Submitted:
27 November 2025
Posted:
27 November 2025
You are already at the latest version
Abstract
Keywords:
Introduction
Results
NUP96-SNAP Tag Does Not Affect the HIV-1 Nuclear Import Kinetics
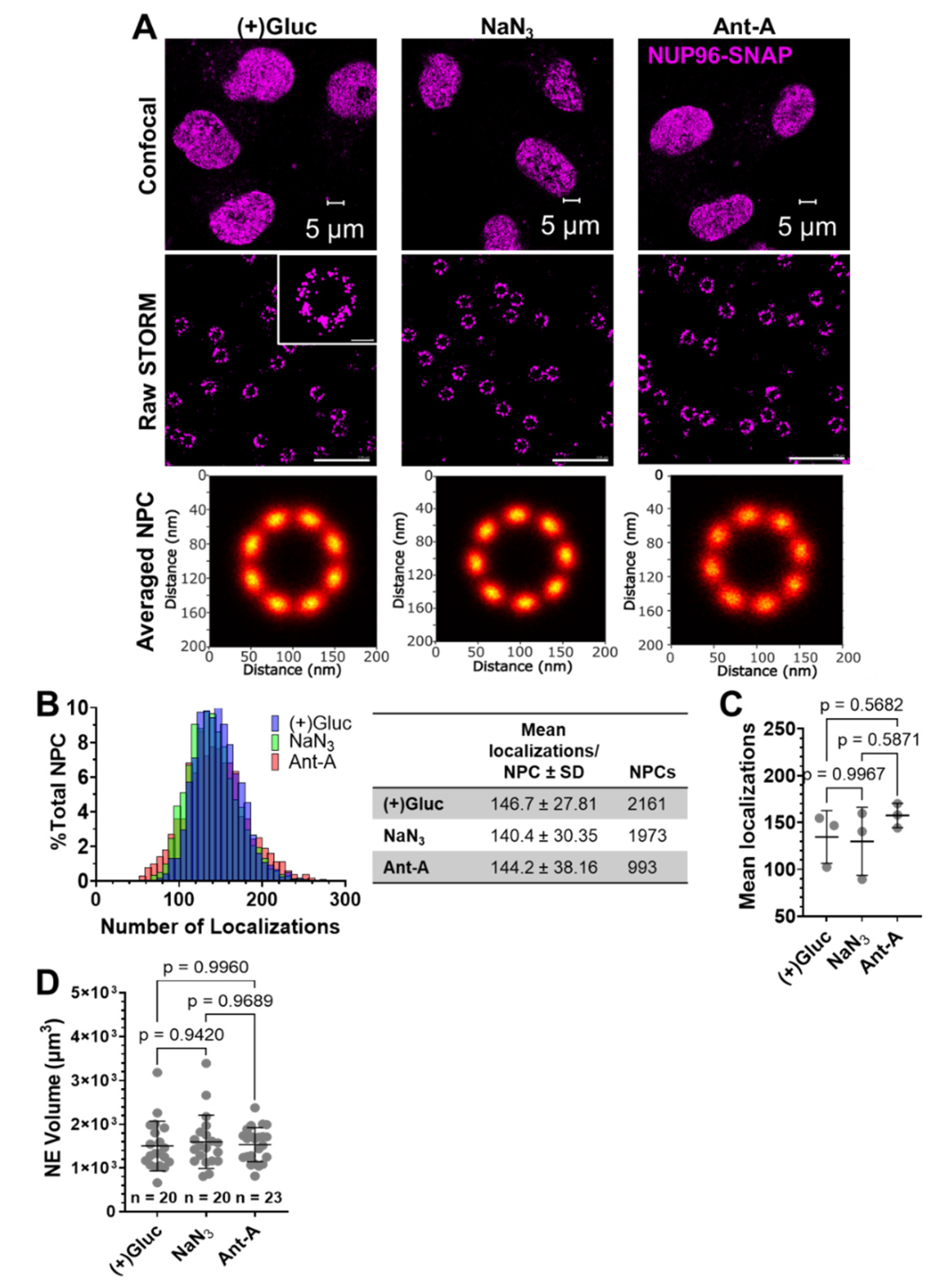
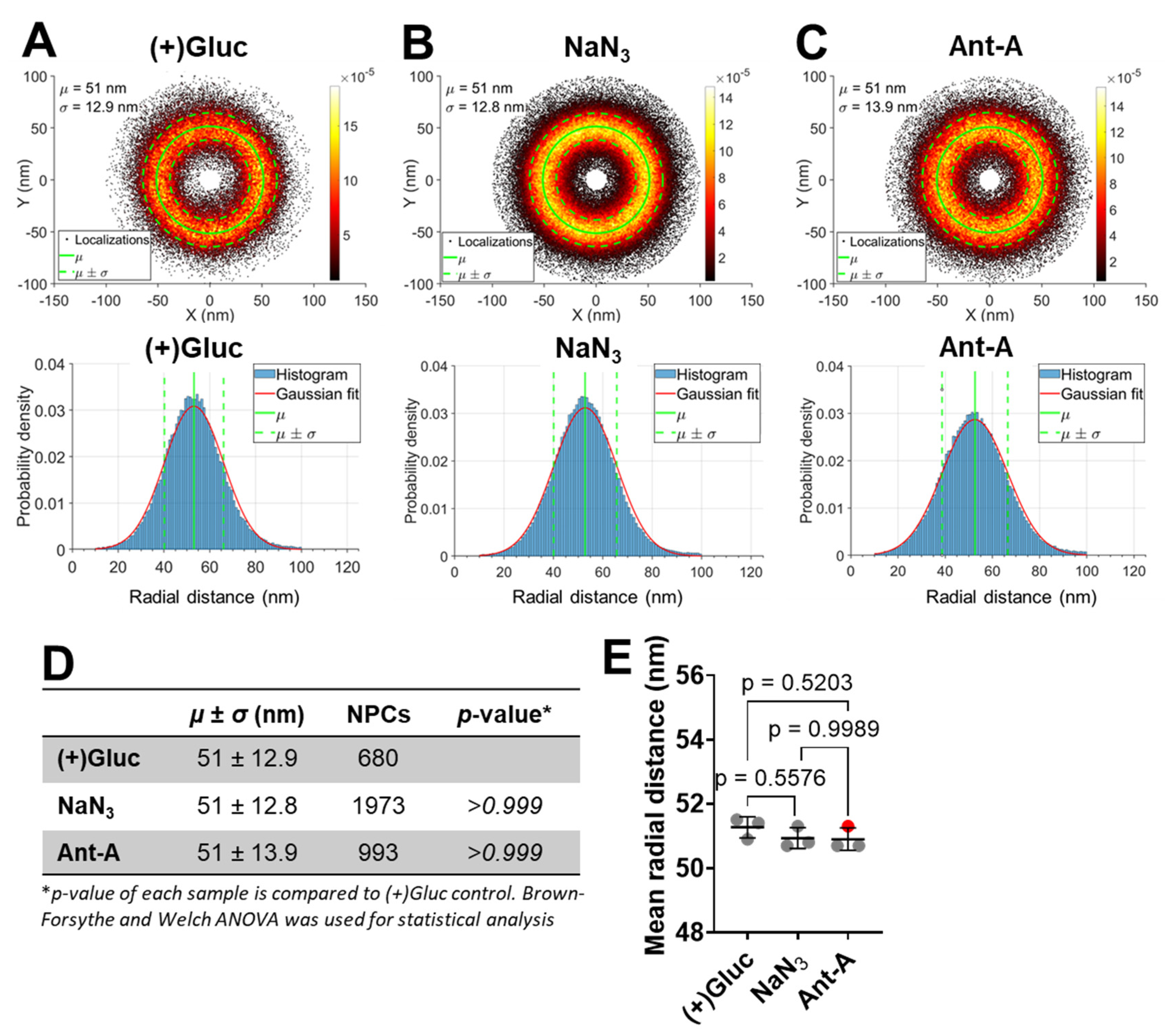
ATP Depletion Does Not Cause Detectable Changes in NPC Size
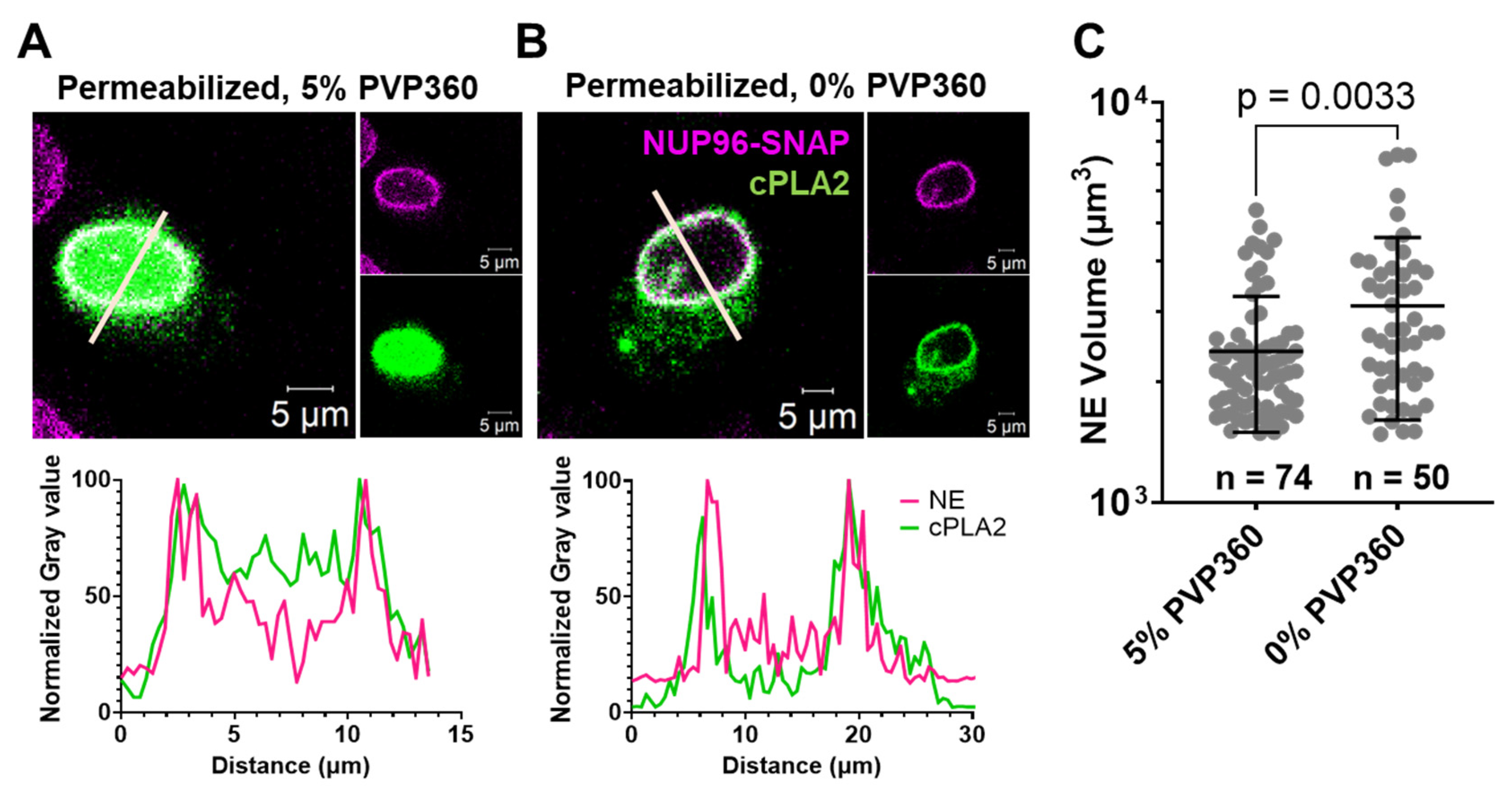
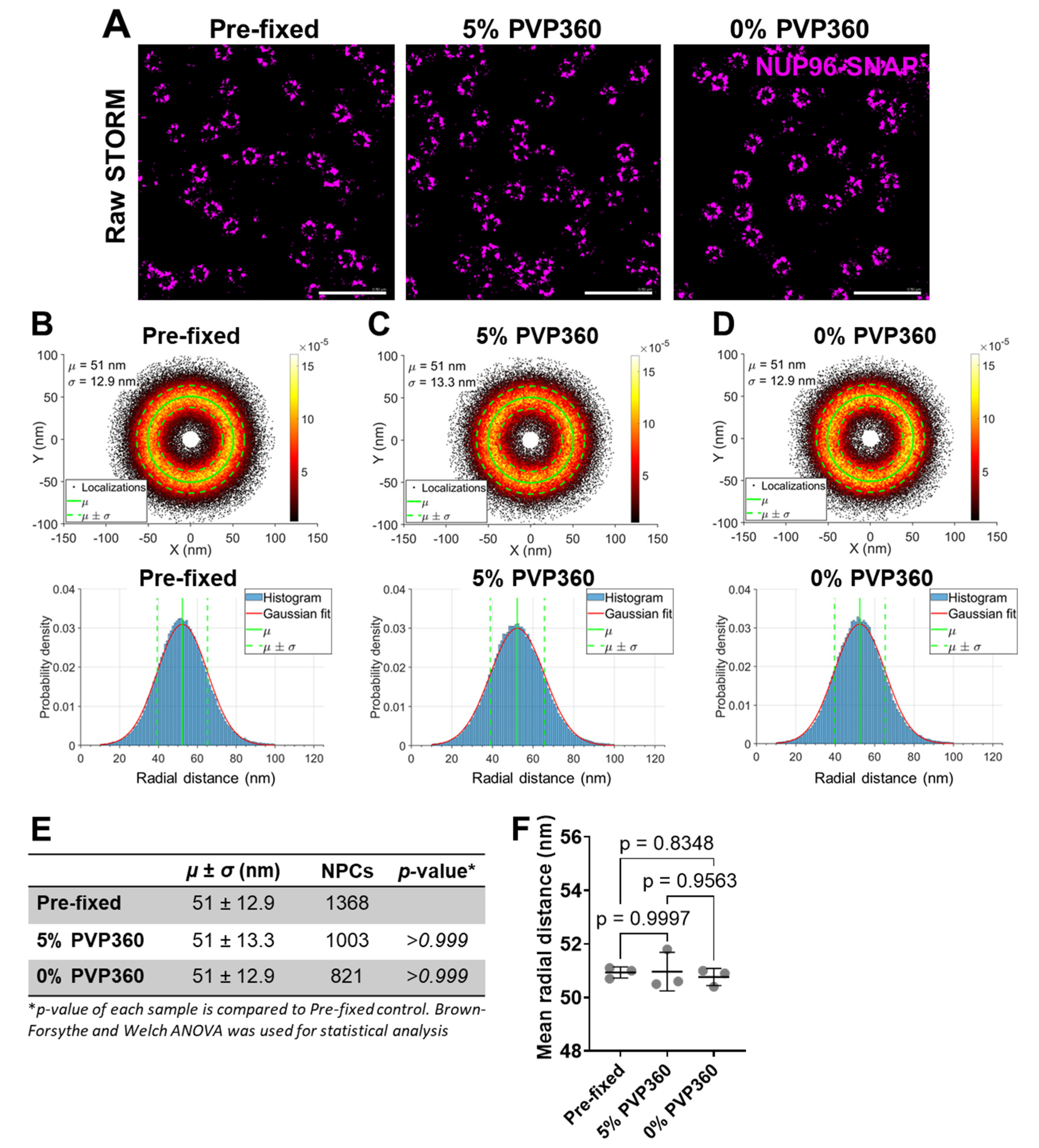
NPC Size Is Resistant to Stretching and Shrinking of the Nuclear Envelope
Discussion
Methods
Cells and Reagents
Production and Characterization of HIV-1 Pseudoviruses
ATP Depletion
Osmotic Stretching and Shrinkage of Nuclear Envelope
Preparation of Samples for Imaging
Confocal and STORM Imaging
Image Analysis and Statistics
Supplementary Materials
Acknowledgements
References
- M. Beck, E. Hurt, The nuclear pore complex: understanding its function through structural insight. Nat Rev Mol Cell Biol 18, 73–89 (2017).
- K. E. Knockenhauer, T. U. Schwartz, The Nuclear Pore Complex as a Flexible and Dynamic Gate. Cell 164, 1162–1171 (2016).
- D. H. Lin, A. Hoelz, The Structure of the Nuclear Pore Complex (An Update). Annu. Rev. Biochem. 88, 725–783 (2019).
- G. Paci, J. Caria, E. A. Lemke, Cargo transport through the nuclear pore complex at a glance. Journal of Cell Science 134, jcs247874 (2021).
- K. H. Bui, A. von Appen, A. L. DiGuilio, A. Ori, L. Sparks, M.-T. Mackmull, T. Bock, W. Hagen, A. Andrés-Pons, J. S. Glavy, M. Beck, Integrated Structural Analysis of the Human Nuclear Pore Complex Scaffold. Cell 155, 1233–1243 (2013).
- G. Celetti, G. Paci, J. Caria, V. VanDelinder, G. Bachand, E. A. Lemke, The liquid state of FG-nucleoporins mimics permeability barrier properties of nuclear pore complexes. Journal of Cell Biology 219, e201907157 (2020).
- S. Frey, R. P. Richter, D. Görlich, FG-Rich Repeats of Nuclear Pore Proteins Form a Three-Dimensional Meshwork with Hydrogel-Like Properties. Science 314, 815–817 (2006).
- N. Pante, M. Kann, Nuclear Pore Complex Is Able to Transport Macromolecules with Diameters of ϳ39 nm. Molecular Biology of the Cell 13, 10 (2002).
- G. Paci, T. Zheng, J. Caria, A. Zilman, E. A. Lemke, Molecular determinants of large cargo transport into the nucleus. eLife 9, e55963 (2020).
- V. Zila, E. Margiotta, B. Turoňová, T. G. Müller, C. E. Zimmerli, S. Mattei, M. Allegretti, K. Börner, J. Rada, B. Müller, M. Lusic, H.-G. Kräusslich, M. Beck, Cone-shaped HIV-1 capsids are transported through intact nuclear pores. Cell 184, 1032-1046.e18 (2021).
- A. P. Schuller, M. Wojtynek, D. Mankus, M. Tatli, R. Kronenberg-Tenga, S. G. Regmi, P. V. Dip, A. K. R. Lytton-Jean, E. J. Brignole, M. Dasso, K. Weis, O. Medalia, T. U. Schwartz, The cellular environment shapes the nuclear pore complex architecture. Nature 598, 667–671 (2021).
- J. Mahamid, S. Pfeffer, M. Schaffer, E. Villa, R. Danev, L. Kuhn Cuellar, F. Förster, A. A. Hyman, J. M. Plitzko, W. Baumeister, Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351, 969–972 (2016).
- C. E. Zimmerli, M. Allegretti, V. Rantos, S. K. Goetz, A. Obarska-Kosinska, I. Zagoriy, A. Halavatyi, G. Hummer, J. Mahamid, J. Kosinski, M. Beck, Nuclear pores dilate and constrict in cellulo. Science 374, eabd9776 (2021).
- K. J. Morgan, E. Carley, A. N. Coyne, J. D. Rothstein, C. P. Lusk, M. C. King, Visualizing nuclear pore complex plasticity with pan-Expansion Microscopy. [Preprint] (2024). [CrossRef]
- R. Taniguchi, C. Orniacki, J. P. Kreysing, V. Zila, C. E. Zimmerli, S. Böhm, B. Turoňová, H.-G. Kräusslich, V. Doye, M. Beck, Nuclear pores safeguard the integrity of the nuclear envelope. Nat Cell Biol, doi: 10.1038/s41556-025-01648-3 (2025).
- J. Sellés, M. Penrad-Mobayed, C. Guillaume, A. Fuger, L. Auvray, O. Faklaris, F. Montel, Nuclear pore complex plasticity during developmental process as revealed by super-resolution microscopy. Sci Rep 7, 14732 (2017).
- A. Löschberger, S. van de Linde, M.-C. Dabauvalle, B. Rieger, M. Heilemann, G. Krohne, M. Sauer, Super-resolution imaging visualizes the eightfold symmetry of gp210 proteins around the nuclear pore complex and resolves the central channel with nanometer resolution. Journal of Cell Science 125, 570–575 (2012).
- Z. Hou, Y. Shen, S. Fronik, J. Shen, J. Shi, J. Xu, L. Chen, N. Hardenbrook, C. Thompson, S. Neumann, A. N. Engelman, C. Aiken, P. Zhang, Correlative In Situ Cryo-ET Reveals Cellular and Viral Remodeling Associated with Selective HIV-1 Core Nuclear Import. [Preprint] (2025). [CrossRef]
- J. P. Kreysing, M. Heidari, V. Zila, S. Cruz-León, A. Obarska-Kosinska, V. Laketa, L. Rohleder, S. Welsch, J. Köfinger, B. Turoňová, G. Hummer, H.-G. Kräusslich, M. Beck, Passage of the HIV capsid cracks the nuclear pore. Cell 188, 930-943.e21 (2025).
- Z. Hou, S. Fronik, Y. Shen, L. Chen, C. Thompson, S. Neumann, P. Zhang, Direct visualization of HIV-1 core nuclear import and its interplay with the nuclear pore. EMBO Rep, doi: 10.1038/s44319-025-00567-6 (2025).
- V. J. Sabinina, M. J. Hossain, J.-K. Hériché, P. Hoess, B. Nijmeijer, S. Mosalaganti, M. Kueblbeck, A. Callegari, A. Szymborska, M. Beck, J. Ries, J. Ellenberg, Three-dimensional superresolution fluorescence microscopy maps the variable molecular architecture of the nuclear pore complex. MBoC 32, 1523–1533 (2021).
- X. Ye, M. Guan, Y. Guo, X. Liu, K. Wang, T. Chen, S. Zhao, L. Chen, Live-cell super-resolution imaging unconventional dynamics and assemblies of nuclear pore complexes. swwlxb 9, 206 (2023).
- A. Von Appen, J. Kosinski, L. Sparks, A. Ori, A. L. DiGuilio, B. Vollmer, M.-T. Mackmull, N. Banterle, L. Parca, P. Kastritis, K. Buczak, S. Mosalaganti, W. Hagen, A. Andres-Pons, E. A. Lemke, P. Bork, W. Antonin, J. S. Glavy, K. H. Bui, M. Beck, In situ structural analysis of the human nuclear pore complex. Nature 526, 140–143 (2015).
- B. Koch, B. Nijmeijer, M. Kueblbeck, Y. Cai, N. Walther, J. Ellenberg, Generation and validation of homozygous fluorescent knock-in cells using CRISPR–Cas9 genome editing. Nat Protoc 13, 1465–1487 (2018).
- J. V. Thevathasan, M. Kahnwald, K. Cieśliński, P. Hoess, S. K. Peneti, M. Reitberger, D. Heid, K. C. Kasuba, S. J. Hoerner, Y. Li, Y.-L. Wu, M. Mund, U. Matti, P. M. Pereira, R. Henriques, B. Nijmeijer, M. Kueblbeck, V. J. Sabinina, J. Ellenberg, J. Ries, Nuclear pores as versatile reference standards for quantitative superresolution microscopy. Nat Methods 16, 1045–1053 (2019).
- S. Rankovic, R. Ramalho, C. Aiken, I. Rousso, PF74 Reinforces the HIV-1 Capsid To Impair Reverse Transcription-Induced Uncoating. J Virol 92, e00845-18 (2018).
- A. Bhattacharya, S. L. Alam, T. Fricke, K. Zadrozny, J. Sedzicki, A. B. Taylor, B. Demeler, O. Pornillos, B. K. Ganser-Pornillos, F. Diaz-Griffero, D. N. Ivanov, M. Yeager, Structural basis of HIV-1 capsid recognition by PF74 and CPSF6. Proc. Natl. Acad. Sci. U.S.A. 111, 18625–18630 (2014).
- I. Zurnic Bönisch, L. Dirix, V. Lemmens, D. Borrenberghs, F. De Wit, F. Vernaillen, S. Rocha, F. Christ, J. Hendrix, J. Hofkens, Z. Debyser, Capsid-Labelled HIV To Investigate the Role of Capsid during Nuclear Import and Integration. J Virol 94, e01024-19 (2020).
- A. E. Hulme, Z. Kelley, D. Foley, T. J. Hope, Complementary Assays Reveal a Low Level of CA Associated with Viral Complexes in the Nuclei of HIV-1-Infected Cells. J Virol 89, 5350–5361 (2015).
- M. Balasubramaniam, J. Zhou, A. Addai, P. Martinez, J. Pandhare, C. Aiken, C. Dash, PF74 Inhibits HIV-1 Integration by Altering the Composition of the Preintegration Complex. J Virol 93, e01741-18 (2019).
- G. L. Robinson, D. Dinsdale, M. MacFarlane, K. Cain, Switching from aerobic glycolysis to oxidative phosphorylation modulates the sensitivity of mantle cell lymphoma cells to TRAIL. Oncogene 31, 4996–5006 (2012).
- S. Song, E. Hwang, A Rise in ATP, ROS, and Mitochondrial Content upon Glucose Withdrawal Correlates with a Dysregulated Mitochondria Turnover Mediated by the Activation of the Protein Deacetylase SIRT1. Cells 8, 11 (2018).
- C.-S. Zhang, D. G. Hardie, S.-C. Lin, Glucose Starvation Blocks Translation at Multiple Levels. Cell Metabolism 31, 217–218 (2020).
- G. Holzer, W. Antonin, Breaking the Y. PLoS Genet 15, e1008109 (2019).
- M. W. Goldberg, Nuclear pore complex tethers to the cytoskeleton. Seminars in Cell & Developmental Biology 68, 52–58 (2017).
- B. Enyedi, M. Jelcic, P. Niethammer, The Cell Nucleus Serves as a Mechanotransducer of Tissue Damage-Induced Inflammation. Cell 165, 1160–1170 (2016).
- Z. Shen, K. T. Belcheva, M. Jelcic, K. L. Hui, A. Katikaneni, P. Niethammer, A synergy between mechanosensitive calcium- and membrane-binding mediates tension-sensing by C2-like domains. Proc. Natl. Acad. Sci. U.S.A. 119, e2112390119 (2022).
- S. Rajoo, P. Vallotton, E. Onischenko, K. Weis, Stoichiometry and compositional plasticity of the yeast nuclear pore complex revealed by quantitative fluorescence microscopy. Proc. Natl. Acad. Sci. U.S.A. 115 (2018).
- M. Allegretti, C. E. Zimmerli, V. Rantos, F. Wilfling, P. Ronchi, H. K. H. Fung, C.-W. Lee, W. Hagen, B. Turoňová, K. Karius, M. Börmel, X. Zhang, C. W. Müller, Y. Schwab, J. Mahamid, B. Pfander, J. Kosinski, M. Beck, In-cell architecture of the nuclear pore and snapshots of its turnover. Nature 586, 796–800 (2020).
- A. Elosegui-Artola, I. Andreu, A. E. M. Beedle, A. Lezamiz, M. Uroz, A. J. Kosmalska, R. Oria, J. Z. Kechagia, P. Rico-Lastres, A.-L. Le Roux, C. M. Shanahan, X. Trepat, D. Navajas, S. Garcia-Manyes, P. Roca-Cusachs, Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores. Cell 171, 1397-1410.e14 (2017).
- X. Meng, Y. Zhu, H. Tan, B. Daraqel, Y. Ming, X. Li, G. Yang, X. He, J. Song, L. Zheng, The cytoskeleton dynamics-dependent LINC complex in periodontal ligament stem cells transmits mechanical stress to the nuclear envelope and promotes YAP nuclear translocation. Stem Cell Res Ther 15, 284 (2024).
- M. Pizzato, O. Erlwein, D. Bonsall, S. Kaye, D. Muir, M. O. McClure, A one-step SYBR Green I-based product-enhanced reverse transcriptase assay for the quantitation of retroviruses in cell culture supernatants. Journal of Virological Methods 156, 1–7 (2009).
- Nikon, Super Resolution Microscope N-STORM protocol (2013).
- S. Haghparast, S. Stallinga, B. Rieger, Detecting continuous structural heterogeneity in single-molecule localization microscopy data. Sci Rep 13, 19800 (2023).
- W. Wang, H. Heydarian, T. A. P. M. Huijben, S. Stallinga, B. Rieger, Joint registration of multiple point clouds for fast particle fusion in localization microscopy. Bioinformatics 38, 3281–3287 (2022).
- R. P. J. Nieuwenhuizen, K. A. Lidke, M. Bates, D. L. Puig, D. Grünwald, S. Stallinga, B. Rieger, Measuring image resolution in optical nanoscopy. Nat Methods 10, 557–562 (2013).

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




