1. Introduction
The phenomenon of biological aging is underpinned by a complex cascade of molecular alterations that progressively impair cellular function and augment susceptibility to disease [
1]. Gene expression profiling offers a quantitative methodology to dissect these age-related molecular signatures. Human mesenchymal stromal cells (hMSCs) represent a valuable in vitro model for aging research, given their accessibility and well-documented age-dependent functional decline [
1].
The Ensembl gene identification system (ENSG) provides stable, unique identifiers which are independent of nomenclature fluctuations. This identifier corresponds to the
NEFH gene, which encodes the Neurofilament Heavy protein. Neurofilaments are intermediate filament heteropolymers that comprise the axoskeleton and are essential for maintaining neuronal caliber and intracellular transport [
3].
NEFH is commonly used as a biomarker for neuronal damage, and mutations in the gene have been associated with susceptibility to amyotrophic lateral sclerosis (ALS) [
3].
Previous investigations have implicated numerous genes with age-dependent expression patterns in critical cellular processes [
2,
4,
5]. This study was initiated to rigorously examine the expression pattern of
NEFH in hMSCs across a spectrum of donor ages to evaluate its potential utility as a biomarker of biological aging in this cell type.[
9]
2. Materials and Methods
2.1. Acquisition of Transcriptomic Data
Gene expression data were procured from the Stemformatics repository (Dataset: Alves et al., 2012). This dataset comprises microarray expression profiles from human mesenchymal stromal cell (hMSC) samples with documented chronological donor ages. The dataset was accessed via the Stemformatics platform and curated for subsequent quantitative analysis.
2.2. Data Citation
The primary dataset utilized for this analysis is: Alves H, van Ginkel J, Groen N, Hulsman M, Mentink A, Reinders M, van Blitterswijk C, de Boer J. A mesenchymal stromal cell gene signature for donor age.
PLoS One. 2012;7(8):e42908. doi: 10.1371/journal.pone.0042908 [
1].
2.3. Gene Nomenclature
The Ensembl Gene ID system (ENSG) is employed to provide stable and unique identifiers for genetic loci. The identifier ENSG00000100285 corresponds to the genetic locus for the NEFH gene (Neurofilament Heavy). This nomenclature maintains consistency across annotation releases, enabling reliable data integration and longitudinal analysis.
2.4. Quantitative Correlation and Regression Analyses
All statistical analyses were performed using Python and R via Rstudio and Google Collab. The analysis was performed on data from a cohort of 61 donors. A Pearson correlation analysis was executed to ascertain the magnitude and directionality of the relationship between NEFH transcript expression and chronological age. Subsequently, a simple linear regression model was applied to model the age-dependent expression trend. Statistical significance was defined at an alpha level of . Descriptive statistics, including mean, standard deviation (SD), minimum, and maximum expression values, were calculated.
3. Results
3.1. Correlation Analysis
Transcript levels of
NEFH (ENSG00000100285) exhibited a statistically significant negative correlation with chronological age (Pearson’s r = -0.4316, p = 0.0005). The normalized expression values ranged from 1.5963 to 2.8567, with a mean of 2.0792 and a standard deviation of 0.2634. A statistical summary is presented in
Table 1.
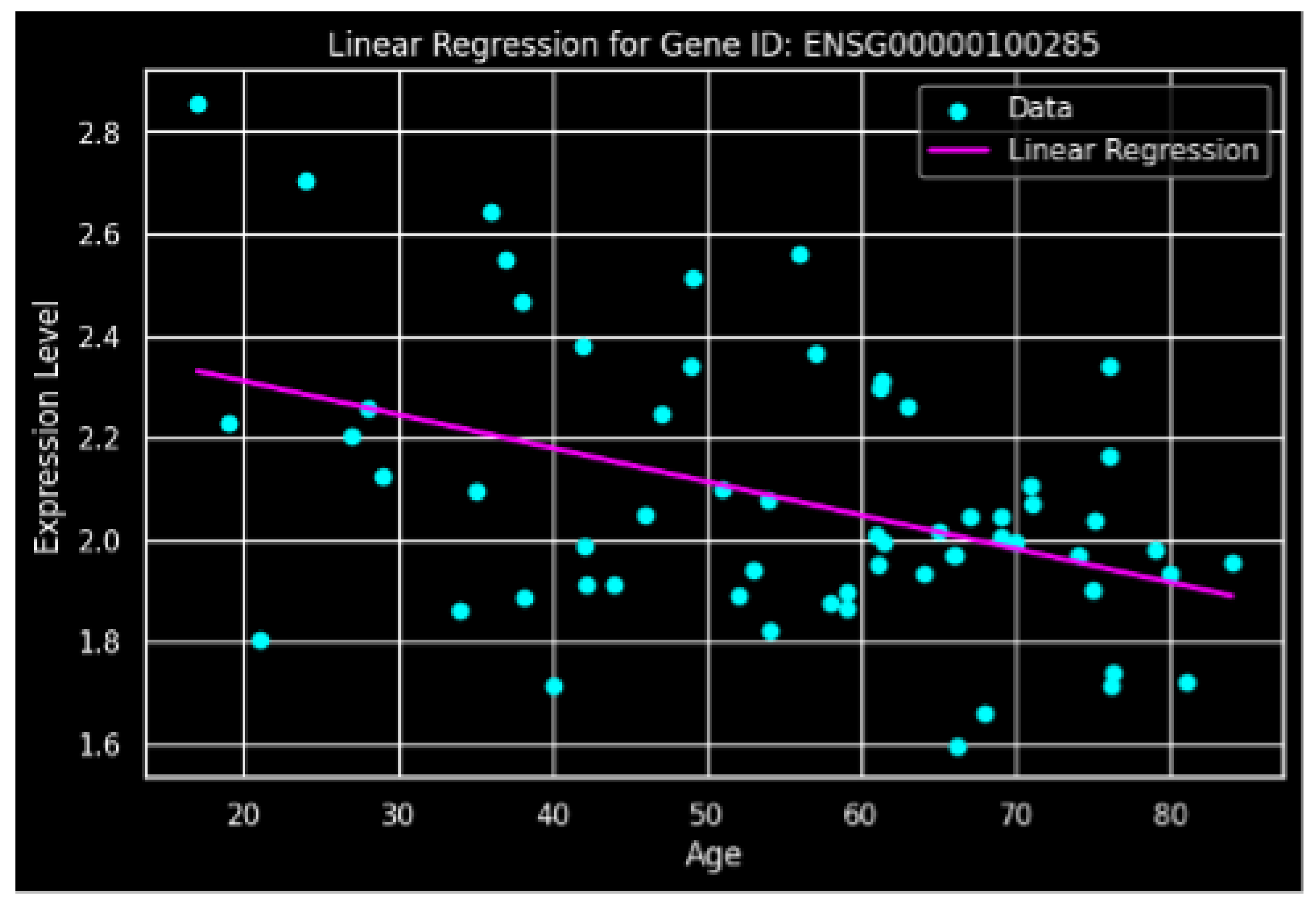
Figure 1.
Linear Regression analysis of gene (ENSG00000100285) expression level as a function of age. The scatter plot shows the individual expression data points, and the magenta line represents the fitted linear regression, indicating a slight negative correlation between gene expression and increasing age.
Figure 1.
Linear Regression analysis of gene (ENSG00000100285) expression level as a function of age. The scatter plot shows the individual expression data points, and the magenta line represents the fitted linear regression, indicating a slight negative correlation between gene expression and increasing age.
3.2. Linear Regression Analysis
Linear regression modeling was employed to quantify the relationship. This analysis yielded an intercept of 2.4429 and a slope coefficient of -0.0066, confirming a consistent age-dependent decrease in
NEFH expression. The resulting regression equation is:
This model demonstrates that NEFH expression decreases by approximately 0.0066 normalized units for each chronological year of age.
3.3. Heatmap Analysis
Figure 2.
Heatmap visualizing the relationship between gene (ENSG00000100285) expression level and age. The color intensity represents the -transformed expression level (ranging from approximately 1.5 to 2.5), with higher values indicating greater expression, across different ages (in years).
Figure 2.
Heatmap visualizing the relationship between gene (ENSG00000100285) expression level and age. The color intensity represents the -transformed expression level (ranging from approximately 1.5 to 2.5), with higher values indicating greater expression, across different ages (in years).
4. Discussion
The present analysis elucidates a significant age-dependent downregulation of NEFH (ENSG00000100285) transcript abundance in human mesenchymal stromal cells. The moderate negative correlation (r = -0.4316) and the consistent linear decline (slope = -0.0066) support the candidacy of NEFH as a quantitative biomarker of biological aging in this specific cell population.
The observed downregulation aligns with broader, established patterns of age-related transcriptional modulation [
2,
4,
5]. However, the finding is particularly intriguing given that
NEFH is canonically known as a structural protein of the neuronal axoskeleton and a biomarker for neuronal damage [
3].
Mechanistically, the presence and modulation of
NEFH in non-neuronal MSCs may be linked to their multipotent differentiation capacity. MSCs are known to possess neurogenic potential, and the expression of neuronal markers can be indicative of this state. The observed age-dependent downregulation of
NEFH in undifferentiated hMSCs may, therefore, reflect an age-related decline in this baseline neurogenic potential or plasticity.[
8]
The consistency of the linear relationship (slope = -0.0066) is fundamental for biomarker applications. However, the moderate correlation coefficient (
) indicates that chronological age accounts for approximately 18.6% of the variance in
NEFH expression. This suggests that other factors, such as epigenetic modifications or the progression of cellular senescence, significantly influence its expression levels.[
9]
4.1. Limitations and Future Directions
This analysis is constrained by its reliance on cross-sectional data from a singular, albeit relevant, cell type (hMSCs). Validation in longitudinal cohorts and investigation across diverse human tissues (particularly neuronal and non-neuronal) are imperative to establish the generalizability of NEFH as a systemic aging biomarker.
Future functional studies are required to elucidate the precise mechanistic role of
NEFH downregulation in MSC aging. Investigating whether this decline correlates with a functional impairment in neurogenic differentiation capacity would be a valuable avenue of inquiry.[
8,
9]
5. Conclusions
The expression of the NEFH gene (ENSG00000100285) exhibits a significant and consistent age-dependent downregulation in human mesenchymal stromal cells (r = -0.4316, p = 0.0005). These findings establish NEFH as a viable candidate biomarker for biological aging in MSCs, potentially reflecting an age-related decline in cellular plasticity. Further validation and mechanistic elucidation are warranted.
References
- Alves H, van Ginkel J, Groen N, Hulsman M, Mentink A, Reinders M, van Blitterswijk C, de Boer J. A mesenchymal stromal cell gene signature for donor age. PLoS One. 2012;7(8):e42908. doi: 10.1371/journal.pone.0042908.
- Fernández RJR. Unraveling the role of RNA Binding Protein with Multiple Splicing (RBPMS) in Ovarian Cancer Cells. Doctoral dissertation. 2023. Available from: ProQuest Dissertations & Theses Global.
- National Center for Biotechnology Information (NCBI). Gene: NEFH neurofilament heavy. [provided by RefSeq, Oct 2008].
- Yu H, Nagi SS, Usoskin D, Hu Y, Kupari J, et al. Leveraging deep single-soma RNA sequencing to explore the neural basis of human somatosensation. Nature Neuroscience. 2024;27:1198-1213. doi: 10.1038/s41593-024-01794-1.
- Zhu R, Yang X, Guo W, Xu XJ, Zhu L. An eight-mRNA signature predicts the prognosis of patients with bladder urothelial carcinoma. PeerJ. 2019;7:e7836. doi: 10.7717/peerj.7836.
- Farah K, Damien C, Pascale B. Neurofilaments in health and Charcot-Marie-Tooth disease. Front. Cell Dev. Biol. 2023;11:1275155. doi: 10.3389/fcell.2023.1275155.
- Jacquier, A., Delorme, C., Belotti, E., Juntas-Morales, R., Solé, G., Dubourg, O., Giroux, M., Maurage, C.-A., Castellani, V., Rebelo, A., Abrams, A., Zuchner, S., Stojkovic, T., Schaeffer, L., & Latour, P. (2017). Cryptic amyloidogenic elements in mutant NEFH causing Charcot-Marie-Tooth 2 trigger aggresome formation and neuronal death. Acta Neuropathologica Communications, 5(1), 63. doi: 10.1186/s40478-017-0457-1.
- Millington-Ward, S., Chadderton, N., Berkeley, M., et al. (2020). Novel 199 base pair NEFH promoter drives expression in retinal ganglion cells. Scientific Reports, 10(1), 16515. doi: 10.1038/s41598-020-73257-z.
- Scaber, J., Thomas-Wright, I., Clark, A. J., Xu, Y., Vahsen, B. F., Carcolé, M., Dafinca, R., Farrimond, L., Isaacs, A. M., Bennett, D. L., & Talbot, K. (2024). Cellular and axonal transport phenotypes due to the C9ORF72 HRE in iPSC motor and sensory neurons. Stem Cell Reports. Published online June 13, 2024. doi: 10.1016/j.stemcr.2024.05.008.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).