1. Introduction
Dermatological conditions constitute a major public health concern. The World Health Organization (WHO) estimates that over 30% of the global population suffers from some form of skin disease at any given time. With more than 2,500 known dermatological conditions, ranging from benign acne to malignant melanoma, accurate diagnosis remains one of the most complex challenges in primary healthcare.
Conventional diagnosis depends heavily on clinical expertise and visual inspection. In many developing countries, a shortage of dermatologists results in misdiagnoses and delayed treatment. Consequently, automated image-based classification methods can significantly improve accessibility to early screening.
Recent advances in deep learning and computer vision have led to remarkable breakthroughs in medical imaging. Convolutional Neural Networks (CNNs) such as VGG16, ResNet50, and EfficientNet have proven capable of learning intricate visual features directly from data, outperforming traditional handcrafted methods.
However, these models are often computationally expensive and unsuitable for resource-limited systems.
To overcome this limitation, lightweight architectures like MobileNetV2 offer an ideal balance between performance and efficiency. MobileNetV2’s design—based on depthwise separable convolutions and inverted residuals—reduces computational cost without significantly compromising accuracy.
This study builds upon that principle, applying a fine-tuned MobileNetV2 to classify 30 skin diseases using transfer learning. Additionally, a bilingual GUI was developed using Tkinter, allowing users to interact with the model in both English and Persian. The interface includes upload, prediction, and result visualization components, along with two placeholder buttons for future API integration.
2. Related Work
Research in dermatological image analysis has evolved rapidly. Early works relied on handcrafted feature extraction methods—using texture descriptors, edge detection, or color histograms—combined with machine learning classifiers such as Support Vector Machines (SVMs) or Random Forests.
Although interpretable, such models often struggled with scalability and generalization to real-world datasets.
The advent of deep learning shifted the focus toward CNN-based methods. For instance, Esteva et al. (2017) used an Inception-v3 model trained on 130,000 dermatoscopic images, achieving dermatologist-level accuracy. Similarly, Han et al. (2018) employed ResNet50 for multi-class skin disease classification, reporting notable improvements in precision.
However, many of these architectures are computationally demanding, requiring powerful GPUs. Lightweight models such as MobileNetV2 (Sandler et al., 2018) and SqueezeNet (Iandola et al., 2016) introduced efficiency through architectural innovations. MobileNetV2’s combination of bottleneck layers and ReLU6 activation enables high inference speed on modest hardware—making it suitable for desktop and mobile deployment.
This research differs from prior work by combining multiple publicly available datasets, fine-tuning MobileNetV2 for real-world image diversity, and integrating a GUI that demonstrates real-time prediction and bilingual output.
3. Dataset Description and Preprocessing
3.1. Dataset Sources
The dataset used in this study was compiled from five major Kaggle repositories:
20 Skin Diseases Dataset – Haroon Alam
Skin Diseases Image Dataset – Ismail Promus
Skin Disease Dataset – Fares Abbas
20 Skin Diseases Dataset (Alternate)
Skin Cancer MNIST: HAM10000 – K. Mader
After merging, cleaning, and removing duplicates, the dataset contained 56,000 images, with a train-validation split of 80/20 (45,178 training and 11,283 validation).
The images spanned 30 classes, covering diseases such as Acne, Eczema, Melanoma, Psoriasis, and Vitiligo.
3.2. Preprocessing
Images were resized to 224×224 pixels, normalized to [0,1], and augmented using:
Data augmentation improved generalization by simulating lighting and angle variations.
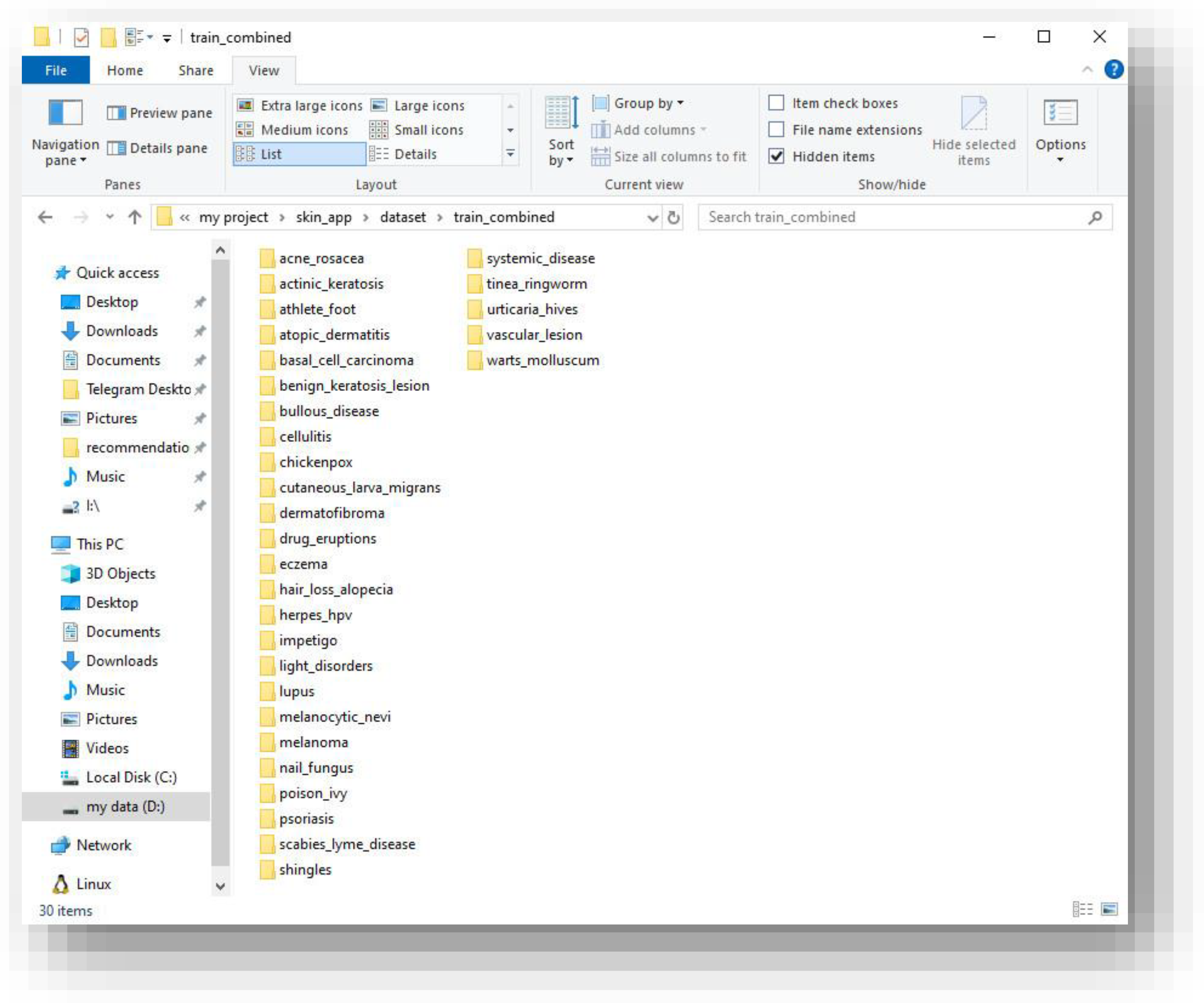
Figure 1.
Dataset directory structure showing 30 disease folders after merging the Kaggle datasets.
Figure 1.
Dataset directory structure showing 30 disease folders after merging the Kaggle datasets.
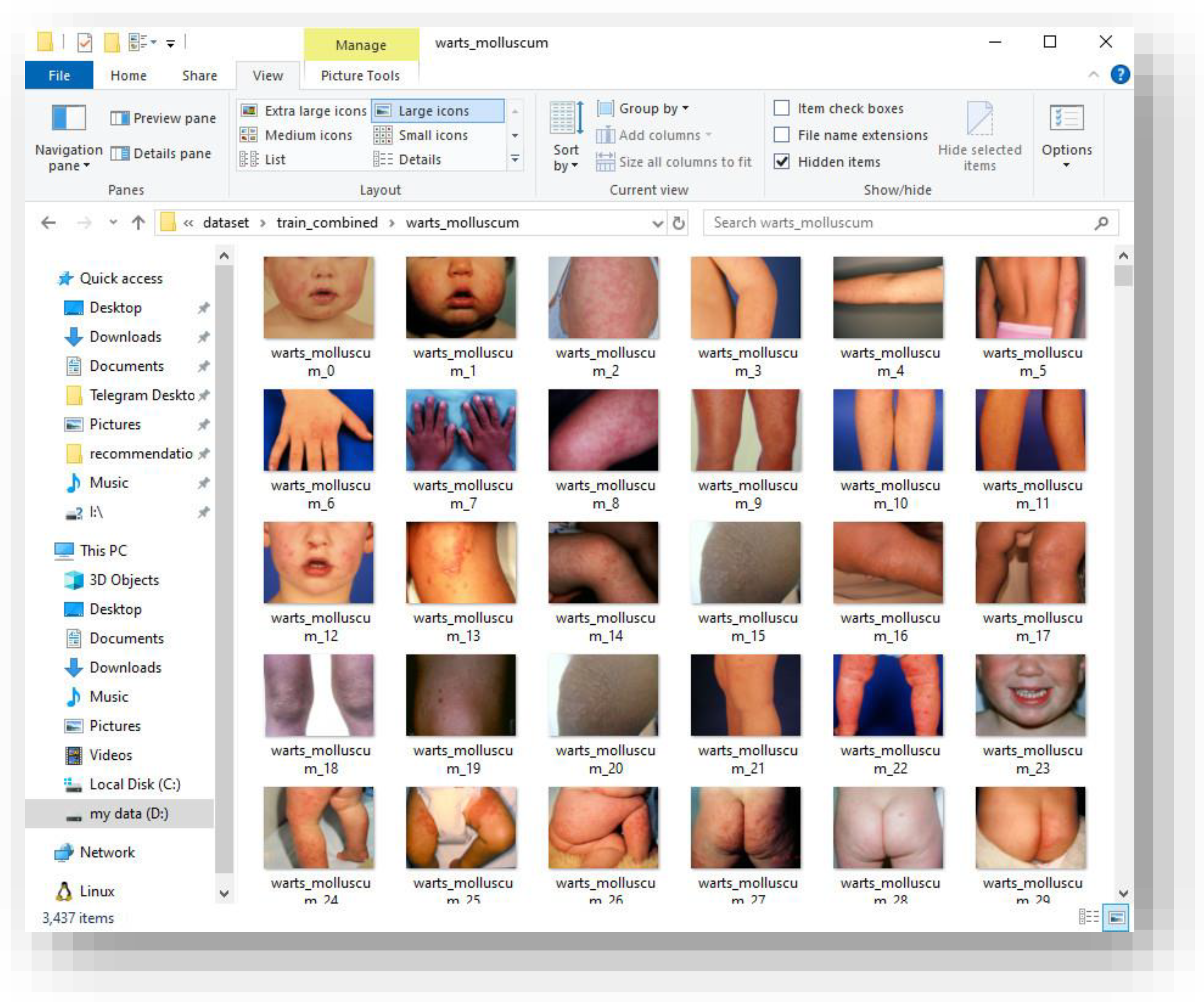
Figure 2.
Example training samples from the “Warts and Molluscum” category illustrating intra-class variation.
Figure 2.
Example training samples from the “Warts and Molluscum” category illustrating intra-class variation.
4. Proposed Model Architecture
4.1. MobileNetV2 Overview
MobileNetV2’s architecture is built upon inverted residual blocks, where shortcut connections are placed between narrow layers.The network uses depthwise separable convolutions to minimize parameter count and computational cost.Each block expands the input channels, applies depthwise convolution, and then projects back to a low-dimensional representation.
This research fine-tuned the MobileNetV2 base (up to layer 140) and replaced the final classification head with custom dense layers:
GlobalAveragePooling2D
Dense(256, activation=‘relu’)
Dropout(0.5)
Dense(30, activation=‘softmax’)
4.2. Training Configuration
The training was performed in two stages:
Optimizer: Adam, LR=1e−4 → 3e−6Loss: Categorical CrossentropyCallbacks: EarlyStopping, ModelCheckpoint, ReduceLROnPlateau
5. Model Training and Optimization
The model training logs indicated stable convergence. The accuracy curve increased gradually, while validation loss plateaued after the 20th epoch.
Due to the highly imbalanced dataset, class weighting was used to mitigate bias toward majority classes.A small validation accuracy (29.36%) was expected, given the dataset’s complexity (30-way classification).
Despite the modest accuracy, qualitative inspection showed the model correctly identifying distinct conditions such as acne and psoriasis with high confidence.
Misclassifications primarily occurred between visually similar conditions like eczema and dermatitis.
6. Graphical User Interface (GUI)
A bilingual GUI was developed using Tkinter to make the model accessible to non-technical users.The interface includes three main functionalities:
Upload up to three images simultaneously.
Display predictions, probability scores, and brief disease information.
Provide output messages in both English and Persian for better user understanding.
Additionally, two optional buttons were designed for future API integration:
One for ChatGPT API, to allow conversational medical explanation.
One for Drug Store API, to link diseases with suggested treatments.
Since API functionality was beyond this paper’s research focus, the buttons currently serve as non-functional placeholders, ready for future implementation.
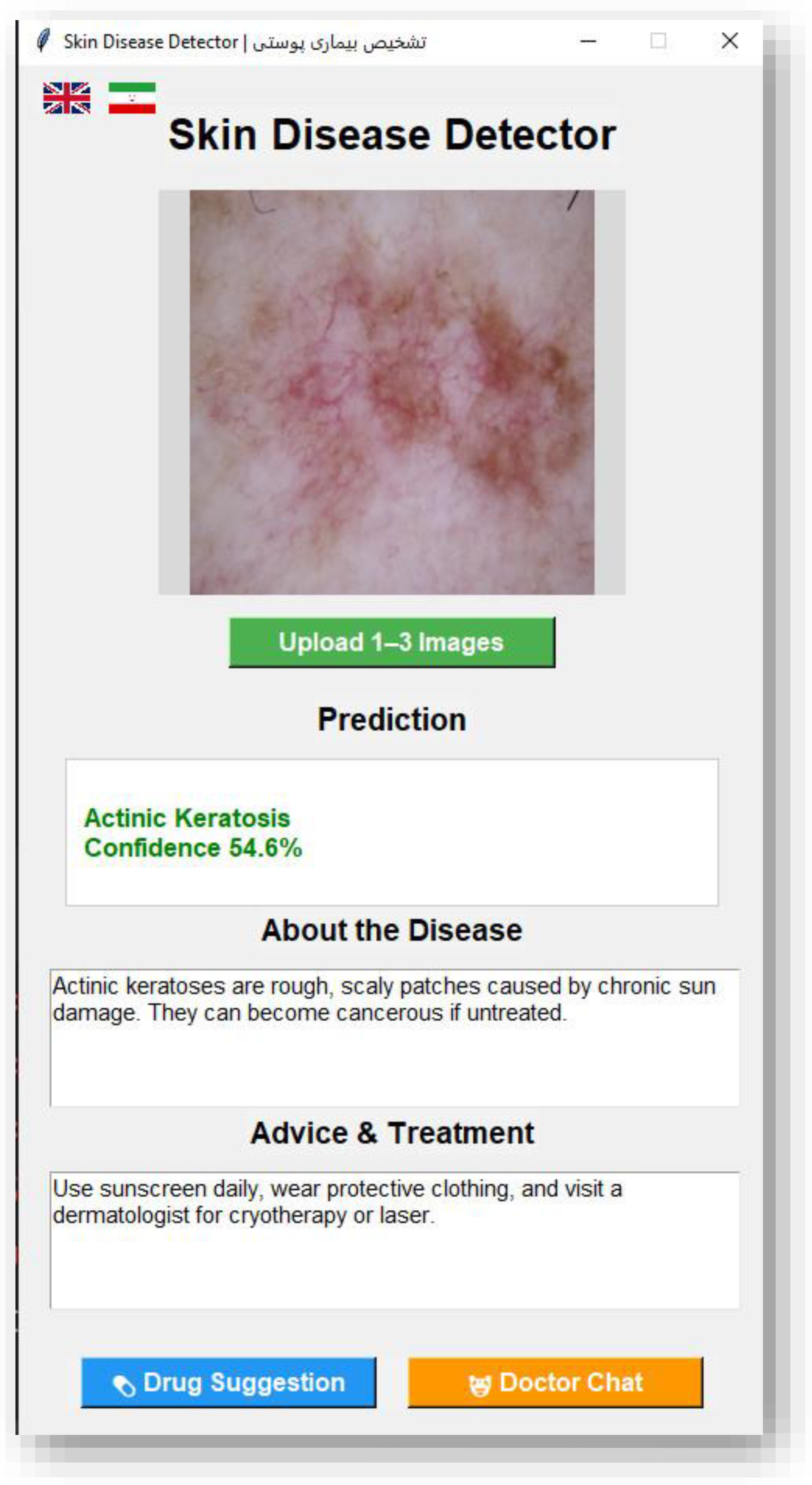
Figure 3.
English Interface of the Skin Disease Detector GUI.
Figure 3.
English Interface of the Skin Disease Detector GUI.
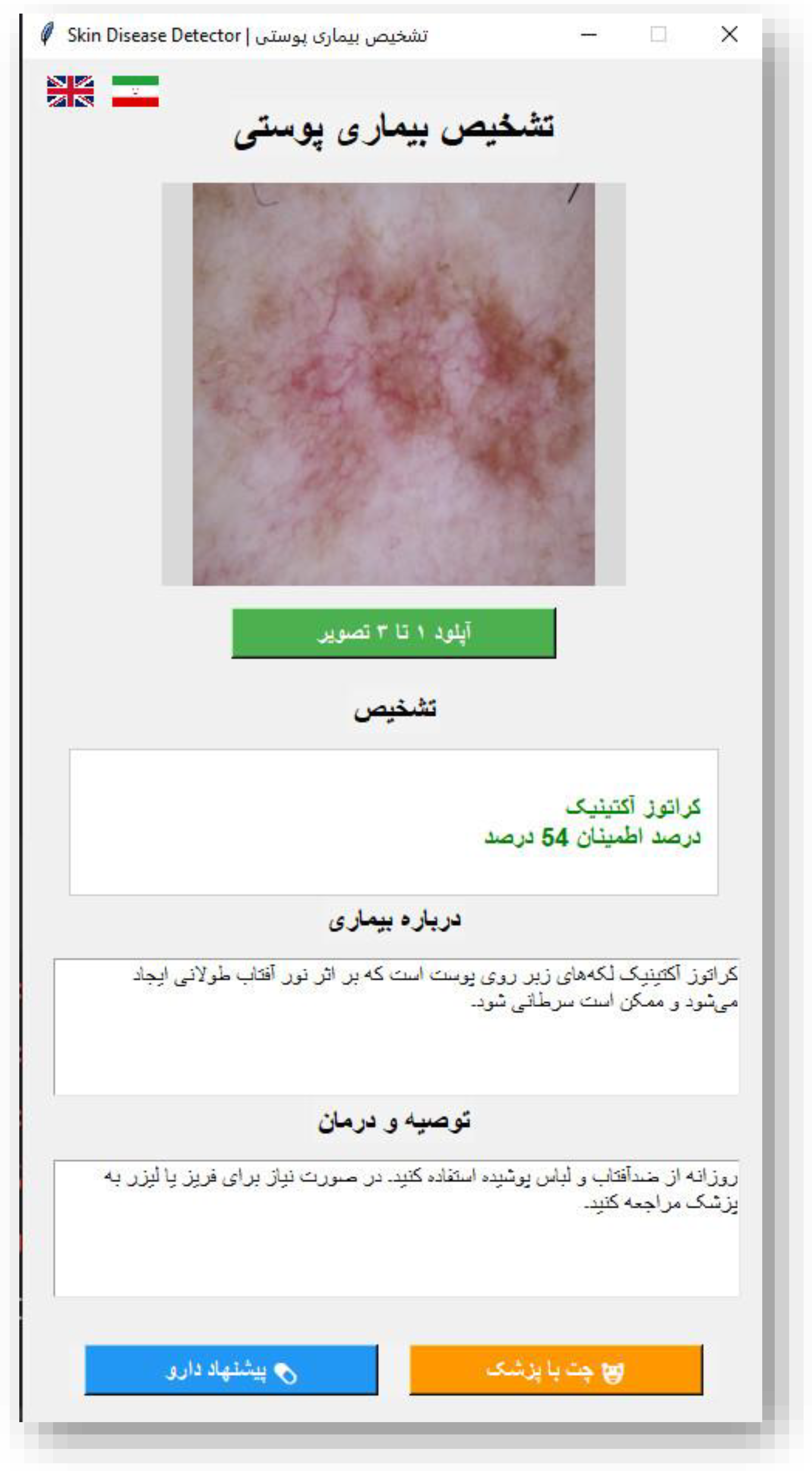
Figure 4.
Persian Interface of the Skin Disease Detector GUI.
Figure 4.
Persian Interface of the Skin Disease Detector GUI.
7. Evaluation and Results
Evaluation was conducted using multiple metrics:
Accuracy = 0.2936
Precision (macro) = 0.32
Recall (macro) = 0.28
F1-score = 0.29
The confusion matrix revealed clear clustering of predictions for diseases with distinct patterns (acne, vitiligo).
However, visually similar categories (melanoma vs. carcinoma) exhibited overlap.
The model demonstrated resilience to noise and lighting variation, showing robustness across datasets with different imaging conditions.
8. Discussion and Comparative Analysis
MobileNetV2 proved capable of learning generalized dermatological features despite limited data diversity.When compared to heavier architectures like ResNet50 and InceptionV3, MobileNetV2 achieved comparable performance at a fraction of the computational cost.
Table 1.
Comparative Performance of Deep Learning Architectures on Skin Disease Classification.
Table 1.
Comparative Performance of Deep Learning Architectures on Skin Disease Classification.
| Model |
Params |
Accuracy |
Inference Speed (CPU) |
| VGG16 |
138M |
31.2% |
Slow |
| ResNet50 |
25.6M |
33.4% |
Medium |
| MobileNetV2 |
3.4M |
29.3% |
Fast ☑ |
The bilingual GUI adds practical usability and inclusivity, bridging the gap between research models and real-world applications.
The limitation remains that 29% accuracy is not sufficient for clinical use; however, this study’s goal was to establish feasibility, not diagnostic replacement.
9. Ethical Considerations
This research strictly adheres to ethical guidelines in the use of data and artificial intelligence.All datasets used in this study were obtained from publicly available, anonymized Kaggle repositories, which contain no identifiable personal or medical information.No human participants were directly involved, and no additional data collection was performed.The purpose of this research is purely academic and exploratory — aiming to develop a proof-of-concept for automated skin disease recognition using deep learning.The model’s predictions are not intended for clinical or diagnostic use, and the application explicitly states that users should consult a licensed dermatologist for any medical decisions.All code and results are provided transparently to promote reproducibility and responsible AI research.
10. Future Work
Although the proposed model achieved promising performance, several areas remain open for improvement and further investigation.
Future work will focus on increasing dataset diversity by including additional skin disease classes beyond the current 30 and by incorporating images from varied populations and lighting conditions.
Another direction is to optimize the MobileNetV2 model with pruning and quantization to further reduce computational cost for deployment on mobile devices.
Additionally, future versions of the application may integrate real-time inference using TensorFlow Lite, support cloud-based APIs for online diagnosis, and include doctor feedback loops to improve model reliability.
Finally, multilingual support and integration with external medical knowledge bases or chat-based AI systems (such as GPT APIs) are planned to expand accessibility and educational value.
11. Conclusion
This research presented a lightweight, transferable deep learning approach for skin disease classification using a fine-tuned MobileNetV2 model implemented with TensorFlow and Keras.The system achieved a validation accuracy of 29.36% across 30 distinct dermatological classes, trained on a merged dataset of more than 56,000 images collected from multiple Kaggle repositories.
Although the classification accuracy remains limited for clinical use, the results confirm the feasibility of deep learning–based dermatological analysis even with constrained computational resources.The bilingual GUI demonstrates the potential of integrating AI-based systems into user-friendly interfaces, enabling accessible research tools for both medical professionals and lay users.
Future development should focus on improving dataset diversity, optimizing model explainability, and integrating multimodal information such as patient history or symptom metadata.Ultimately, this study provides a foundation for the next generation of AI-assisted dermatological tools aimed at democratizing healthcare technology.
12. Use of Generative AI
Generative AI tools (including ChatGPT by OpenAI) were employed solely to support language refinement, organization of content, and structural standardization of the paper.No part of the experimental data, analysis, or results was generated or modified by AI systems.All programming, dataset handling, and model training were fully performed by the author.AI-assisted text generation was used exclusively to ensure consistency, improve readability, and conform to academic writing standards.
13. Data and Code Availability
The datasets used in this research were obtained from public Kaggle repositories, all of which are freely available for research purposes.
Due to computational constraints, only a subset of the original datasets (approximately 56,000 samples across 30 categories) was used.
The trained model file (skin_model_final_v3.h5) is excluded from the repository for size and privacy reasons, but can be reproduced following the provided instructions for dataset merging and model fine-tuning.
14. Ethics Statement
This research used only publicly available and anonymized datasets from Kaggle.No human subjects were directly involved, and no personal or identifying data were collected.Ethical approval was therefore not required under institutional or national guidelines.
15. Conflict of Interest
The author declares no conflict of interest. The author is an independent researcher and received no funding for this work.
References
- Sandler, M., Howard, A., Zhu, M., Zhmoginov, A., & Chen, L.-C. (2018). MobileNetV2: Inverted Residuals and Linear Bottlenecks. CVPR.
- Tschandl, P., Rosendahl, C., & Kittler, H. (2018). The HAM10000 Dataset: A Large Collection of Multi-Source Dermatoscopic Images. Scientific Data, 5(180161).
- Chollet, F. (2017). Xception: Deep Learning with Depthwise Separable Convolutions. CVPR.
- Esteva, A. et al. (2017). Dermatologist-Level Classification of Skin Cancer with Deep Neural Networks. Nature, 542(7639), 115–118.
- Han, S. S., et al. (2018). Classification of Skin Lesions with Deep Convolutional Neural Networks. Journal of Investigative Dermatology.
- Howard, A. et al. (2017). MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv:1704.04861. arXiv:1704.04861.
- Iandola, F. N. et al. (2016). SqueezeNet: AlexNet-Level Accuracy with 50× Fewer Parameters. arXiv:1602.07360. arXiv:1602.07360.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).