1. Introduction
Sea buckthorn (
Hippophae rhamnoides L.) is a valuable crop tree. Its berries are rich in bioactive compounds, including valuable unsaturated fatty acids [
1,
2,
3,
4,
5]. Sea buckthorn fruit pulp contains high amounts of the rare palmitoleic acid (16:1), and its seeds are rich in linoleic acid (18:2) and linolenic acid (18:3) [
3,
6,
7]. Studies on the investigation of genes involved in the synthesis of fatty acids in sea buckthorn fruits identified genes that encode β-ketoacyl-acyl carrier protein synthase (KAS), oleoyl-acyl carrier protein thioesterase 1 (FATA), palmitoyl-acyl carrier protein thioesterase (FATB), stearoyl-ACP desaturase (SAD), and fatty acid desaturase (FAD), as well as evaluated their expression levels [
6,
8,
9,
10,
11,
12]. In our previous study [
11], we identified and characterized four
KAS II, eight
FAT, nine
SAD, and 12
FAD genes in the
H. rhamnoides genome CNP0001846 (
https://db.cngb.org, accessed on 1 November 2025) [
13]. However, there were errors in the following genes in the CNP0001846 genome:
FATB (28610),
SAD (18766),
FAD2 (12459),
FAD2 (21624),
FAD3 (24146). This issue is likely associated with mistakes in the genome assembly or annotation. The present study aimed to create an annotation for a contiguous and accurate genome assembly of the variety Triumf and obtain the correct sequences of genes involved in fatty acid synthesis.
2. Materials and Methods
For the transcriptomic analysis, plant material was collected from the sea buckthorn variety Triumf in three biological replicates. The cuttings were collected in the Federal Altai Scientific Center of Agrobiotechnologies (Barnaul, Russia). They were cut into 13-15 cm pieces and placed in containers with a Kornevin (IMK) solution (AgroSintez, Russia). After 24 hours, the cuttings were transferred to containers filled with water. The water was replaced daily, and samples were taken at necessary intervals as the plants grew. The samples included roots, wood, and bark, buds, leaves, and flowers. Buds were sampled at four stages of development: dormant bud, beginning of swelling, swollen/unfolding bud, and fully opened bud. Leaves were collected whole and separated into base, tip, marginal zone, and central vein parts. The plant material was placed in 1.5-ml tubes, immediately frozen in liquid nitrogen, and stored in a freezer at -70 °C until RNA extraction. Thirteen samples of the plant material in two replicates were selected for further transcriptomic studies.
Prior to RNA extraction, the samples were ground into a fine powder using disposable homogenization pestles (Helicon, Russia) attached to the DeWALT DCD701D2 cordless drill (DeWALT, USA) in 1.5-ml tubes placed in liquid nitrogen to prevent thawing. RNA extraction was performed based on a modified CTAB buffer protocol [
14]. Total RNA was then purified using the CleanRNA Standard Kit (Evrogen, Russia) according to the manufacturer’s protocol with an addition of a DNase I (Magen, China) treatment step. The quality and concentration of RNA samples were assessed using the horizontal agarose gel electrophoresis and Qubit 4.0 fluorometer (Thermo Fisher Scientific, USA). Only undegraded RNA samples with a concentration of at least 25 ng/μL were used for further analysis.
We prepared cDNA libraries for sequencing the transcriptomes of various organs/tissues of the variety Triumf on the Illumina platform. For each sample, we used 500 ng of total RNA. The following kits were used to prepare the samples: MagicPure mRNA Kit (TransGen, China), TransNGS Fast RNA-Seq Library Prep Kit for Illumina (TransGen), and MagicPure Size Selection DNA Beads (TransGen). The kits were used according to the manufacturer’s protocols.
We used the Qsep1-Plus capillary electrophoresis system (BiOptic, Taiwan) and horizontal electrophoresis in a 2% agarose gel to assess the quality of the resulting cDNA libraries. We determined the concentration of the libraries using the Qubit 4.0 fluorometer (Thermo Fisher Scientific). Then we mixed the libraries equimolarly and sequenced them on NextSeq 2000 (Illumina, USA) with 100+100 bp reads.
We annotated the genome of the sea buckthorn variety Triumf with masked repeats using BRAKER3 v3.0.8 [
15], applying Triumf’s transcriptomic data obtained in this study and in the previous study [
12], as well as the Viridiplantae OrthoDB [
16] protein database.
We used the tool developed by us to identify and analyze sea buckthorn genes of the
KAS,
FATA,
SAD, and
FAD families, which are associated with fatty acid synthesis. Representative protein sequences of
Arabidopsis thaliana for the target gene families were downloaded from the Arabidopsis Information Resource (TAIR,
https://www.arabidopsis.org/, accessed on 1 November 2025). These
A. thaliana protein sequences were aligned against the translated transcripts of the sea buckthorn genome assemblies of the variety Triumf and Chinese variety under ID CNP0001846 (CNGB,
https://db.cngb.org, accessed on 1 November 2025) using local BLASTP v2.17.0. This resulted in a list of candidate proteins, which were then manually assessed and filtered based on relative alignment scores. In general, proteins with identity of less than 30% to the corresponding
A. thaliana sequences were excluded. Candidate proteins and reference
A. thaliana sequences were used as queries in HMMER 3.0 (
http://hmmer.org/, accessed on 1 November 2025) against the Pfam database of conserved domains [
17]. Candidate sequences lacking the relevant conserved domains were manually filtered out. Domain structure of the analyzed sequences was then re-assessed using the NCBI batch CD-search against the default NCBI conserved domain database [
18]. In addition, the exon-intron structures of the candidate genes were assessed and compared with the corresponding
A. thaliana gene models. Both domain and exon-intron structures were visualized using the ggplot2 v.4.0.1 R package. The protein sequences were aligned using MAFFT v7.525, and phylogenetic trees were constructed from the resulting multiple alignment using RapidNJ 2.3.2 with the Neighbor Joining method and 1000 bootstrap replicates. Phylogenetic trees were visualized using the ggtree R package [
19] and iTOL v7.3 [
20].
3. Results and Discussion
We performed sequencing of 26 cDNA libraries obtained from various organs/tissues of the sea buckthorn variety Triumf using the Illumina platform. On average, we generated 2.6 million reads for each sample. These data along with the previously obtained data for Triumf’s seeds and pulp at four fruit development stages (NCBI SRA, BioProject PRJNA1163394) were used to annotate the Triumf genome assembly. Structural annotation predicted 25,915 gene models and 30,527 transcript models.
Using the in-house developed tool, we identified genes of the
KAS,
FAT,
SAD, and
FAD families in the annotated genome assemblies of the variety Triumf and Chinese variety under ID CNP0001846 (CNGB). We have previously used the latter genome assembly to identify genes of the studied families [
11]. In the present study, the following genes of the examined families were identified in the two sea buckthorn genome assemblies: three
KAS I genes (Triumf), six
KAS II genes (Triumf) and four
KAS II genes (CNP0001846); two
KAS III genes (both genomes); one
mtKAS gene (both genomes); three
FATA genes (Triumf) and two
FATA genes (CNP0001846); five
FATB genes (Triumf) and six
FATB genes (CNP0001846); eight
SAD genes (Triumf) and nine
SAD genes (CNP0001846); three
FAD2 genes (both genomes); four
FAD3 genes (Triumf) and five
FAD3 genes (CNP0001846); one
FAD4 gene (both genomes); one
FAD6 gene (both genomes); and five
FAD7/8 genes (Triumf) and three
FAD7/8 genes (CNP0001846).
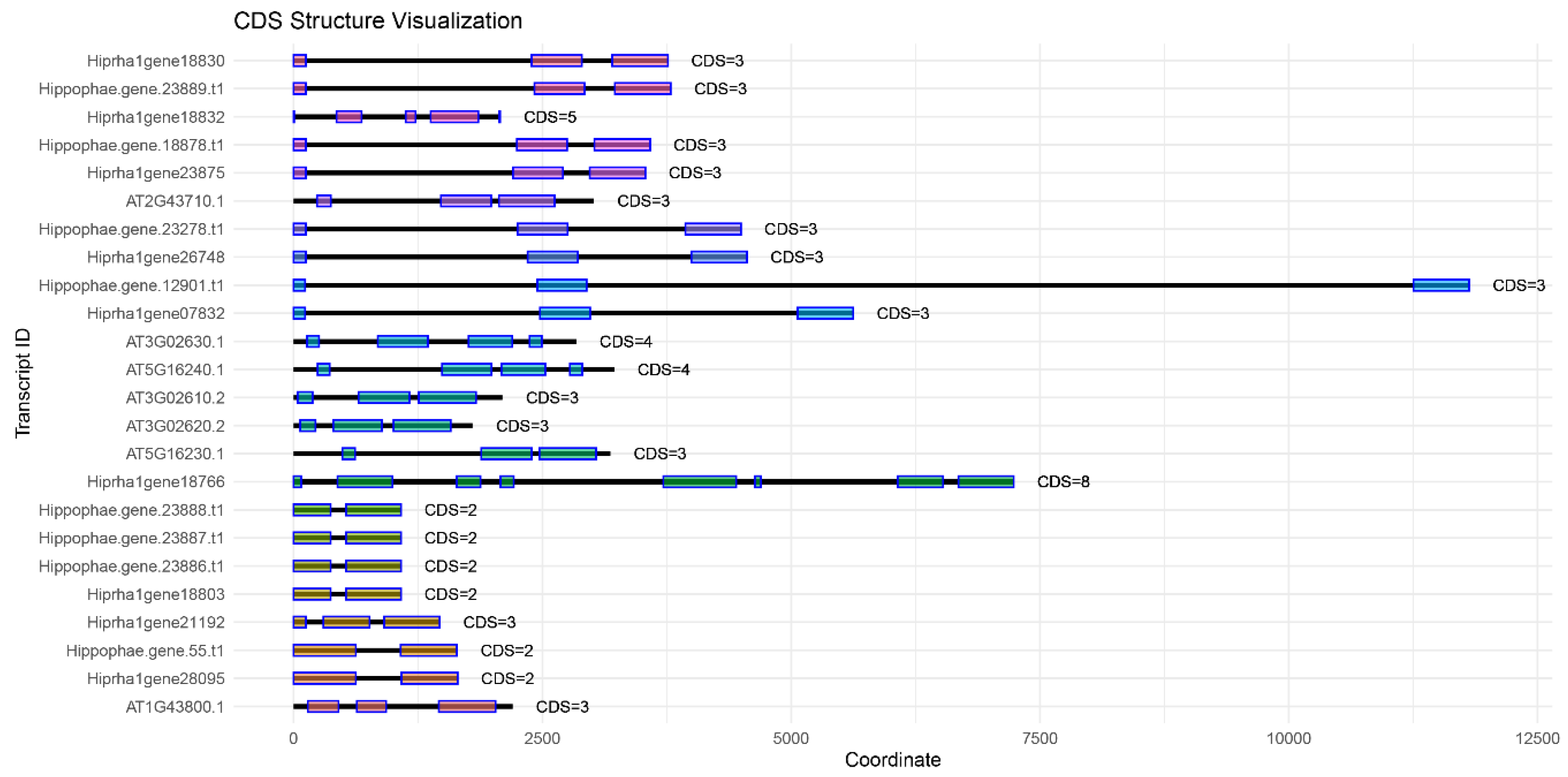
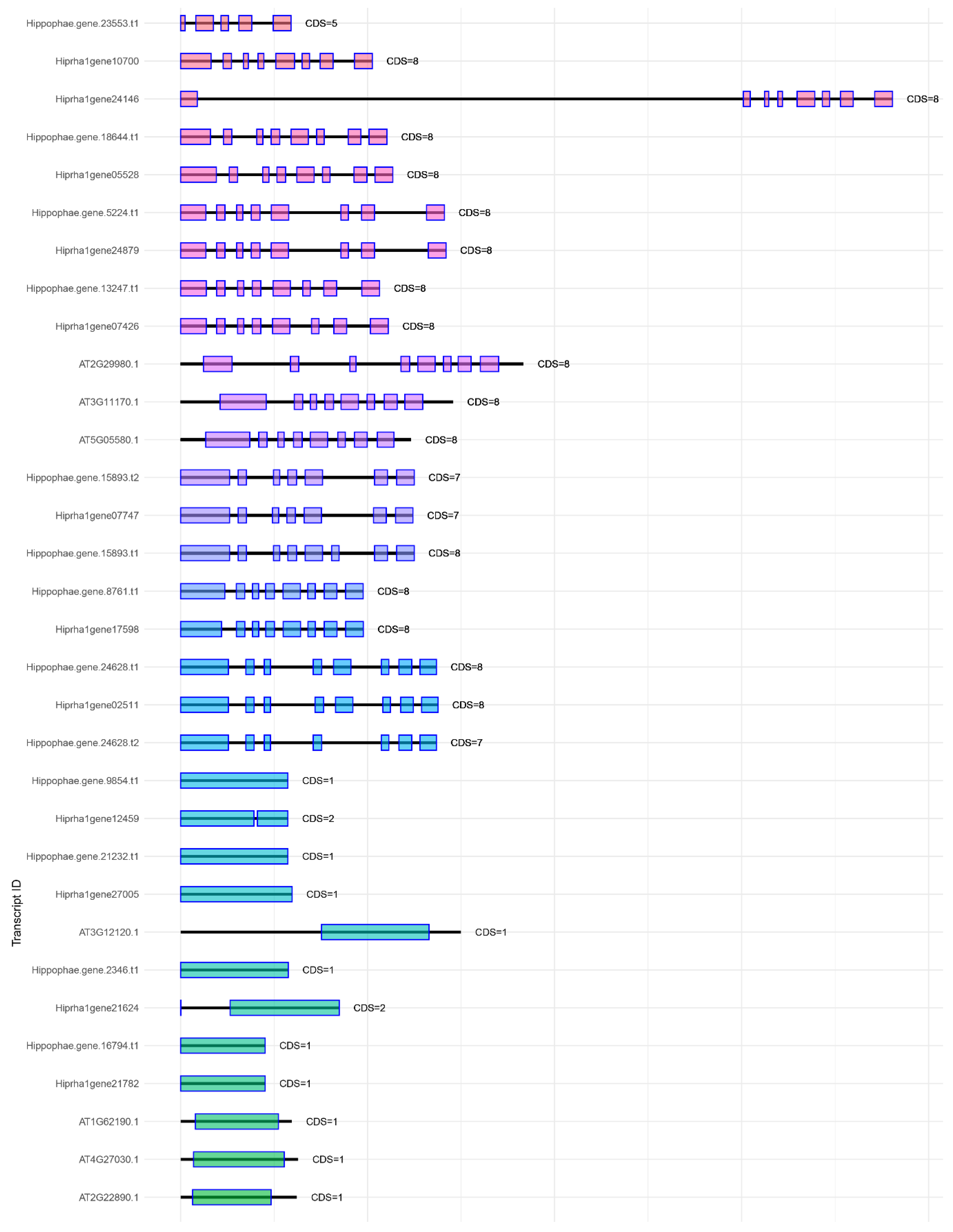
Figure 1,
Figure 2,
Figure 3 and
Figure 4 show a comparison of exon-intron structures of the identified genes of the
KAS, FAT, SAD, and
FAD families.
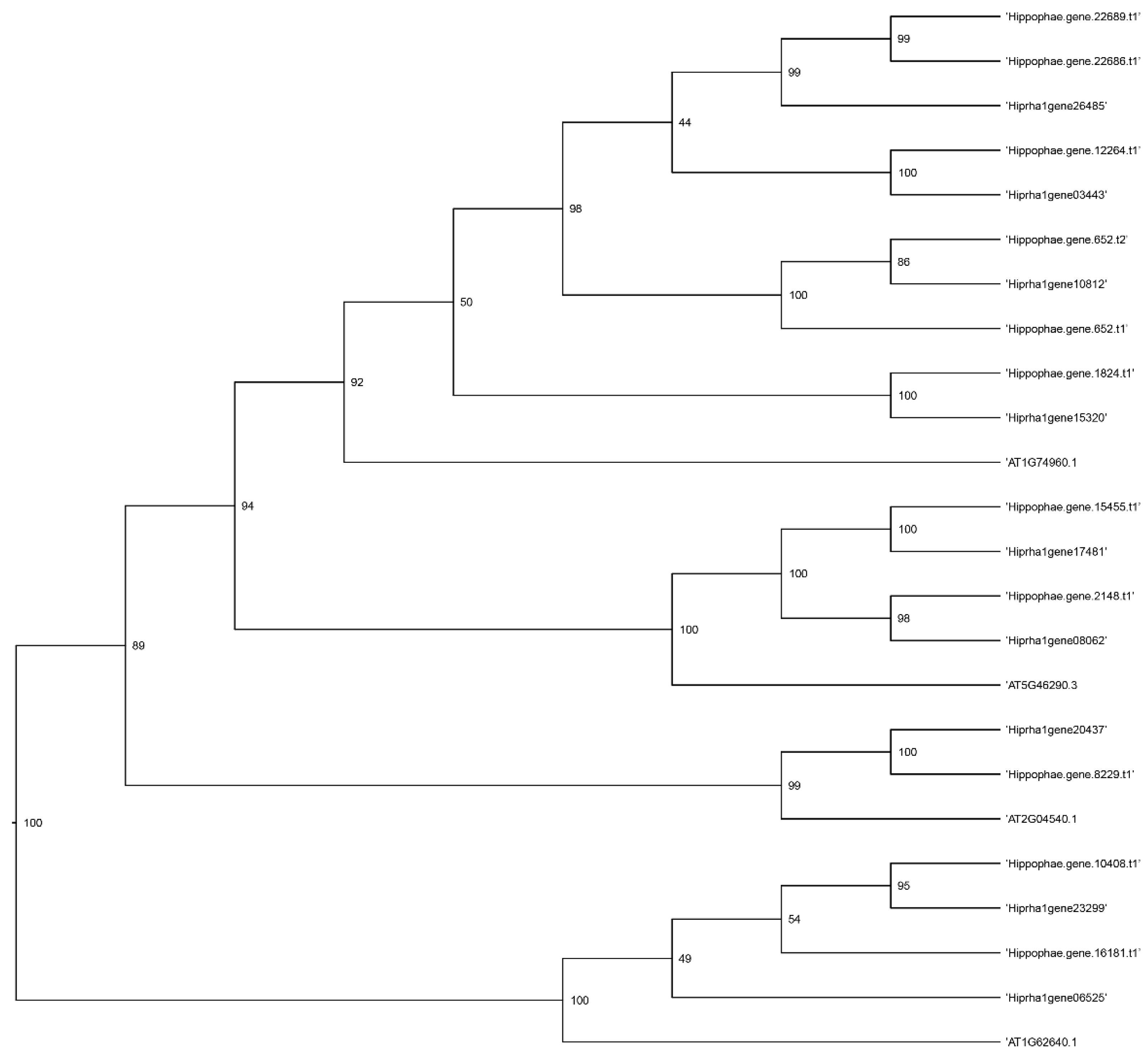
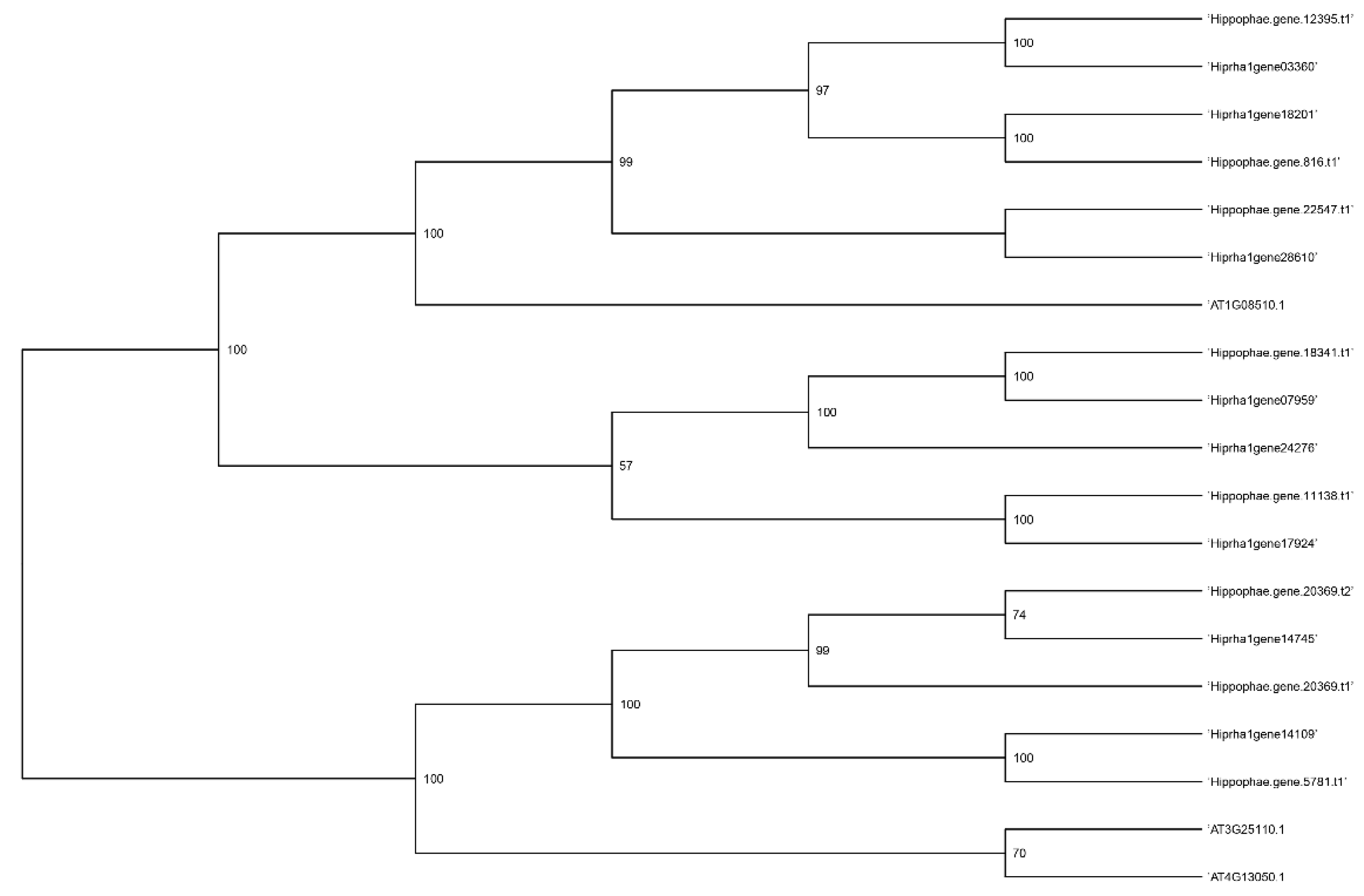
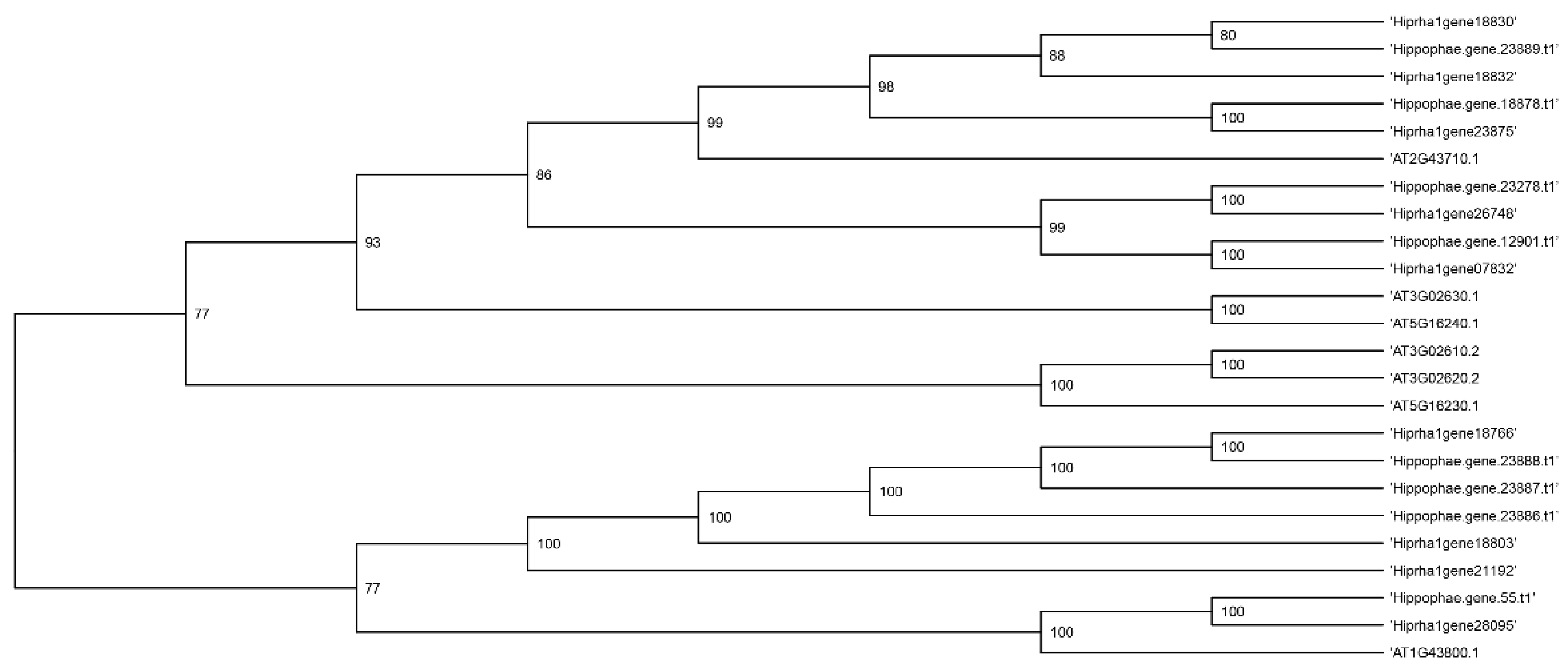
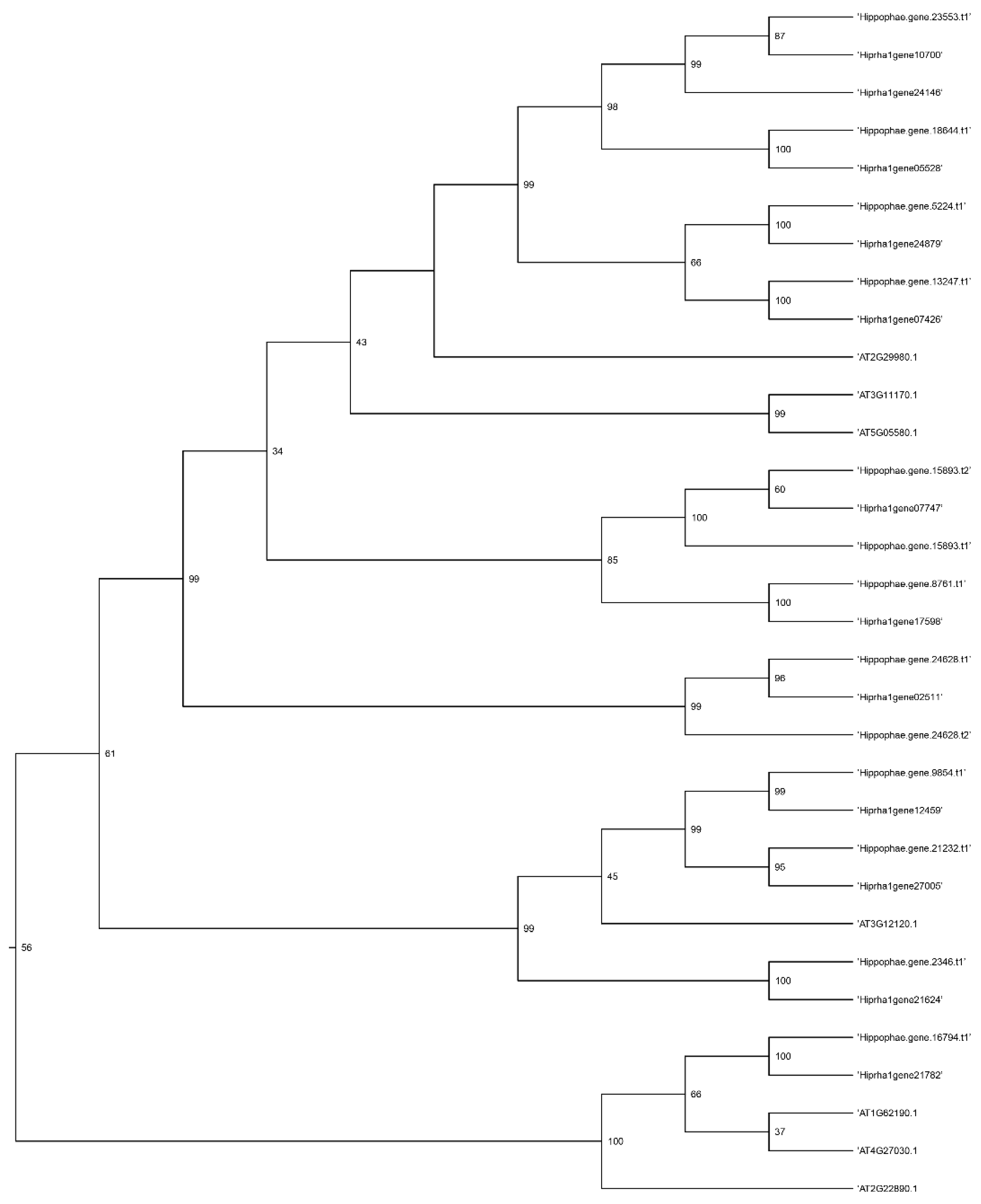
Figure 5,
Figure 6,
Figure 7 and
Figure 8 present a phylogenetic analysis of the identified genes of the
KAS,
FAT,
SAD, and
FAD families.
Annotating the Triumf genome assembly enabled us to identify and characterize the
KAS,
FAT,
SAD, and
FAD gene families in sea buckthorn. We also obtained correct gene sequences corresponding to
FATB (28610),
SAD (18766),
FAD2 (12459),
FAD2 (21624), and
FAD3 (24146) genes in the sea buckthorn genome assembly CNP0001846, which contained errors [
11]. This is an important step in investigating the molecular mechanisms of fatty acid synthesis in sea buckthorn fruits and creating varieties with improved oil composition. Furthermore, the obtained genome annotation is a valuable tool for genetic studies of sea buckthorn, including the identification and analysis of other significant gene families.
Author Contributions
Conceptualization, N.V.M. and A.A.D.; performing experiments, E.N.P., V.L.K., D.A.K., and N.M.B.; data analysis, A.A.A., F.D.K., Y.A.Z., N.V.M., and A.A.D.; writing, E.N.P., A.A.A., N.V.M., and A.A.D. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the Russian Science Foundation, grant № 23-46-00026, https://rscf.ru/project/23-46-00026/.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data generated in this study are available at NCBI under the BioProject accession number PRJNA1177110.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Jasniewska, A.; Diowksz, A. Wide Spectrum of Active Compounds in Sea Buckthorn (Hippophae rhamnoides) for Disease Prevention and Food Production. Antioxidants 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Ciesarova, Z.; Murkovic, M.; Cejpek, K.; Kreps, F.; Tobolkova, B.; Koplik, R.; Belajova, E.; Kukurova, K.; Dasko, L.; Panovska, Z.; et al. Why is sea buckthorn (Hippophae rhamnoides L.) so exceptional? A review. Food research international 2020, 133, 109170. [Google Scholar] [CrossRef] [PubMed]
- Sola Marsinach, M.; Cuenca, A.P. The impact of sea buckthorn oil fatty acids on human health. Lipids in health and disease 2019, 18, 145. [Google Scholar] [CrossRef] [PubMed]
- Zuchowski, J. Phytochemistry and pharmacology of sea buckthorn (Elaeagnus rhamnoides; syn. Hippophae rhamnoides): progress from 2010 to 2021. Phytochemistry reviews: proceedings of the Phytochemical Society of Europe 2023, 22, 3–33. [Google Scholar] [CrossRef] [PubMed]
- Gatlan, A.M.; Gutt, G. Sea Buckthorn in Plant Based Diets. An Analytical Approach of Sea Buckthorn Fruits Composition: Nutritional Value, Applications, and Health Benefits. International journal of environmental research and public health 2021, 18. [Google Scholar] [CrossRef] [PubMed]
- Fatima, T.; Snyder, C.L.; Schroeder, W.R.; Cram, D.; Datla, R.; Wishart, D.; Weselake, R.J.; Krishna, P. Fatty Acid Composition of Developing Sea Buckthorn (Hippophae rhamnoides L.) Berry and the Transcriptome of the Mature Seed. PLOS ONE 2012, 7, e34099. [Google Scholar] [CrossRef] [PubMed]
- Cakir, A. Essential oil and fatty acid composition of the fruits of Hippophae rhamnoides L. (Sea Buckthorn) and Myrtus communis L. from Turkey. Biochemical Systematics and Ecology 2004, 32, 809–816. [Google Scholar] [CrossRef]
- Yu, L.; Diao, S.; Zhang, G.; Yu, J.; Zhang, T.; Luo, H.; Duan, A.; Wang, J.; He, C.; Zhang, J. Genome sequence and population genomics provide insights into chromosomal evolution and phytochemical innovation of Hippophae rhamnoides. Plant biotechnology journal 2022, 20, 1257–1273. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Ruan, C.; Du, W.; Guan, Y. RNA-seq data reveals a coordinated regulation mechanism of multigenes involved in the high accumulation of palmitoleic acid and oil in sea buckthorn berry pulp. BMC plant biology 2019, 19, 207. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Ruan, C.; Guan, Y.; Li, H.; Du, W.; Lu, S.; Wen, X.; Tang, K.; Chen, Y. Nontargeted metabolomic and multigene expression analyses reveal the mechanism of oil biosynthesis in sea buckthorn berry pulp rich in palmitoleic acid. Food Chemistry 2022, 374, 131719. [Google Scholar] [CrossRef] [PubMed]
- Arkhipov, A.A.; Dvorianinova, E.M.; Turba, A.A.; Novakovskiy, R.O.; Zubarev, Y.A.; Predushchenko, P.A.; Sigova, E.A.; Zhernova, D.A.; Borkhert, E.V.; Pushkova, E.N.; et al. Identification and Analysis of KAS II, FAT, SAD, and FAD Gene Families in Hippophae rhamnoides. Plants 2024, 13. [Google Scholar] [CrossRef] [PubMed]
- Melnikova, N.V.; Arkhipov, A.A.; Zubarev, Y.A.; Novakovskiy, R.O.; Turba, A.A.; Yablokov, A.G.; Vladimirov, G.N.; Osipenko, S.V.; Bashilov, A.A.; Kostyukevich, Y.I.; et al. Gene Expression and Fatty Acid Composition in Sea Buckthorn Seeds and Pulp During Fruit Development of Different Varieties. International journal of molecular sciences 2025, 26. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Chen, H.; Pan, Y.; Feng, H.; Fang, D.; Yang, J.; Wang, Y.; Yang, J.; Sahu, S.K.; Liu, J.; et al. Genome of Hippophae rhamnoides provides insights into a conserved molecular mechanism in actinorhizal and rhizobial symbioses. The New phytologist 2022, 235, 276–291. [Google Scholar] [CrossRef] [PubMed]
- Pushkova, E.N.; Povkhova, L.V.; Dvorianinova, E.M.; Novakovskiy, R.O.; Rozhmina, T.A.; Gryzunov, A.A.; Sigova, E.A.; Zhernova, D.A.; Borkhert, E.V.; Turba, A.A. Expression of FAD and SAD genes in developing seeds of flax varieties under different growth conditions. Plants 2024, 13, 956. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, L.; Bruna, T.; Hoff, K.J.; Ebel, M.; Lomsadze, A.; Borodovsky, M.; Stanke, M. BRAKER3: Fully automated genome annotation using RNA-seq and protein evidence with GeneMark-ETP, AUGUSTUS and TSEBRA. bioRxiv: the preprint server for biology 2024. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, D.; Tegenfeldt, F.; Manni, M.; Seppey, M.; Berkeley, M.; Kriventseva, E.V.; Zdobnov, E.M. OrthoDB v11: annotation of orthologs in the widest sampling of organismal diversity. Nucleic acids research 2023, 51, D445–D451. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic acids research 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic acids research 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.-Y. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic acids research 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).