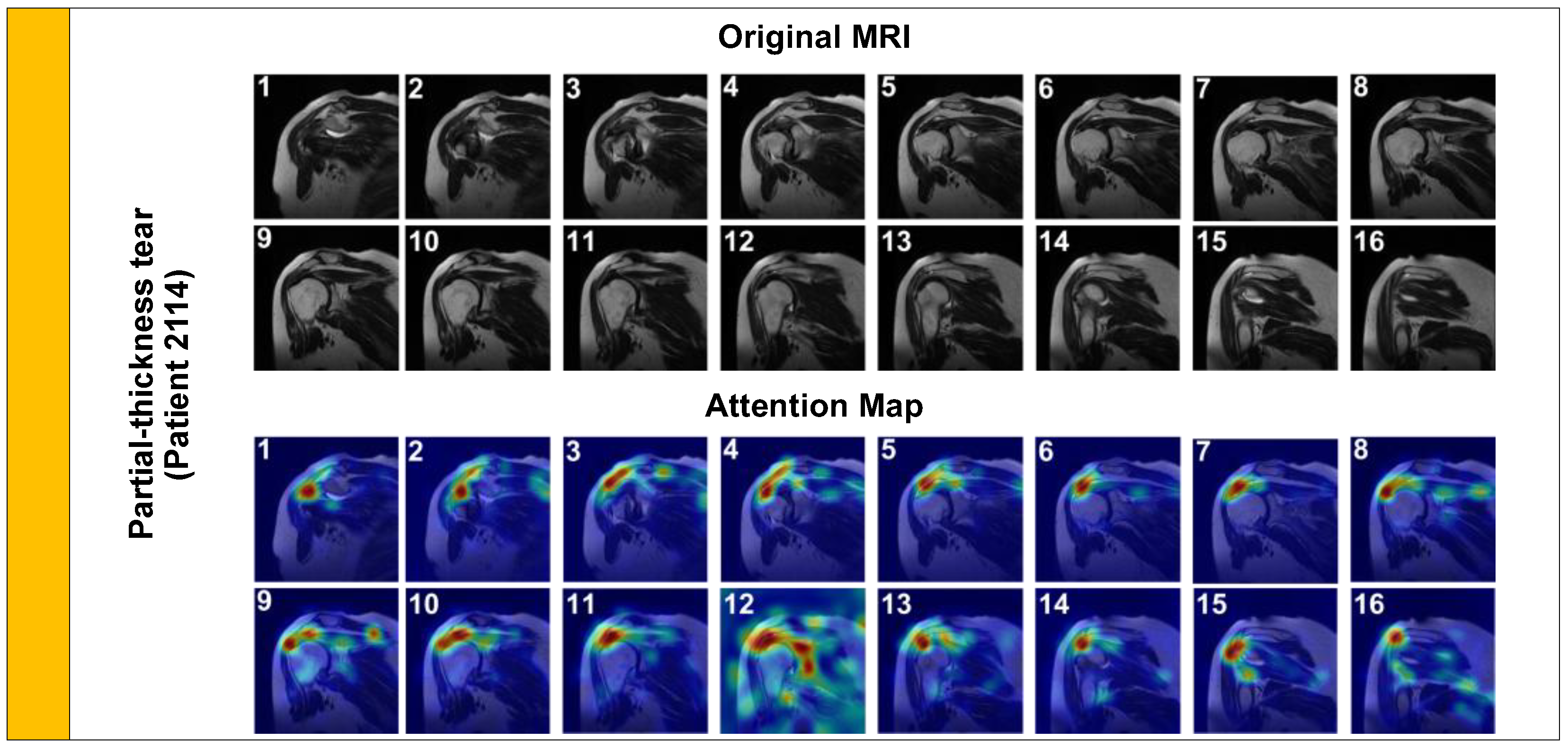
We first analyzed a Partial-Thickness Tear (
Figure 5, Patient 2114). Clinically, these tears are characterized by focal signal hyperintensity on the articular or bursal side of the tendon, without complete disruption of the fibers.
As shown in
Figure 5, the model’s attention is tightly focalized on the articular side of the supraspinatus tendon footprint. This precise localization confirms that the model is detecting the subtle ‘footprint’ lesion rather than general joint inflammation, aligning with expert radiological assessment.
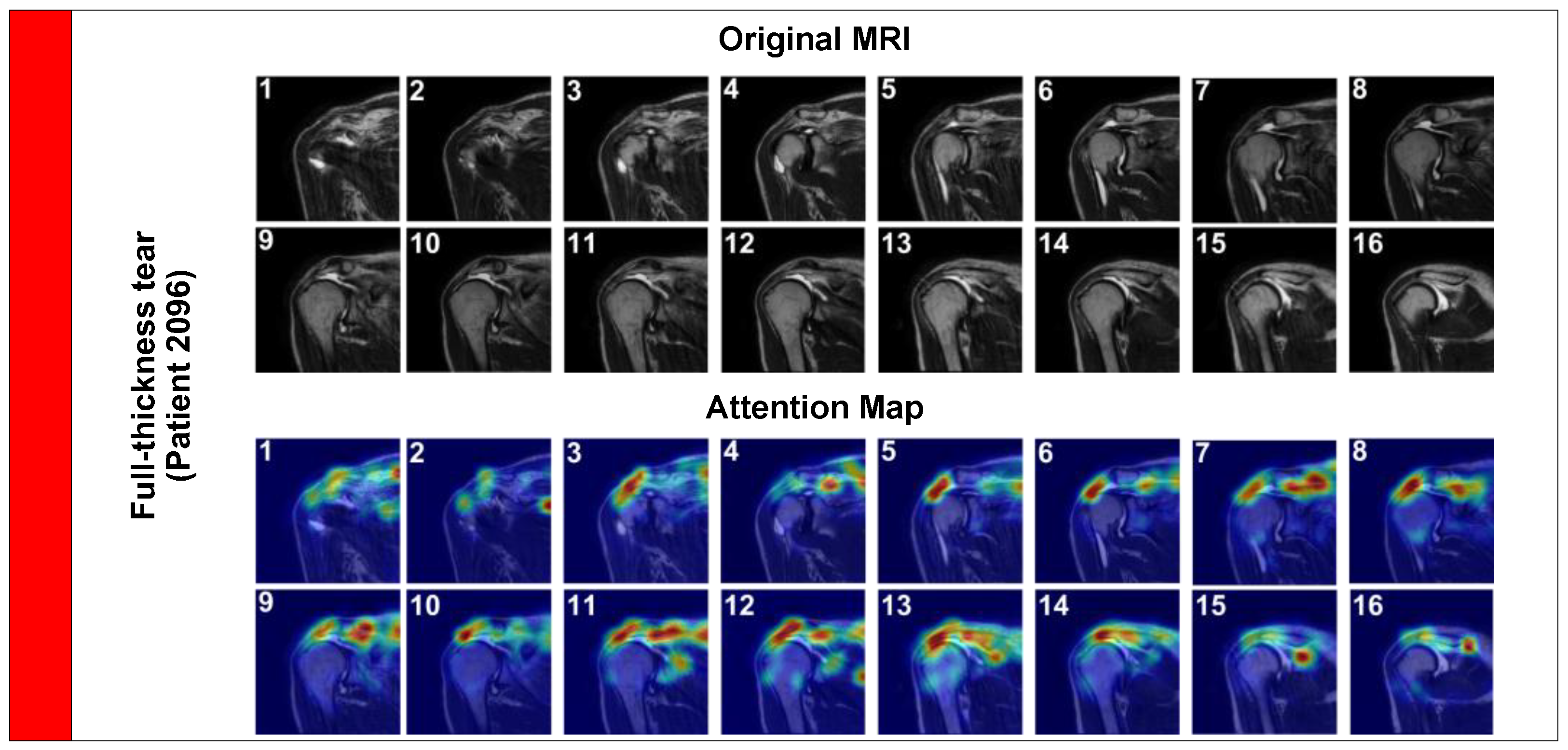
Next, we examined a Full-Thickness Tear (
Figure 6, Patient 2096). These are morphologically distinct, involving the complete detachment of the tendon from the humeral head, often accompanied by retraction and fluid filling the gap.
In this case, the attention map expands significantly to cover the gap created by the retracted tendon. The model accurately tracks the fluid signal (hyperintensity) filling the space between the humeral head and the acromion. This effectively replicates the ‘fluid sign’ used by radiologists to diagnose full tears, demonstrating the model’s ability to identify complex pathological signs.
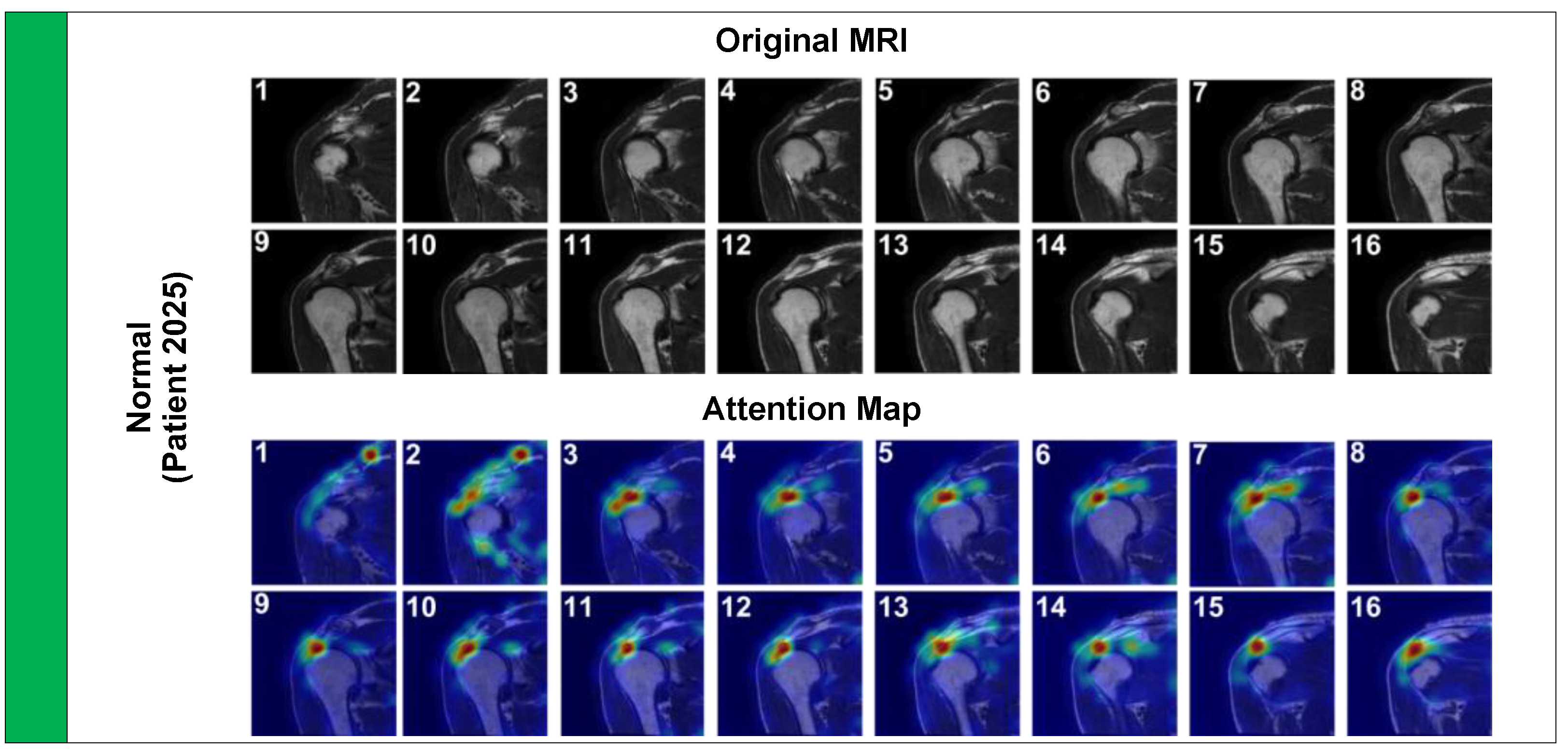
Finally, we analyzed a Normal case (
Figure 7, Patient 2025) to ensure the model does not produce false positive activations on healthy anatomy.
The attention pattern here is markedly different; it is diffuse and spreads along the entire length of the intact tendon-bone interface. This diffuse attention suggests the model is verifying the structural continuity of the tendon rather than latching onto a focal pathology. Collectively, these visual explanations provide strong qualitative evidence that the high quantitative performance of the Pa-ViT model is underpinned by a robust, medically grounded understanding of shoulder anatomy and pathology.
4. Discussion
This study introduces the Patient-Aware Vision Transformer (Pa-ViT), a novel deep learning framework that fundamentally redefines the automated diagnosis of Rotator Cuff Tears (RCT) by shifting the computational paradigm from local, slice-level convolution to global, patient-level attention. By achieving a high accuracy of 91% and a macro-averaged F1-score of 0.91 on a challenging, highly imbalanced dataset, our model demonstrates that the inductive biases of Transformers—specifically their ability to model long-range dependencies via Multi-Head Self-Attention—are inherently superior to traditional Convolutional Neural Networks (CNNs) for volumetric medical imaging tasks [
4,
15,
25].
The most significant finding of our work is the substantial performance margin between our Pa-ViT model and the baseline VGG-16 model proposed by Kim et al. [
24], which achieved 87% accuracy using a weighted linear combination of 2D slices. While CNN architectures like VGG-16, ResNet, or DenseNet effectively detect local textures, they struggle to contextualize features across large spatial distances. This capability is critical for RCT diagnosis, where the pathological signal requires understanding the geometric relationship between the supraspinatus tendon footprint, the humeral head, and the acromion. Furthermore, recent studies employing 3D CNNs, such as the Voxception-ResNet by Shim et al. (92.5% accuracy) [
22] and multi-input CNNs by Yao et al. (AUC 0.94) [
20], have attempted to capture this volumetric context. However, these 3D architectures are often computationally prohibitive and data-hungry. Our Pa-ViT model offers a competitive alternative by leveraging the efficiency of 2D slice processing while achieving 3D-like global understanding through the Transformer’s attention mechanism, mirroring the “global processing” cognitive model utilized by expert radiologists.
A critical advancement of this framework lies in bridging the diagnostic gap for Partial-Thickness Tears, which represent the most challenging category in automated shoulder diagnosis due to their subtle radiological presentation and high inter-observer variability. Previous attempts using CNNs have yielded suboptimal results for this class; notably, the baseline study by Kim et al. [
24] reported a classification accuracy of approximately 38% for partial tears, with a Precision-Recall area of only 0.45. Similarly, Yao et al. [
20] reported a sensitivity of only 72.5% for partial-thickness tears compared to 100% for full-thickness tears, highlighting this class as a universal bottleneck in AI diagnostics. In contrast, our Pa-ViT framework achieved a classification accuracy of 51.61% and an ROC AUC of 0.903 for this minority class. This substantial improvement is attributable to the synergy between our Weighted Cross-Entropy Loss, which effectively counteracted the extreme class imbalance (6.4% prevalence), and the high spatial fidelity of the ViT patch embeddings. These embeddings preserve fine-grained anatomical details often lost during the aggressive pooling operations of deep CNNs. Furthermore, the confusion matrix reveals that the majority of misclassifications for partial tears were “over-estimations” into the Full-Thickness class rather than “under-estimations” into the Normal class. In a clinical decision support context, this bias is preferable as it ensures that patients with potential pathology remain within the pathway for specialist review rather than being falsely cleared, aligning with the conservative management strategies often employed for equivocal cases.
The efficacy of our approach is further underpinned by the formulation of the diagnostic task as a Weakly-Supervised Multiple Instance Learning (MIL) problem with Global Mean Pooling. Rotator cuff tears are inherently volumetric 3D defects [
24]; a signal hyperintensity that appears in only a single slice might be an artifact (e.g., magic angle effect), whereas a signal persisting across contiguous slices is likely genuine pathology [
16]. The success of our permutation-invariant aggregation strategy confirms that the model successfully learns to filter out slice-specific noise and relies on inter-slice consistency to form a prediction. This renders the Pa-ViT framework robust to stochastic variations in slice thickness or patient positioning—issues that frequently plague 2D slice-based classifiers. Moreover, unlike full 3D segmentation models that require labor-intensive pixel-level annotations [
1,
26], our MIL approach eliminates the need for dense supervision, making our solution more feasible for deployment in clinical environments with limited hardware resources and annotated data.
Finally, the interpretability provided by our Attention Rollout visualizations addresses the “black box” skepticism often associated with medical AI. Unlike prior CNN-based Class Activation Maps (CAM), which often produce coarse heatmaps, our visualizations demonstrate precise anatomical reasoning: identifying the articular footprint in partial tears, tracking the fluid gap in full-thickness tears, and verifying tendon continuity in normal cases. This aligns with the findings of Fazal Gafoor et al. [
16], who emphasized the high correlation between MRI findings and arthroscopy, suggesting that our model’s attention maps could serve as a reliable “second reader” to highlight subtle lesions that might be overlooked in a busy clinical workflow.
Despite these successes, several limitations of our study must be acknowledged. First, the distinction between high-grade partial tears and small full-thickness tears remains a source of ambiguity for the model, reflecting the inherent subjectivity in radiological grading and the continuum of tendon degeneration described in previous literature. Second, our current mean pooling aggregation treats slices as an unordered set, potentially discarding valuable spatial continuity information along the z-axis. Future iterations could incorporate 3D positional embeddings or sequence modeling (e.g., RNNs or Transformers over slices) to better capture the volumetric shape of the tear. Third, while we utilized a robust dataset, the study is retrospective and single-center. External validation on multi-center datasets with varying MRI protocols (1.5T vs 3.0T) is essential to confirm generalizability, as field strength can influence diagnostic accuracy. Lastly, integrating quantitative radiomics features or T2 mapping data into the Pa-ViT framework could further enhance its sensitivity to early tendinopathy before macroscopic tearing occurs.
In conclusion, the Pa-ViT framework represents a significant step forward in the automated diagnosis of rotator cuff pathology, offering a clinically viable, accurate, and interpretable solution that outperforms traditional CNN baselines, particularly in the challenging diagnosis of partial-thickness tears.
5. Conclusions
In this study, we introduced the Patient-Aware Vision Transformer (Pa-ViT), a unified framework designed to overcome the limitations of traditional CNN-based architectures in the volumetric diagnosis of rotator cuff tears. By integrating a Vision Transformer backbone with a Weakly-Supervised Multiple Instance Learning paradigm, our approach successfully captures global anatomical context and effectively aggregates volumetric data, addressing critical gaps in automated shoulder MRI analysis.
The experimental validation on a large-scale dataset demonstrated that Pa-ViT achieves strong performance with an accuracy of 91% and an F1-score of 0.91, significantly outperforming conventional baselines. Clinically, the most impactful contribution of this framework is its superior sensitivity in detecting partial-thickness tears, a diagnosis that is historically challenging for both automated systems and varying levels of radiological expertise. Furthermore, the generation of precise, anatomically aligned attention maps bridges the gap between “black-box” deep learning and clinical trust, confirming that the model relies on genuine pathological features rather than spurious correlations.
Ultimately, this work provides a methodological template for future research in medical image analysis, proving the efficacy of self-attention mechanisms over standard convolutions for musculoskeletal pathology. Future directions will focus on enhancing the framework’s clinical utility by integrating multi-modal data—such as patient history and demographics—and extending the architecture to multi-task learning for simultaneous tear detection and segmentation.
Author Contributions
Conceptualization, M.A.; methodology, S.A. and A.Y.; software, S.A.; validation, M.A. and İ.O.; formal analysis, S.A. and M.A.; investigation, M.A., S.A., A.Y., and İ.O.; data curation, S.A.; writing—original draft preparation, M.A., S.A., and A.Y.; writing—review and editing, M.A., S.A., and A.Y.; visualization, S.A.; supervision, M.A., A.Y., and İ.O.; project administration, M.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of Non-Interventional Clinical Research of Bilecik Şeyh Edebali University (Decision No. 9, 10th Meeting, dated 30 October 2025).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data used in this study were obtained upon request from a previously conducted study and are not publicly available due to ethical, legal, and privacy restrictions. Access to the data may be considered by the data owners upon reasonable request.
Acknowledgments
The authors would like to thank the institution responsible for the original study for kindly providing access to the dataset upon request, as well as all patients whose data were used in this research.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Riem, L.; Feng, X.; Cousins, M.; Ducharme, O.; Leitch, E.B.; Werner, B.C.; Sheean, A.J.; Hart, J.; Antosh, I.J.; Blemker, S.S. A Deep Learning Algorithm for Automatic 3D Segmentation of Rotator Cuff Muscle and Fat from Clinical MRI Scans. 2023, 5. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.J.; Schwier, M.; Geiger, B.; Raithel, E.; Von Busch, H.; Fritz, J.; Kline, M.; Brooks, M.; Dunham, K.; Shukla, M.; et al. Deep Learning Diagnosis and Classification of Rotator Cuff Tears on Shoulder MRI. Invest Radiol 2023, 58, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.R.; Olson, J.J.; Sefko, J.A.; Steger-May, K.; Teefey, S.A.; Middleton, W.D.; Keener, J.D. Does Surgical Intervention Alter the Natural History of Degenerative Rotator Cuff Tears? Comparative Analysis from a Prospective Longitudinal Study. J Shoulder Elbow Surg 2025, 34, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Xia, X.; Wang, J.; Li, X.; Huang, M.; Miao, S.; Hao, D.; Li, J. Fully Automated Approach for Diagnosis of Supraspinatus Tendon Tear on Shoulder MRI by Using Deep Learning. Acad Radiol 2024, 31, 994–1002. [Google Scholar] [CrossRef]
- Lee, K.C.; Cho, Y.; Ahn, K.S.; Park, H.J.; Kang, Y.S.; Lee, S.; Kim, D.; Kang, C.H. Deep-Learning-Based Automated Rotator Cuff Tear Screening in Three Planes of Shoulder MRI. Diagnostics 2023, Vol. 13 13, 3254. [Google Scholar] [CrossRef]
- Giai Via, A.; De Cupis, M.; Spoliti, M.; Oliva, F. Clinical and Biological Aspects of Rotator Cuff Tears. Muscles Ligaments Tendons J 2013, 3, 70. [Google Scholar] [CrossRef]
- Dickinson, R.N.; Kuhn, J.E. Nonoperative Treatment of Rotator Cuff Tears. Phys Med Rehabil Clin N Am 2023, 34, 335–355. [Google Scholar] [CrossRef]
- Velasquez Garcia, A.; Hsu, K.L.; Marinakis, K. Advancements in the Diagnosis and Management of Rotator Cuff Tears. The Role of Artificial Intelligence. J Orthop 2024, 47, 87–93. [Google Scholar] [CrossRef]
- Ro, K.; Kim, J.Y.; Park, H.; Cho, B.H.; Kim, I.Y.; Shim, S.B.; Choi, I.Y.; Yoo, J.C. Deep-Learning Framework and Computer Assisted Fatty Infiltration Analysis for the Supraspinatus Muscle in MRI. Scientific Reports 2021, 11 11, 15065. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372. [Google Scholar] [CrossRef]
- Key, S.; Demir, S.; Gurger, M.; Yilmaz, E.; Barua, P.D.; Dogan, S.; Tuncer, T.; Arunkumar, N.; Tan, R.S.; Acharya, U.R. ViVGG19: Novel Exemplar Deep Feature Extraction-Based Shoulder Rotator Cuff Tear and Biceps Tendinosis Detection Using Magnetic Resonance Images. Med Eng Phys 2022, 110, 103864. [Google Scholar] [CrossRef] [PubMed]
- Hahn, S.; Yi, J.; Lee, H.-J.; Lee, Y.; Lim, Y.-J.; Bang, J.-Y.; Kim, H.; Lee, J.; Hahn, S.; Yi, J.; et al. Image Quality and Diagnostic Performance of Accelerated Shoulder MRI With Deep Learning–Based Reconstruction. https://www.ajronline.org/ 2021, 218, 506–516. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, J.P.; Droppelmann, G.; García, N.; Jorquera, C.; Feijoo, F. High-Accuracy Detection of Supraspinatus Fatty Infiltration in Shoulder MRI Using Convolutional Neural Network Algorithms. Front Med (Lausanne) 2023, 10, 1070499. [Google Scholar] [CrossRef] [PubMed]
- Zhan, J.; Liu, S.; Dong, C.; Ge, Y.; Xia, X.; Tian, N.; Xu, Q.; Jiang, G.; Xu, W.; Cui, J. Shoulder MRI-Based Radiomics for Diagnosis and Severity Staging Assessment of Surgically Treated Supraspinatus Tendon Tears. European Radiology 2023, 2023 33 33, 5587–5593. [Google Scholar] [CrossRef]
- Rodriguez, H.C.; Rust, B.; Hansen, P.Y.; Maffulli, N.; Gupta, M.; Potty, A.G.; Gupta, A. Artificial Intelligence and Machine Learning in Rotator Cuff Tears. Sports Med Arthrosc Rev 2023, 31, 67–72. [Google Scholar] [CrossRef]
- Fazal Gafoor, H.; Jose, G.A.; Mampalli Narayanan, B. Role of Magnetic Resonance Imaging (MRI) in the Diagnosis of Rotator Cuff Injuries and Correlation With Arthroscopy Findings. Cureus 2023, 15. [Google Scholar] [CrossRef]
- Lalehzarian, S.P.; Gowd, A.K.; Liu, J.N. Machine Learning in Orthopaedic Surgery. World J Orthop 2021, 12, 685. [Google Scholar] [CrossRef]
- Lisacek-Kiosoglous, A.B.; Powling, A.S.; Fontalis, A.; Gabr, A.; Mazomenos, E.; Haddad, F.S. Artificial Intelligence in Orthopaedic Surgery EXPLORING ITS APPLICATIONS, LIMITATIONS, AND FUTURE DIRECTION. Bone Joint Res 2023, 12, 47–454. [Google Scholar] [CrossRef]
- Esfandiari, M.A.; Fallah Tafti, M.; Jafarnia Dabanloo, N.; Yousefirizi, F. Detection of the Rotator Cuff Tears Using a Novel Convolutional Neural Network from Magnetic Resonance Image (MRI). Heliyon 2023, 9, e15804. [Google Scholar] [CrossRef]
- Yao, J.; Chepelev, L.; Nisha, Y.; Sathiadoss, P.; Rybicki, F.J.; Sheikh, A.M. Evaluation of a Deep Learning Method for the Automated Detection of Supraspinatus Tears on MRI. Skeletal Radiology 2022 51:9 2022, 51, 1765–1775. [Google Scholar] [CrossRef]
- Ni, M.; Zhao, Y.; Zhang, L.; Chen, W.; Wang, Q.; Tian, C.; Yuan, H. MRI-Based Automated Multitask Deep Learning System to Evaluate Supraspinatus Tendon Injuries. European Radiology 2023, 2023 34 34, 3538–3551. [Google Scholar] [CrossRef]
- Shim, E.; Kim, J.Y.; Yoon, J.P.; Ki, S.Y.; Lho, T.; Kim, Y.; Chung, S.W. Automated Rotator Cuff Tear Classification Using 3D Convolutional Neural Network. Scientific Reports 2020, 10:1 10, 15632. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, J.H.; Oh, K.S.; Yoon, J.P.; Seo, A.; Jeong, Y.J.; Chung, S.W. Automated 3-Dimensional MRI Segmentation for the Posterosuperior Rotator Cuff Tear Lesion Using Deep Learning Algorithm. PLoS One 2023, 18, e0284111. [Google Scholar] [CrossRef]
- Kim, M.; Kim, J.Y.; Kim, S.H.; Van Hoeke, S.; De Neve, W.; Kim, M.; Park, H.-M. MRI-Based Diagnosis of Rotator Cuff Tears Using Deep Learning and Weighted Linear Combinations. Proc Mach Learn Res 2020, 126, 292–308. [Google Scholar]
- Zhan, H.; Teng, F.; Liu, Z.; Yi, Z.; He, J.; Chen, Y.; Geng, B.; Xia, Y.; Wu, M.; Jiang, J. Artificial Intelligence Aids Detection of Rotator Cuff Pathology: A Systematic Review. Arthroscopy: The Journal of Arthroscopic & Related Surgery 2024, 40, 567–578. [Google Scholar] [CrossRef]
- Kim, H.; Shin, K.; Kim, H.; Lee, E.S.; Chung, S.W.; Koh, K.H.; Kim, N. Can Deep Learning Reduce the Time and Effort Required for Manual Segmentation in 3D Reconstruction of MRI in Rotator Cuff Tears? PLoS One 2022, 17, e0274075. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).