1. Introduction
Lectins play crucial roles in innate immunity as pattern-recognition molecules that detect pathogen-associated molecular patterns (PAMPs) as well as endogenous danger-associated molecular patterns [
1]. Mammals possess mannose-binding lectin (MBL) and ficolins, oligomeric serum lectins with N-terminal collagenous stalks that function as pathogen sensors to trigger the lectin pathway of the complement system, a major humoral effector of innate immunity [
2]. The lectin pathway is particularly important for antibacterial defense in newborns and young children prior to the establishment of adaptive immunity [
3].
MBL and ficolin recognize target carbohydrates via a C-type lectin domain and a fibrinogen-like domain, respectively [
2]. MBL binds mannose, N-acetyl-D-glucosamine (GlcNAc), glucose, and fucose, whereas ficolin recognizes GlcNAc and N-acetyl-D-galactosamine (GalNAc) through acetyl-group interactions [
4]. Despite their overlapping monosaccharide specificities, the two lectins exhibit distinct recognition spectra toward natural microbial ligands: MBL preferentially binds bacterial peptidoglycan, whereas ficolin binds more strongly to fungal β-1,3-glucans, suggesting complementary functions in broad microbial surveillance [
5].
Both MBL and ficolin associate with a set of serine proteases—MBL-associated serine proteases (MASP)-1, -2, and -3—and their truncated non-catalytic isoforms such as Map19 and Map44, via the collagenous region. These MASPs connect pathogen recognition by MBL and ficolin to proteolytic activation of complement components C4, C2, and C3 [
6,
7].
From a phylogenetic perspective, MBL, ficolin, and MASP-1/3 have been identified in some invertebrates together with complement C3, indicating that the lectin pathway represents an ancient and fundamental mechanism of complement activation that predates the antibody-dependent classical pathway [
8,
9].
Bony fish, one of the most ancient vertebrate groups, possess a well-developed complement system that includes the lectin, classical, alternative, and cytolytic pathways [
10,
11]. Components of the lectin pathway identified in teleosts include two MBL homologues (MBL and galactose-binding lectin, GalBL), MASP2, MASP3, and MRP (later renamed Map44 in mammals) [
12,
13,
14].
Notably, despite extensive searches of the whole-genome sequences of pufferfish, zebrafish, medaka, carp, and other species, no orthologue of ficolin has been identified in bony fish. Because ficolin is present in ascidians—the closest invertebrate relatives of vertebrates—and in amphibians [
9,
15], ficolin appears to have been lost specifically in the bony fish lineage [
16].
In the present study, we attempted to isolate a putative ficolin-like lectin from the serum of the common carp (Cyprinus carpio), a teleost species, and identified it as a homologue of mammalian microfibrillar-associated protein 4 (MFAP4). We also discuss potential innate immune functions of teleost MFAP4 as an innate immune factor.
2. Materials and Methods
2.1. Carp Serum
Carp (approximately 1 kg) were purchased from a local fish farm. Serum was collected as previously described [
17]. Briefly, blood from the caudal vessels was allowed to clot for 30 min at room temperature and 30 min on ice, followed by centrifugation at 2,500 rpm for 10 min at 4 °C. The resulting serum was frozen in liquid nitrogen and stored at −80 °C until use.
2.2. Purification of a Ficolin-like Lectin from Carp Serum
A purification protocol for carp MBL [
14] was followed with minor modifications based on a reported procedure for human ficolin [
6]. Thawed serum was cleared by centrifugation (9,000 rpm, 15 min, 4 °C) and adjusted to 7% polyethyleneglycol (PEG) 4000. The 7% PEG precipitate was collected, dissolved in SB (50 mM Tris-HCl, pH 7.8, containing 200 mM NaCl and 10 mM CaCl₂), and applied to a GlcNAc-agarose (Sigma) column (1.2 x 5 cm) equilibrated with SB. After washing, bound proteins were sequentially eluted with SB containing 40 mM mannose, 400 mM mannose, and finally 150 mM GlcNAc.
2.3. SDS-PAGE and Western Blotting
SDS-PAGE was performed on 10% gels as described [
18]. Proteins were stained with Coomassie Brilliant Blue R-250. For western blotting, proteins were transferred to PVDF membranes and blocked with 5% skim milk in PBS. Membranes were incubated with anti-carp MFAP4 (1:500; see below) followed by HRP-conjugated anti-rabbit IgG (1:2000). After washing, antigen bands were visualized using 4-chloro-1-naphthol and hydrogen peroxide.
2.4. Preparation of Anti-Carp MFAP4 Polyclonal Antibody
Purified MFAP4 was separated by SDS-PAGE and visualized by imidazole/zinc negative staining [
19]. The 33-kDa bands were excised, washed with PBS, homogenized, emulsified in FCA, and injected subcutaneously into rabbits seven times at weekly intervals. Antiserum was collected one week after the final immunization.
2.5. N-Terminal Amino Acid Sequence Analysis
MFAP4 separated by SDS-PAGE was transferred to PVDF, stained with CBB, and subjected to automated Edman-degradation on a PPSQ-21 protein sequencer (Shimadzu).
2.6. Molecular Mass Estimation by Gel-Filtration
Purified MFAP4 was analyzed on a Superdex 200 column equilibrated with 10 mM Tris-HCl (pH 7.5) containing 150 mM NaCl and eluted at 0.5 ml/min. Fractions were analyzed by SDS-PAGE. Apoferritin (443 kDa), b-amylase (200 kDa), and ovalbumin (45 kDa) were used as molecular mass markers.
2.7. Determination of Specific Inhibitory Monosaccharides for Carp MFAP4
Purified MFAP4 (2 μg) was incubated with GlcNAc-agarose beads in SB containing monosaccharides (50–200 mM) at room temperature for 30 min. Supernatants were analyzed by SDS-PAGE to detect unbound MFAP4.
2.8. Preparation of Microbial Suspensions
The following bacteria were used for assay: Gram-positive bacteria (Micrococcus luteus, Staphylococcus aureus, Staphylococcus epidemidis, Streptococcus iniae, Streptococcus parauberis type I, Streptococcus parauberis type II, Streptococcus pyogenes, and Enterococcus faecalis), Gram-negative bacteria (Vibrio anguillarum, Aeromonas hydrophila, Aeromonas salmonicida, Edwarsiella tarda, Escherichia coli , and Klebsiella pneumoniae) and zymosan A.
Vibrio anguillarum was cultured in 1.5% NaCl-HI broth, genus Streptococcus was cultured in Tryptic soy broth, and all the other bacteria were cultured in HI broth. The bacterial suspension was killed by UV irradiation (70 J/cm2, 3 min, 20 times) in a petri dish, washed three times with sterile PBS. Zymosan A (Sigma) were suspended in SB at 10 mg/ml.
2.9. Binding Assay of MFAP4 to Microbial Targets
Purified MFAP4 was freed of inhibiting sugar (GlcNAc) by diafiltration through Amicon-Ultra 0.5 repeated five times using TBS and concentrated to 50 µg/ml. The MFAP4 solution (100 µl) was incubated with the same volume of microbe suspension (1 x 109 cells/ml) and zymosan (10 mg/ml) in SB at 25˚C for 1 h, followed by centrifugation at 10,000 rpm for 5 min. The target particles were washed three times with the same buffer, and bound proteins were eluted with 15 µl of SDS-sample buffer containing 50 mM dithiothreitol at 25˚C for 10 min. After centrifugation, the supernatant was collected, heated at 100˚C for 3 min, and applied to SDS-PAGE gels, followed by western blotting.
2.10. Cloning and Sequence Analysis
Reverse-transcription PCR (RT-PCR) was conducted using ExTaq polymerase (Takara) and primers shown in
Table 1. The amplified product was gel-purified and sequenced with MFAP4-Fw and MFAP4-Rv primers (
Table 1).
3. Results
3.1. Purification of a Ficolin-like GlcNAc-Binding Lectin from Carp Serum
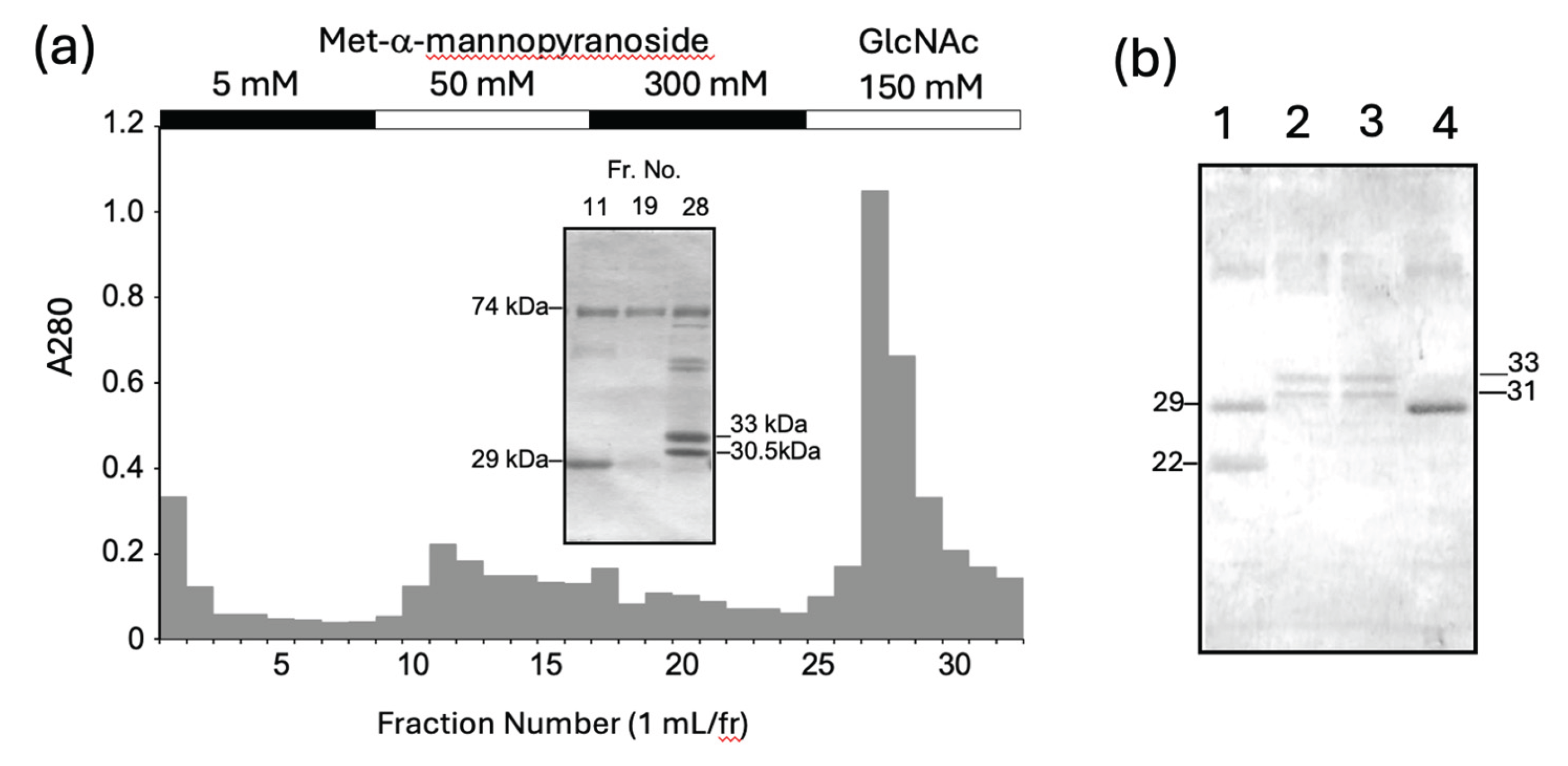
A GlcNAc-binding lectin was purified from carp serum, following the published protocol developed for mammalian MBL and L-ficolins/ficolin-2. After elution of MBL (29 kDa) from the GlcNAc-agarose column with mannose-containing buffer, a ficolin-like protein was eluted with 150 mM GlcNAc. The final preparation showed doublet major bands of molecular masses of 33 kDa and 31 kDa on SDS-PAGE under reducing conditions (
Figure 1a). Relative intensities of the 33- and 31-kDa bands showed variation among carp individuals. IgM, likely natural anti- GlcNAc antibody, was a major contaminant. The IgM content also varied among individuals (data not shown). Edman degradation of the 33 and 31-kDa polypeptides yielded identical N-terminal sequences, ITDGHDVDKPVDTSXVYKSG (X = undetermined). No collagen-like sequence motif (Gly-X-Y) was detected in the N-terminal region. In addition, digestion with collagenase type III (Worthington Biochemical) did not leave the protein (
Figure 1b).
3.2. Identification of Carp Ficolin-like Serum Lectin as a Novel MFAP4 Homologue
BLASTP search of non-redundant protein databases (nr) using the N-terminal amino acids sequence of purified carp ficolin-like lectin, limiting target sequences from teleost species, yielded top hits of MFAP4-like sequences of carp (XP_018962721.1). Based on the nucleotide sequence encoding this amino acid sequence, PCR primers were designed in 5‘- and 3‘- untranslated resion as listed in
Table 1. RT-PCR with the primer set from carp hepatopancreas RNA yielded a single amplicon of 948 bp with a nucleotide sequence including 735-bp open reading frame (deposited in DDBJ database under the accession number LC903834). Its deduced amino acid sequence contains a sequence stretch, CDCGHDVDKPVDCSDVYKSG, which is very close to the N-terminal sequence of the purified ficolin-like serum lectin (
Figure 2). BLASTP search against a teleost subset of ‘nr-cluster-seq’ protein database, using NCBI Blast server yielded several MFAP4-like sequences of carp and other cyprinid species as top hits. As shown in their multiple alignment (
Figure 2), the carp fciolin-like sequence showed higher similarity with KTF77344.1 sequence than with other sequences. In addition, only KTF77344.1 sequence shares the sequence corresponding to the protein N-terminal sequence with the carp ficolin-like sequence. Therefore, the ficolin-like lectin sequence isolated here is identified as a novel carp homologue of MFAP4, designated as carp MFAP4-lectin (MFAP4Lec).
3.3. Oligomeric Structure of Carp MFAP4Lec
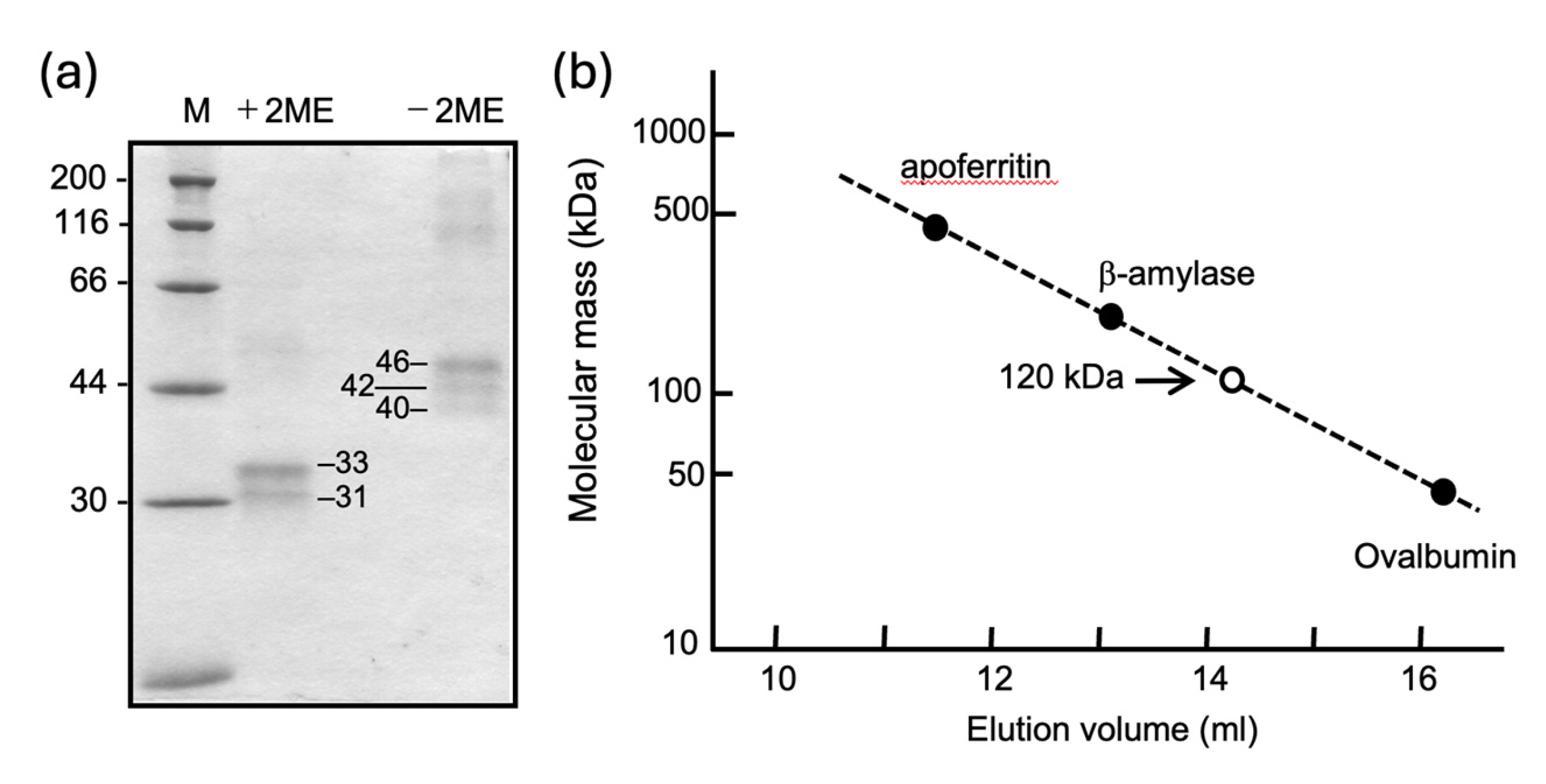
Purified carp MFAP4Lec protein gave doublet bands of 33 and 31 kDa under reducing conditions, and triplet bands of apparent molecular masses of 40, 42, and 46 kDa. This suggests a disulfide-linked dimeric structure of carp MFAP4. Upon gel-filtration through a Superdex 200 column, carp MFAP4Lec was eluted at the volume corresponding to molecular mass of about 120 kDa (
Figure 3). Taken those results together, it is suggested that native MFAP4Lec in the serum is composed of two disulfide-linked dimers that are noncovalently associated.
3.4. Monosaccharide Binding Specificity of Carp MFAP4Lec
Binding of carp MFAP4Lec to GlcNAc-agarose was inhibited by GlcNAc and GalNAc in a dose-dependent manner but not by mannose, glucose or galactose (
Figure 4). These results indicate that carp MFAP4Lec is a GlcNAc/GalNAc-specific lectin as is ficolin. Unlike mammalian ficolin, MFAP4Lec binding was not inhibited by EDTA, indicating its Ca
2⁺-independent sugar recognition (Data not shown).
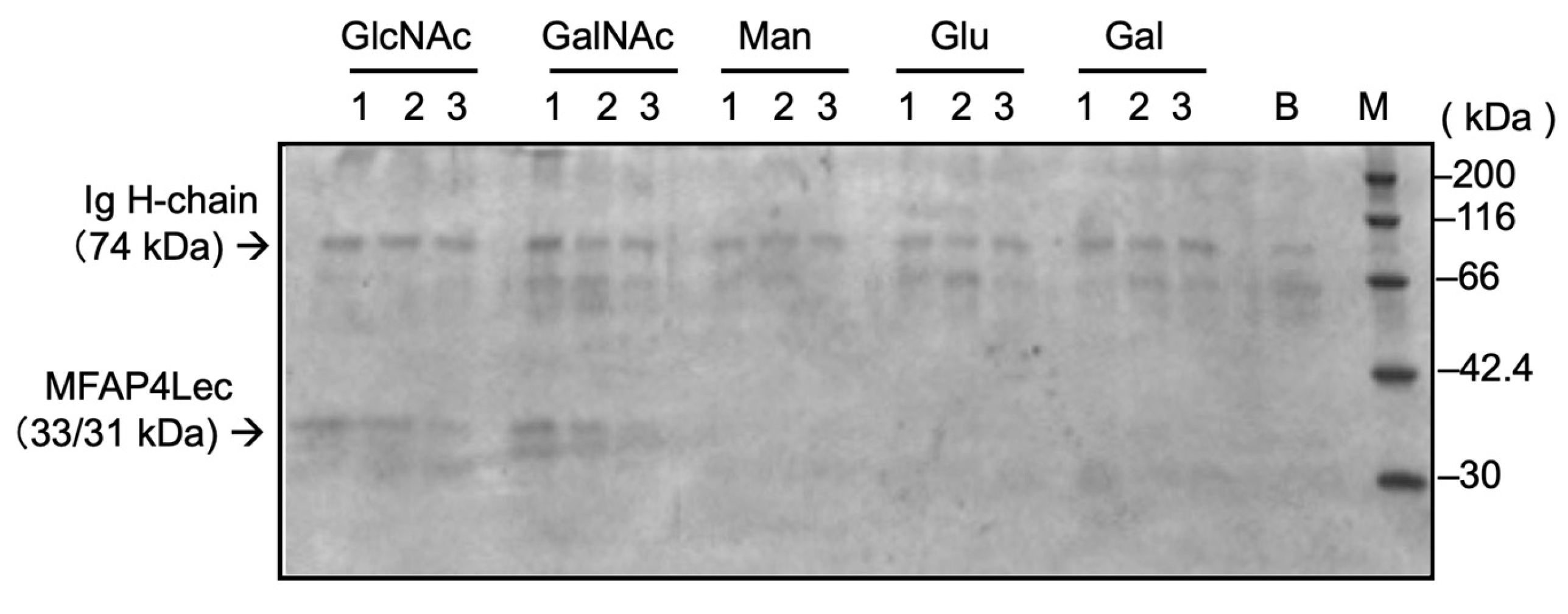
3.5. Microbial-Binding Specificity
Binding of carp MFAP4Lec was tested against a panel of microbes. Western blotting of bound fractions showed specific binding to
Vibrio anguillarum, a Gram-negative fish pathogen, whereas no detectable binding was observed for other bacteria or zymosan (
Figure 4A). Binding to
Vibrio was inhibited by GlcNAc but not by EDTA, consistent with carbohydrate-binding results (
Figure 4B).
Figure 5.
Binding characteristics of MFAP4Lec in carp serum to microbial targets. Carp serum was incubated with various species of UV-killed bacteria, and bound proteins were eluted with SDS-containing buffer and analyzed by western blotting using anti-carp MFAP4Lec antibody. (a) Specificity towards various bacteria. Lane M, marker protein; lane 1, carp serum as a positive control; lane 2, V. anguillarum; lane 3, E. tarda; lane 4, A. hydrophila; lane 5, E. coli; lane 6, S. aureus; lane 7, zymosan. (b) Carp serum was incubated with V. anguillarum in the presence or absence of EDTA and GlcNAc, and bound proteins were analyzed by western blotting as above. Lane M, marker proteins; lane 1, purified MFAP4Lec as a positive control; lane 2, serum + Vibrio; lane 3, serum + Virbio + EDTA; lane 4, serum + Vibrio + GlcNAc; lane 5, buffer + Vibrio as negative control. The migrated position of MFAP4Lec (31 kDa) is shown on the left of each panel. Note that this lot of carp serum did not contain 33 kDa band of MFAP4Lec.
Figure 5.
Binding characteristics of MFAP4Lec in carp serum to microbial targets. Carp serum was incubated with various species of UV-killed bacteria, and bound proteins were eluted with SDS-containing buffer and analyzed by western blotting using anti-carp MFAP4Lec antibody. (a) Specificity towards various bacteria. Lane M, marker protein; lane 1, carp serum as a positive control; lane 2, V. anguillarum; lane 3, E. tarda; lane 4, A. hydrophila; lane 5, E. coli; lane 6, S. aureus; lane 7, zymosan. (b) Carp serum was incubated with V. anguillarum in the presence or absence of EDTA and GlcNAc, and bound proteins were analyzed by western blotting as above. Lane M, marker proteins; lane 1, purified MFAP4Lec as a positive control; lane 2, serum + Vibrio; lane 3, serum + Virbio + EDTA; lane 4, serum + Vibrio + GlcNAc; lane 5, buffer + Vibrio as negative control. The migrated position of MFAP4Lec (31 kDa) is shown on the left of each panel. Note that this lot of carp serum did not contain 33 kDa band of MFAP4Lec.

4. Discussion
In this study, we purified a lectin from carp serum using a protocol analogous to that employed for mammalian L-ficolin. Contrary to ficolin, however, the purified protein lacked a collagen-like domain and consisted solely of a fibrinogen-like domain, identifying it as a novel homologue of MFAP4. This result indicates that teleost fish possess an MFAP4-homologue, rather than ficolin, as a GlcNAc-specific serum lectin. This conclusion is further supported by teleost whole-genome database analyses, which have revealed no bona fide ficolin genes comprising an N-terminal collagen-like domain followed by a C-terminal fibrinogen-like domain [
16].
The alignment (
Figure 2) of carp MFAP4-like sequences indicates the presence of multiple isoforms of MFAP4 homologues in carp genome, in agreement with the findings in catfish [
20]. It is intriguing to note that MFAP4Lec cloned in this study showed closest similarity with the ‘hypothetical protein (KTF77344.1)’ of carp, sharing the sequence stretch corresponding to N-terminal protein sequence over 20 residues. Only a marked difference between MFAP4Lec and the ‘hypothetical protein’ is a 24-residues indel, predicting molecular mass difference by 3137 Da. The long and short isoforms might encode 33 kDa and 31 kDa polypeptides of purified carp MFAP4Lec. It is unknown if the long and short forms are products of alternative splicing or from distinct alleles.
In mammals, MFAP4 is an extracellular matrix protein that plays essential roles in elastic fiber homeostasis, integrin-mediated signaling, and cancer. It is highly expressed in elastic fiber–rich tissues such as skin, arteries, lungs, and heart, where it stabilizes elastic fibers by promoting elastin self-assembly. MFAP4 also modulates cellular behavior through RGD-dependent integrins, including the promotion of vascular smooth muscle cell proliferation and migration, as reviewed in [
16].
In contrast, the physiological functions of MFAP4 in teleosts remain insufficiently characterized. Previous studies have suggested an immunological role, in addition to homeostatic functions [
21,
22]. For example, MFAP4 expression is upregulated in the liver, kidney, and spleen of catfish and tilapia following bacterial infection [
20,
23]. Moreover, recombinant tilapia MFAP4 expressed in
E. coli has been shown to recognize and agglutinate the fish pathogens
Streptococcus agalactiae and
Aeromonas hydrophila [
23], as well as opsonic activity as in human ficolin [
24].
Here, we demonstrated that carp MFAP4Lec is present in serum as a lectin and can recognize the Gram-negative fish pathogen
V. anguillarum in a Ca
2⁺-independent manner. Together with previous findings in catfish and tilapia [
20,
23], these results strongly suggest that some MFAP4 homologues can serve as a serum pattern-recognition molecule in the innate immune system of teleosts. However, in contrast to the reported agglutinating activity of recombinant tilapia MFAP4 [
23], purified native carp MFAP4Lec showed no agglutination toward the bacterial species tested in this study (data not shown). Whether this discrepancy reflects differences between recombinant and native proteins or structural divergence among MFAP4 orthologs from different fish species remains unresolved.
The functional significance of possessing MFAP4Lec—rather than ficolin—in carp innate immunity requires careful consideration. Both ficolins and MFAP4 recognize GlcNAc and GalNAc (
Figure 4 and reference [
4]). Regarding PAMP specificity, human L-ficolin binds b-1,3-glucan and functions as a fungal recognition molecule, complementing MBL, which recognizes bacteria via peptidoglycan [
5]. In contrast, carp MFAP4Lec did not bind zymosan, suggesting that its PAMP specificity differs substantially from that of human ficolin. In addition, the downstream effector mechanisms following ligand recognition are expected to diverge between the two proteins. Ficolin associates with MASPs through their collagen-like domains and activates the lectin complement pathway, particularly via MASP-2 [
6]. Because MFAP4Lec lacks a collagen-like domain, it is highly unlikely to activate complement, and the mechanisms by which MFAP4Lec mediates the elimination of its target ligands remain to be elucidated. Determining whether native MFAP4Lec exhibits the opsonic activities reported for recombinant tilapia MFAP4 will be crucial for clarifying its immunological roles, in addition to identification of receptors for MFAP4Lec.
In conclusion, this study provides evidence—for the first time using natural purified protein—that MFAP4Lec may partly replaces ficolin as a serum lectin in teleosts and may function as a microorganism-recognition molecule in their innate immune defense.
Author Contributions
Conceptualization, M.N. and M.K.; methodology, M.K., A.I., D.H., M.N.; investigation, M.K., T.S., T.N., M.N.; data curation, M.K., M.N.; writing—original draft preparation, M.K., M.N.; writing—review and editing, M.N.; supervision, T.S., T.N., M.N.; funding acquisition, M.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by MEXT Grants-in-Aid for Scientific Research (8760182 and 16380135) to M.N.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable
Data Availability Statement
Not applicable
Acknowledgments
The authors are grateful to Mrs. Akito Ichiki and Daisaku Hatanaka for their technical assistance, and to Ms. Suzuna Fujisaki, a QFC-SP scholar of Kyushu University for a help in RT-PCR.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| MFAP4 |
Microfibrillar-associated protein 4 |
| RGD |
Arginine-Glycine-Aspartic acid motif |
| PAMPs |
Pathogen-associated molecular patterns |
References
- Peiffer, A.L. , Dugan, A.E., Kiessling, L.L. Soluble human lectins at the host-microbe interface. Annu Rev Biochem. 2024, 93, 565–601. [Google Scholar] [CrossRef]
- Fujita, T. , Matsushita, M., Endo, Y. The lectin-complement pathway–its role in innate immunity and evolution. Immunol Rev. 2004, 198, 185–202. [Google Scholar] [CrossRef]
- Eisen, D.P. , Minchinton, R.M. Impact of mannose-binding lectin on susceptibility to infectious diseases. Clin Infect Diseases. 2003, 37, 1496–1505. [Google Scholar] [CrossRef]
- Krarup, A. , Thiel, S., Hansen, A., Fujita, T., Jensenius, J.C., L-ficolin is a pattern recognition molecule specific for acetyl groups. J Biol Chem. 2004, 279, 47513–47519. [Google Scholar] [CrossRef]
- Ma, Y.G. , Cho, M.Y., Zhao, M., Park, J.W., Matsushita, M., Fujita, T., Lee, B.L. Human mannose-binding lectin and L-ficolin function as specific pattern recognition proteins in the lectin activation pathway of complement. J Biol Chem. 2004, 279, 25307–25312. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, M. , Endo, Y., Fujita, T. Cutting edge: complement-activating complex of ficolin and mannose-binding lectin-associated serine protease. J Immunol. 2000, 164, 2281–2284. [Google Scholar] [CrossRef] [PubMed]
- Degn, S.E. , Hansen, A.G., Steffensen, R., Jacobsen, C., Jensenius, J.C., Thiel, S. MAp44, a human protein associated with pattern recognition molecules of the complement system and regulating the lectin pathway of complement activation. J Immunol. 2009, 183, 7371–7378. [Google Scholar] [CrossRef]
- Sekine, H. , Kenjo, A., Azumi, K., Ohi, G., Takahashi, M., Kasukawa, R., Ichikawa, N., Nakata, M., Mizuochi, T., Matsushita, M., Endo, Y., Fujita, T. An ancient lectin-dependent complement system in an ascidian: novel lectin isolated from the plasma of the solitary ascidian, Halocynthia roretzi. J Immunol. 2001, 167, 4504–4510. [Google Scholar] [CrossRef] [PubMed]
- Kenjyo, A. , Takahashi, M., Matsushita, M., Endo, Y., Nakata, M., Mizuochi, T., Fujita, T. Cloning and characterization of novel ficolins form the solitary ascidian, Halocynthia roretzi. J Biol Chem. 2001, 276, 19959–19965. [Google Scholar] [CrossRef]
- Nakao, M. , Tsujikura, M., Ichiki, S., Vo, T.K., Somamoto, T. The complement system in teleost fish: progress of post-homolog-hunting researches. Dev. Comp. Immunol, 1296. [Google Scholar] [CrossRef]
- Li, M.F. , Zhang, H.Q. An overview of complement systems in teleosts. Dev. Comp. Immunol. 2022, 137, 104520. [Google Scholar] [CrossRef]
- Endo, Y. , Takahashi, M., Nakao, M., Saiga, H., Sekine, H., Matsushita, M., Nonaka, M., Fujita, T. Two lineages of mannose-binding lectin-associated serine protease (MASP) in vertebrates. J Immunol. 1998, 161, 4924–4930. [Google Scholar] [CrossRef] [PubMed]
- Nagai, T. , Mutsuro, J., Kimura, M., Kato, Y., Fujiki, K., Yano, T., Nakao, M. A novel truncated isoform of the mannose-binding lectin-associated serine protease (MASP) from the common carp (Cyprinus carpio). Immunogenetics. 2000, 51, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Nakao, M. , Kajiya, T., Sato, Y., Somamoto, T., Kato-Unoki, Y., Matsushita, M., Nakata, M., Fujita, T., Yano, T. Lectin pathway of bony fish complement: identification of two homologs of the mannose-binding lectin associated with MASP2 in the common carp (Cyprinus carpio). J Immunol. 2006, 177, 5471–5479. [Google Scholar] [CrossRef] [PubMed]
- Kakinuma, Y. , Endo, Y., Takahashi, M., Nakata, M., Matsushita, M., Takenoshita, S., Fujita, T. Molecular cloning and characterization of novel ficolins from Xenopas laevis. Immunogenetics. 2003, 55, 29–37. [Google Scholar] [CrossRef]
- Mohammadi, A. , Sorensen, G.L, Pilecki, B. MFAP4-mediated effects in elastic fiber homeostasis, integrin signaling and cancer, and its role in teleost fish. Cells. 2022, 11, 2115. [Google Scholar] [CrossRef]
- Yano, T. , Nakao, M. Isolation of a carp complement protein homologous to mammalian factor D. Mol Immunol. 1994, 31, 337–342. [Google Scholar]
- Laemmli, UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Fernandez-Patron, C. , Castellanos-Serra, L., Rodriguez, P. Reverse staining of sodium dodecyl sulfate polyacrylamide gels by imidazole-zinc salts: sensitive detection of unmodified proteins. Biotechniques. 1992, 12, 564–573. [Google Scholar]
- Zakrzewska, A. , Cui, C., Stockhammer, O.W., Benard, E.L., Spaink, H.P., Meijer, A.H. Macrophage-specific gene functions in Spi1-directed innate immunity. Blood. 2010, 116, e1–e11. [Google Scholar] [CrossRef]
- Ong, S.L.M. , de Vos, I., Meroshini, M., Poobalan, Y., Dunn, N.R. Microfibril-associated glycoprotein 4 (Mfap4) regulates haematopoiesis in zebrafish. Sci. Rep. 2020, 10, 11801. [Google Scholar] [CrossRef]
- Niu, D. , Peatman, E., Liu, H., Lu, J., Kucuktas, H., Liu, S., Sun, F., Zhang, H., Feng, T., Zhou, Z., Terhune, J., Waldbieser, G., Li, J., Liu, Z. Microfibrillar-associated protein4 (MFAP4) genes in catfish play a novel role in innate immune responses. Dev. Comp. Immunol. 2011, 35, 568–579. [Google Scholar] [CrossRef] [PubMed]
- Wu, H. , Mu, L., Yin, X., Han, K., Yan, F., Zhou, E., Han, B., Guo, Z., Ye, J. A microfibril-associated glycoprotein 4 (MFAP4) from Nile tilapia (Oreochromis niloticus) possesses agglutination and opsonization ability to bacterial pathogens. Fish Shellfish Immunol. 2020, 104, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, M. , Endo, Y., Taira, S., Sato, Y., Fujita, T., Ichikawa, N., Nakata, M., Mizouchi, T. A novel human serum lectin with collagen- and fibrinogen-like domains that functions as an opsonin. J. Biol. Chem., 1996, 271, 2448–2454. [Google Scholar] [CrossRef] [PubMed]
Figure 1.
Purification of ficolin-like GlcNAc-specific lectin from carp serum by affinity chromatography. (a) Elution profile of GlcNAc-agarose affinity chromatography. Fractions eluted with 5, 50, and 300 mM methyl-a-mannopyranoside, and with 150 mM GlcNAc were analyzed by SDS-PAGE. Electrophoregram of fractions 11, 19, and 28 are inserted. (b) SDS-PAGE (12% gel) of collagenase-digest of purified carp MBL and ficolin-like lectin. Lanes 1 and 4, MBL; lanes 2 and 3, carp ficolin-like lectin; lanes 1 and 2, collagenase-treated; lanes 3 and 4, non-treated control.
Figure 1.
Purification of ficolin-like GlcNAc-specific lectin from carp serum by affinity chromatography. (a) Elution profile of GlcNAc-agarose affinity chromatography. Fractions eluted with 5, 50, and 300 mM methyl-a-mannopyranoside, and with 150 mM GlcNAc were analyzed by SDS-PAGE. Electrophoregram of fractions 11, 19, and 28 are inserted. (b) SDS-PAGE (12% gel) of collagenase-digest of purified carp MBL and ficolin-like lectin. Lanes 1 and 4, MBL; lanes 2 and 3, carp ficolin-like lectin; lanes 1 and 2, collagenase-treated; lanes 3 and 4, non-treated control.
Figure 2.
Multiple alignment of deduced amino acid sequences of carp MFAP4-like lectin (MFAP4Lec) isolated in this study with carp MFAP4-like sequences retrieved by BLASTP search (KTF77344.1, hypothetical protein cypCar_00044155 [Cyprinus carpio]; XP_042621506.1, microfibril-associated glycoprotein 4-like [Cyprinus carpio]; XP_042621690.1, microfibril-associated glycoprotein 4-like [Cyprinus carpio]; XP_042621475.1, microfibril-associated glycoprotein 4-like isoform X1 [Cyprinus carpio]; XP_018941414.2, microfibril-associated glycoprotein 4-like [Cyprinus carpio]) Asterisks show residues shared by all the sequences, and colon and period denote highly and moderately conserved residues, respectively. Dashes are gaps introduced for maximum similarity. Residue number is shown on the left of each sequence. N-terminal sequence obtained by Edman-degradation of the purified lectin is also aligned on the top raw, where lower case residues do not match the deduced sequence of carp MFAP4Lec cloned in this study.
Figure 2.
Multiple alignment of deduced amino acid sequences of carp MFAP4-like lectin (MFAP4Lec) isolated in this study with carp MFAP4-like sequences retrieved by BLASTP search (KTF77344.1, hypothetical protein cypCar_00044155 [Cyprinus carpio]; XP_042621506.1, microfibril-associated glycoprotein 4-like [Cyprinus carpio]; XP_042621690.1, microfibril-associated glycoprotein 4-like [Cyprinus carpio]; XP_042621475.1, microfibril-associated glycoprotein 4-like isoform X1 [Cyprinus carpio]; XP_018941414.2, microfibril-associated glycoprotein 4-like [Cyprinus carpio]) Asterisks show residues shared by all the sequences, and colon and period denote highly and moderately conserved residues, respectively. Dashes are gaps introduced for maximum similarity. Residue number is shown on the left of each sequence. N-terminal sequence obtained by Edman-degradation of the purified lectin is also aligned on the top raw, where lower case residues do not match the deduced sequence of carp MFAP4Lec cloned in this study.

Figure 3.
Molecular mass estimation of carp MFAP4Lec by SDS-PAGE and gel-filtration. (a) Purified MFAP4Lec was run on 10% SDS-gel under reducing and non-reducing conditions. Molecular masses of marker proteins are shown on the left. (b) Standard curve of molecular mass estimation by gel-filtration on Superdex 200, obtained using marker proteins (apoferritin, 443 kDa; beta-amylase, 200 kDa, ovalbumin, 45 kDa). Elution volution of carp MFAP4Lec is plotted as an open circle.
Figure 3.
Molecular mass estimation of carp MFAP4Lec by SDS-PAGE and gel-filtration. (a) Purified MFAP4Lec was run on 10% SDS-gel under reducing and non-reducing conditions. Molecular masses of marker proteins are shown on the left. (b) Standard curve of molecular mass estimation by gel-filtration on Superdex 200, obtained using marker proteins (apoferritin, 443 kDa; beta-amylase, 200 kDa, ovalbumin, 45 kDa). Elution volution of carp MFAP4Lec is plotted as an open circle.
Figure 4.
Determination of monosaccharides that inhibit binding of carp MFAP4Lec to GlcNAc-agarose. Purified carp MFAP4Lec was incubated with GlcNAc-agarose in the presence of monosaccharides, GlcNAc, GalNAc, mannose (Man), glucose (Glu) and galactose (Gal). at concentrations of 200 mM (Lanes 1), 100 mM (Lanes 2), and 50 mM (Lanes 3), and after centrifugation, the supernatant was run on a 10% SDS-gel. Lane B is a control incubated with buffer in place of the monosaccharide. Lane M shows marker proteins with their molecular masses on the right.
Figure 4.
Determination of monosaccharides that inhibit binding of carp MFAP4Lec to GlcNAc-agarose. Purified carp MFAP4Lec was incubated with GlcNAc-agarose in the presence of monosaccharides, GlcNAc, GalNAc, mannose (Man), glucose (Glu) and galactose (Gal). at concentrations of 200 mM (Lanes 1), 100 mM (Lanes 2), and 50 mM (Lanes 3), and after centrifugation, the supernatant was run on a 10% SDS-gel. Lane B is a control incubated with buffer in place of the monosaccharide. Lane M shows marker proteins with their molecular masses on the right.
Table 1.
Oligonucleotide Primers used in this study.
Table 1.
Oligonucleotide Primers used in this study.
| Name |
Sequence |
| MFAP4-Fw |
TCGTCTACAGCTGGAGAACACC |
| MFAP4-Rv |
TCTAGGAGTTTACTGAAAGTTTATTTAGTGAAG |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).