1. Introduction
Disease progression can significantly alter antibody properties through distinct mechanisms. First, post-translational modifications, such as shifts in glycosylation patterns during diseases, can alter both antigen binding affinity and effector functions by altering Fc receptor interactions [
1]. Current evidence indicates that IgG glycosylation profiles are altered in cancer, autoimmune, infectious and other diseases affecting both the total pool and certain antigen-specific antibodies [
2]. Second, the context of persistent infection itself can drive the formation of polyreactive antibodies, a potential precursor to autoimmunity [
3]. Finally, the hypothesis of molecular mimicry posits that structural homology between pathogen proteins and self-antigens can misdirect the immune response, leading to the production of autoreactive antibodies [
4]. Consequently, investigating antibody glycosylation patterns, polyreactivity, and cross-reactivity is crucial for elucidating the mechanisms that initiate and drive disease progression. However, such analysis is severely constrained by the masking effect of the abundant, non-specific immunoglobulin pool in serum, which obscures critical changes in low-abundance, antigen-specific fractions. The methodology developed in this work directly addresses this bottleneck by enabling the isolation of purified antigen-specific antibodies, thereby providing a critical tool for elucidating their unique structural and functional characteristics in health and disease.
Affinity chromatography represents the benchmark technique for isolating specific proteins from complex biological mixtures. This method is particularly valuable for purifying serum antibodies, as it yields isolates that faithfully retain the native immunoglobulin classes, glycosylation profiles, and antigen-binding avidity. Despite its conceptual advantages, conventional column affinity chromatography presents significant practical limitations for the isolation of low-abundance specific antibodies. The method demands large quantities of purified, stable antigen for sorbent synthesis, creating a major cost barrier. The subsequent elution step further complicates the process, introducing sample dilution and frequent protein loss during concentration and buffer exchange. Consequently, the column chromatographic isolation of specific antibodies from multiple samples remains a labor-intensive, time-consuming, and resource-heavy endeavor. Alternative approaches have circumvented columns by utilizing other platforms, such as the Western blot membrane strips employed by Kurien et al. [
5] or the immunoassay plates used by Mendis et al. [
6]. Meanwhile, the advancement of magnetic particle-based technologies has facilitated the development of miniaturized methods for protein isolation from complex mixtures, significantly expanding their application potential [
7].
Acidic buffers (pH 2.5-3.0) remain a common choice for eluting antibodies from antigen-affinity matrices due to their simplicity and low cost. However, even with brief exposure and immediate neutralization, the low pH can cause irreversible protein degradation, rendering the antibodies unsuitable for functional activity studies. Beyond degradation and aggregation [
8], a more insidious consequence is the induction of polyreactivity, whereby previously monospecific antibodies can develop non-specific binding to multiple unrelated antigens following acid exposure[
9,
10,
11].
Mild elution agents offer a gentler alternative for dissociating antibodies from affinity matrices. While the common anionic detergent sodium dodecyl sulfate (SDS) is a potent denaturant that irreversibly disrupts protein structures, several of its analogs interact with proteins in a milder, reversible manner [
12]. Detergents like sodium lauryl sarcosinate (Sarkosyl) and sodium lauryl glutamate (SLG) are known for their ability to solubilize proteins from inclusion bodies with minimal denaturation [
13,
14]. It is hypothesized that these milder detergents engage in limited protein penetration, primarily interacting with surface hydrophobic regions to induce transient conformational changes without achieving complete denaturation. When applied to antigen-antibody complexes, this mechanism enables complex dissociation while minimizing damage to the antibody’s native structure. Critically, the subsequent removal of detergent residues following antibody isolation facilitates substantial recovery of bioactivity, representing a key advantage over conventional denaturing elution methods.
This study aimed to develop micro-scale workflow for isolating antigen-specific serum antibodies using array- and bead-based platforms, with anionic detergents employed as a gentle elution strategy to preserve antibody functionality.
2. Materials and Methods
2.1. Serum Sample
Serum samples were collected from a cohort of patients with autoimmune diseases and control subjects [
15]. From this collection, a sample that was positive for IgG against bovine serum albumin (BSA), cytomegalovirus (CMV) protein pp150 or thyroglobulin (Tg) was selected based on a multiplex assay.
2.2. Protein Arrays
Hydrogel arrays with immobilized proteins were manufactured by co-polymerization immobilization, following a previously described protocol [
16].
2.2.1. Single-Antigen Capture Arrays
For Capture array the following proteins were used for immobilization: BSA (A7030, Sigma, St. Louis, MO, USA), recombinant thyroglobulin (8RTG4, HyTest, Turku, Finland), recombinant cytomegalovirus pp150 protein (RE003, Xema Co., Ltd., Moscow, Russia), recombinant human insulin protein (ab123768, Abcam, Cambridge, UK). The array layout was identical for all Capture arrays, regardless of the antigen used (
Figure 1).
For method development, a Capture array with immobilized insulin was employed in combination with Cy5.5 fluorescently labeled monoclonal antibodies against insulin (RC3A6, HyTest, Finland).
2.2.2. Multi-Antigen Array for Multiplex Assay
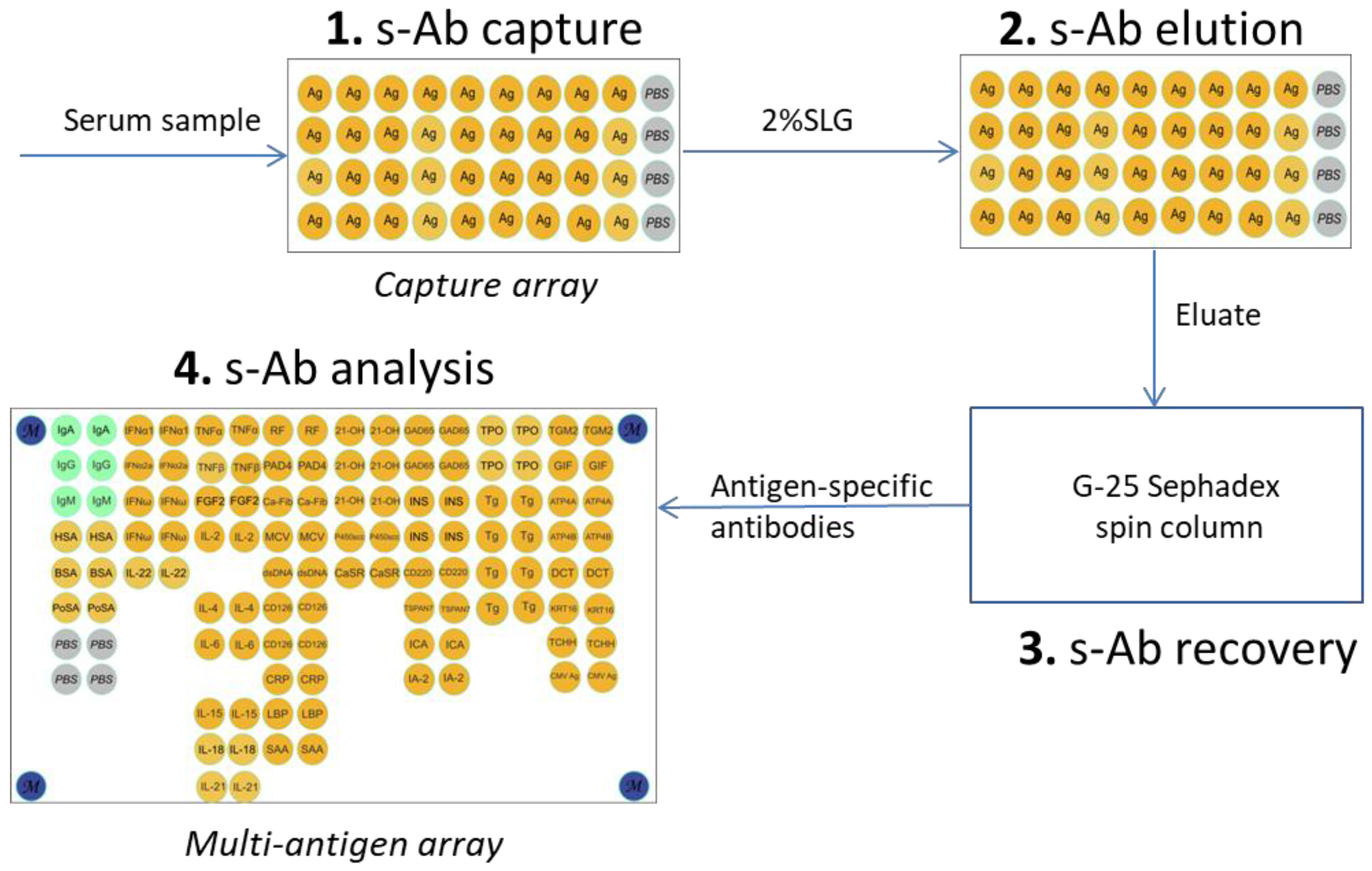
The multi-antigen array contained 120 elements with 58 immobilized proteins, corresponding to 47 unique proteins. The list of proteins for the multi-antigen array is provided in
Table S1. The microarray layout is shown in
Figure 2.
2.3. Magnetic Beads
Total IgG isolation was performed using ready-to-use, commercially available protein G-coated magnetic beads (786-904, Geno Technology, St. Louis, MO USA).
For specific antibodies isolation the following proteins were used for immobilization: BSA (A7030, Sigma, St. Louis, MO, USA), recombinant Thyroglobulin (8RTG4, HyTest, Turku, Finland), recombinant Cytomegalovirus pp150 protein (RE003, Xema Co., Ltd., Moscow, Russia). Protein was covalently immobilized on amino-functionalized magnetic nanoparticles (iron oxide, 300–400 nm; K0501, Sileks, Moscow, Russia) using glutaraldehyde crosslinking, following the manufacturer’s protocol. In brief: 100 μl of magnetic particle solution (5 mg/ml) was washed, resuspended in 250 μl of PBS, 250 μl of 25% glutaraldehyde were added, and incubated in the dark for 3 hours with gentle rocking. After four washes with PBS, the particles were resuspended in 500 µl PBS, supplemented with 100 µl protein (0.2 mg/ml) in PBS containing 0.01% Tween20 (PBSt), incubated overnight with gentle rocking, washed four times with PBS again, and resuspended in 100 µl PBSt.
2.4. Anionic Detergents
The following detergents were used: sodium lauroyl glutamate (sc-495823, Santa Cruz Biotechnology, Inc., Dallas, TX, USA), N-Lauroylsarcosine (Sodium salt) (L-5125, Sigma, St. Louis, MO, USA), sodium dodecyl sulfate (SB-GC204005-01, ServieceBio, Wuhan, China). The preparation of stock solutions differed for each anionic detergent. The protocol for SLG dissolution involved initial addition to milli-Q water (up to 20%) with subsequent gradual titration with a 10 M NaOH solution until clear. SDS was dissolved in water with gentle stirring and heating (~50 °C). Sarkosyl was readily soluble in water. The stock solutions were stored at room temperature and warmed before use if necessary. Working solutions of the detergents (SDS, 0.006–0.5%; Sarcosyl, 0.05–2.0%; SLG, 0.25–10.0%) were prepared fresh in milli-Q water immediately prior to use.
2.5. Micro-Scale Serum Derived Antibodies Isolation
2.5.1. Protein Array Protocol
The serum sample was diluted 1:50 in immunoassay buffer, and 100 µl of the dilution was applied to the array. Following an overnight incubation at 37 °C, the array was washed with PBSt for 20 minutes, rinsed with water, and dried. Subsequently, 100 µl of a freshly prepared eluent containing an anionic detergent was added, followed by incubation for 1 h at 37 °C. The final eluate was then carefully collected by pipette.
2.5.2. Magnetic Beads Protocol
Total IgG Isolation
Patient serum samples (20 µL) were added to tubes containing 50 µL of magnetic beads coated with Protein G (786-904, Geno Technology, St. Louis, MO USA) and incubated for 1 hour at room temperature under end-over-end rotation. Following incubation, the beads were washed three times with 200 µL of binding buffer, with the supernatant removed after each wash. After the final wash, 100 µL of elution buffer was added to the magnetic beads and incubated for 10 minutes, with periodic resuspension. When an acidic elution buffer (0.1 M Gly-HCL, pH 2.5) was used, the eluate was neutralized with 1 M Tris-HCl buffer (pH 8.5) at ratio of 1:7.
The Antigen-Specific Immunoglobulin Isolation
Antibodies were isolated from 20 µL of blood serum using 50 µL of magnetic particle suspension with immobilized protein. Magnetic particles were blocked for 1 hour in 100 μl of EveryBlot Blocking Buffer (Bio-Rad Laboratories, Hercules, CA, USA), washed and resuspended in 500 μl of PBSt, a blood serum sample (20 μl) was added, incubated for 1 hour with gentle rocking, washed three times with 500 μl PBSt and resuspended in 100 μl of PBSt. 100 μl of Elution Buffer (0.1 M Gly-HCL, pH 2.5 or 2% SLG in PBS) were added to the particles, incubated for 10 minutes, periodically mixing by pipetting. In case of acidic elution, the eluate was immediately neutralized with 20 μl of 1 M Tris-HCl (pH 8.5).
2.6. Functional Recovery of Individual Antigen-Specific Immunoglobulins
2.6.1. Gel Filtration
The obtained eluates (100 µL) were applied to spin columns (7326204, Bio-Rad Laboratories, USA) filled with Sephadex G-25 coarse (17-0034-02, GE Healthcare, Chicago, IL, USA) and pre-equilibrated with PBS and centrifuged at 1 500 g. The resulting probe was used for further analysis.
2.6.2. Dialisys
The obtained eluates (100 µL) were dialyzed using Slide-A-Lyzer MINI dialysis devices (3.5 kDa MWCO; Thermo Fisher Scientific, USA) against three changes of PBS (pH 7.2, 300 mL) under constant stirring at RT.
2.7. Analysis of Isolated Immunoglobulins
2.7.1. SDS-PAGE and Western-Blot Analysis
The obtained samples were separated by electrophoresis under denaturing conditions using a 10% resolving gel. Protein bands were visualized by staining with either Coomassie R-250 (786-495, Geno Technology, USA) or silver nitrate (G2080, Servicebio, Wuhan, Hubei, China). The proteins were then transferred to a PVDF membrane (1620262, Bio-Rad Laboratories, USA). Following the transfer, the membrane was blocked overnight with EveryBlot Blocking Buffer (12010020, Bio-Rad Laboratories, USA). The membrane was incubated for one hour with polyclonal goat anti-human IgG antibodies (31163 Invitrogen, Thermo Fisher Scientific, Carlsbad, CA, USA) and then for one hour with horseradish peroxidase-conjugated rabbit anti-goat IgG antibodies (S0010, Affinity Biosciences, Loveland, CO, USA). After washing with PBS containing 0.01% Tween 20, the immune complexes were visualized using the Clarity™ Western ECL substrate kit (1705061, Bio-Rad Laboratories, USA).
2.7.2. Multiplex Immunoassay
For the multiplex immunoassay, both original blood serum samples and specific antibodies isolated from them were used. Serum samples were diluted at a 1:50 ratio, and isolated antibodies were analyzed without dilution. IgG antibodies targeting 60 immobilized proteins were detected using a previously developed assay [
16]. Microarrays were blocked with 1% polyvinyl alcohol (PVA) in phosphate-buffered saline (PBS, pH 7.4) at room temperature (RT) for 1 hour. Samples (100 µL) were applied onto the microarrays. Following an overnight incubation at 37 °C, the arrays were subjected to an intermediate wash with PBS containing 0.1% Tween-20 for 20 minutes. Antigen-antibody complexes were then detected using a fluorescently labeled anti-human IgG antibody (31163, Invitrogen, Thermo Fisher Scientific, Carlsbad, CA, USA) during a two-hour incubation at 37 °C. Finally, the microarrays were washed with PBS containing 0.1% Tween-20 for 30 minutes, rinsed with buffer, and dried prior to scanning.
Fluorescence images of microarrays were obtained using a proprietary laser-excited microarray analyzer [
17]. Signal quantification was performed with proprietary software. For each group of n elements containing identical antigens, the resulting signal (I
n) was calculated as the mean fluorescence intensity of the corresponding spots. Elution efficiency (%) was calculated as the ratio of the median signal from the corresponding array elements after elution to their initial signal. Recovery efficiency (%) was calculated as the ratio of the median signal from the corresponding array elements for isolated specific antibodies to the signal obtained from the same element group during the initial sample analysis.
2.8. Direct Analysis of Fluorescently Labeled Antibodies on the Microarray
Cy5.5-labeled antibodies were diluted in PBS containing 0.14% PVA and 0.14% PVP. A 100 µL volume of the antibody solution (at concentrations ranging from 500 ng/mL to 2.0 µg/mL) was applied to the microarrays. After a two-hour incubation at 37 °C, the microarrays were washed with PBS containing 0.1% Tween 20 for 30 minutes, rinsed with distilled water, and dried. The following antibodies were used: monoclonal anti-insulin (clone RC3A6, HyTest, Finland), polyclonal anti-BSA (A11133, Invitrogen, Thermo Fisher Scientific), and goat anti-human IgG (31163, Invitrogen, Thermo Fisher Scientific).
4. Discussion
Affinity chromatography is a cornerstone technique for the selective purification of antibodies, leveraging the specific and reversible interaction between an immobilized antigen and its target. The process involves two phases: the capture of the target molecule from the solution onto the solid-phase ligand, followed by its elution, which involves disrupting the specific complex to release the purified protein. A critical requirement is that the elution process preserves the protein’s native conformation and biological activity. Various strategies can be employed to disrupt the antigen-antibody complex, including shifts in pH or ionic strength, and the application of chaotropic agents, organic solvents, high-charge eluents, or competitive elution [
18]. The mechanism of the eluting agent is a key determinant, as it can induce either reversible, short-lived unfolding or irreversible denaturation of the antibody.
Acidic buffers, such as glycine-HCl (pH 2.5-3.0), represent a gold standard in laboratory practice for target antibodies elution due to its availability and versatility. The mechanism involves the protonation of charged amino acid residues, primarily histidines, within the antigen-antibody interface. This protonation disrupts electrostatic interactions by shifting the net charge of the molecule and can also compromise hydrogen bonding and alter the isoelectric point. However, this aggressive approach also perturbs the antibody’s tertiary structure, causing partial unfolding and exposure of hydrophobic regions. Such structural alterations frequently lead to a loss of specific antigen-binding activity and can induce undesirable polyreactivity or aggregation [
9,
10,
11]
. These detrimental effects are particularly problematic when the isolated antibodies are intended for functional studies or analytical techniques sensitive to structural integrity.
A critical limitation in current methodologies for the microscale isolation of specific antibodies from serum is the universal reliance on acidic buffers for elution, regardless of the sorbent employed. For example, Madara et al. used polyacrylamide gels to isolate antibodies by electrophoretically separating proteins, immobilizing them in situ with glutaraldehyde, and using the homogenized gel as a microsorbent [
19]. Another approach purified serum autoantibodies using antigen-bound nitrocellulose membranes excised after Western blotting [
5]. Brown et al. developed a high-throughput microscale method to purify antigen-specific antibodies for IgG glycan analysis, utilizing streptavidin-functionalized agarose cartridges conjugated with biotinylated antigens [
20]
. Similarly, Mendis et al. isolated specific autoantibodies from serum using MBP-fusion protein-coated plates [
6]. Although these techniques demonstrate versatility in sorbent design, they share a fundamental drawback: the mandatory use of low-pH elution buffers, which can irreversibly compromise antibody structure and specificity.
To overcome the limitations of conventional elution, we developed a gentle method using anionic detergents to dissociate antibodies from micro-immunosorbents, such as microarrays or magnetic particles. The core of this technique lies in the transient, non-denaturing interaction of the detergent, which, after a subsequent purification via gel filtration, allows for the recovery of antibodies with high purity and minimal loss of native conformation.
Detergents are amphiphilic molecules consisting of a long-chain hydrophobic aliphatic tail and a hydrophilic polar head group. This structure enables their hydrophobic moieties to penetrate and disrupt phospholipid bilayers by displacing membrane lipids, making them highly effective for cell lysis [
21]. Beyond membrane disruption, certain detergents have proven remarkably successful in extracting and refolding recombinant proteins from inclusion bodies. A seminal study by Arakawa et al. demonstrated that SLG could recover up to 100% of native protein, in stark contrast to SDS, which yielded 0% recovery after solubilization [
12]. Inspired by the protein-refolding capabilities of these mild anionic detergents, we investigated their potential as gentle eluents for isolating specific antibodies from human serum.
The anionic detergents evaluated in this study exhibit distinct structural characteristics that govern their interactions with protein structures during antibody elution. While SDS acts as a strong denaturant that typically causes irreversible protein unfolding, both Sarcosyl and SLG function as milder alternatives capable of solubilizing proteins while maintaining their native conformation and biological activity (
Table 1). This fundamental difference in protein-detergent interaction mechanisms directly influences their suitability for antibody isolation applications where preserving structural integrity is paramount.
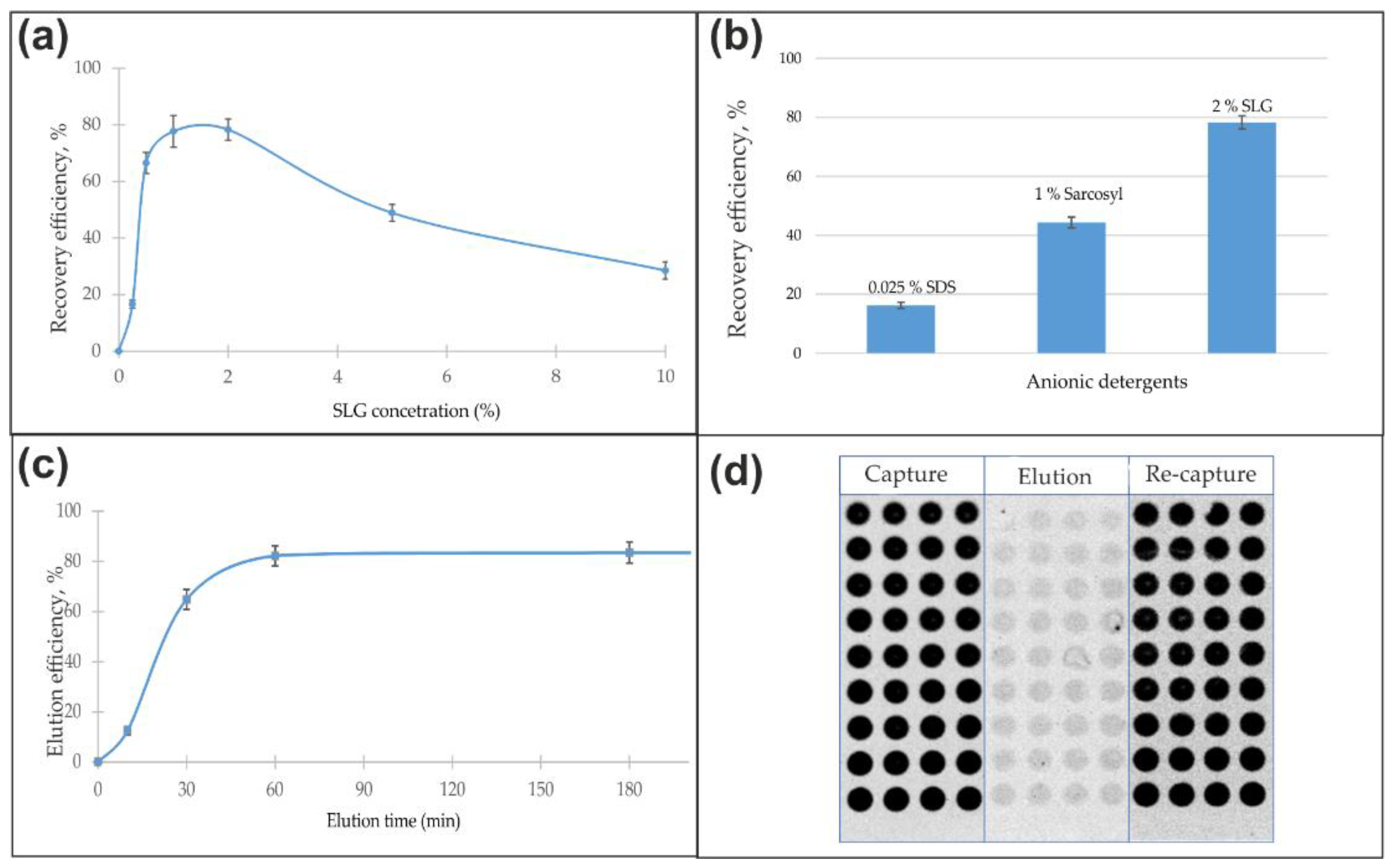
Analysis of antibody recovery as a function of detergent concentration revealed a distinct optimum for each agent, with efficiency declining at both lower and higher concentrations (
Figure 4a). At low concentrations, detergent levels remain below the critical micelle concentration (CMC), preventing effective disruption of the antibody-antigen complex and subsequent antibody release. In contrast, a sharp decline in functional yield observed at concentrations substantially above the CMC is likely attributable to protein denaturation, aggregation, and persistent detergent binding that hinders subsequent purification. Following the identification of these optimal concentrations (0.025% SDS, 1% Sarkosyl, 2% SLG), elution time and temperature were further refined (
Figure 4b). The final recovery of functional antibodies under optimized conditions starkly highlighted the difference in detergent gentleness: SDS yielded only 17%, Sarkosyl 45%, and SLG achieved the highest recovery at 78% (
Figure 4c).
The robustness of the method against matrix interference was confirmed by spiking experiments in blood serum. Neither the elution efficiency nor the specific activity of the recovered antibodies was compromised by the serum components, confirming the method’s applicability to complex biological samples (
Figure 4d).
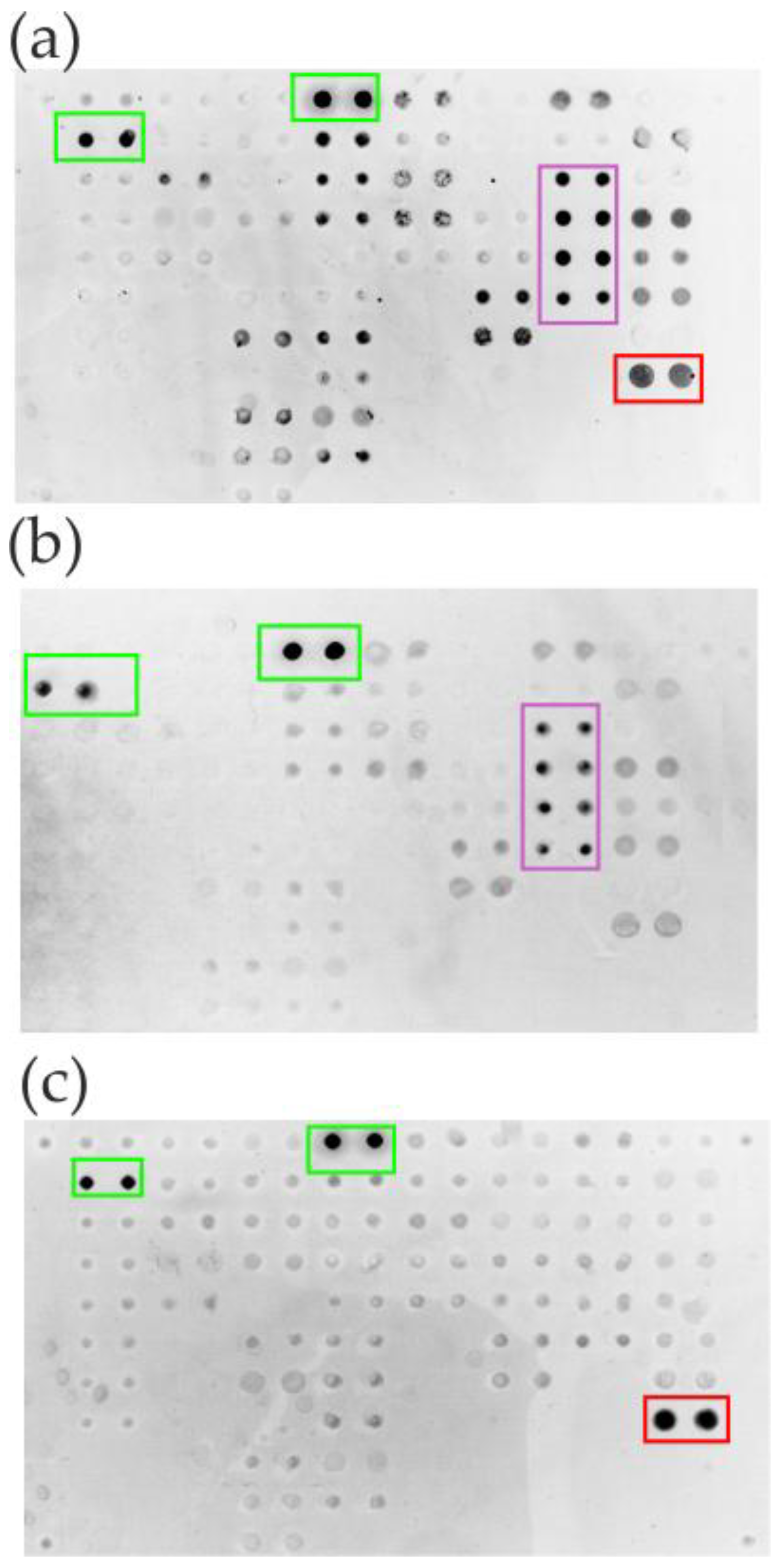
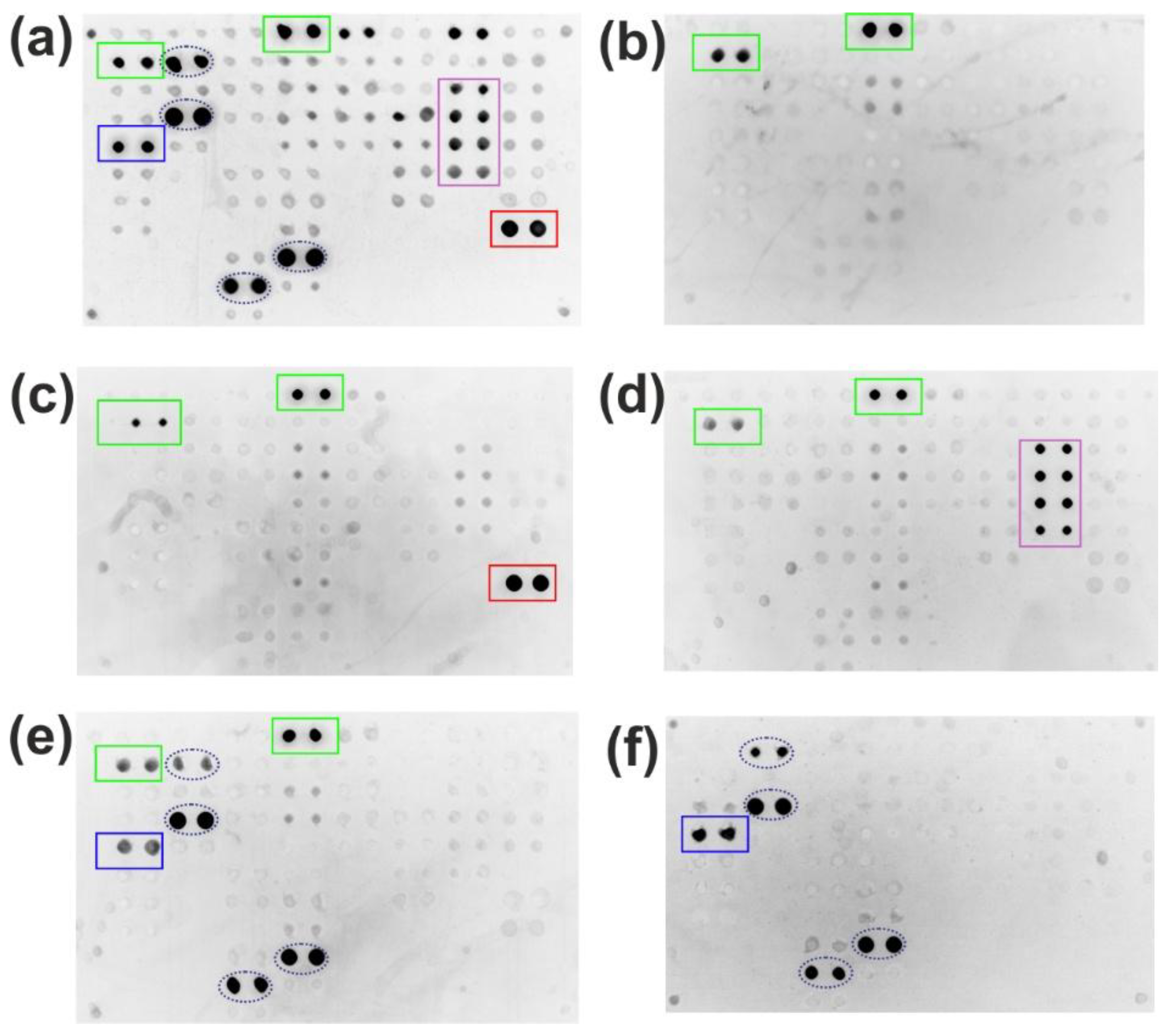
The isolation of specific antibodies from human serum using microarrays confirmed high functional recovery, with 58.5-85.3% of specific bioactivity retained. Comparative analysis between initial serum samples and isolated antibody fractions was performed using a multi-antigen microarray containing numerous immobilized proteins, including CMV pp150 and Tg (
Figure 2). The results demonstrate that the developed method yields sufficient antibody quantities for downstream applications while maintaining structural integrity and antigen-binding capacity (
Figure 5). Specific positive signals from microarray elements containing corresponding antigens confirm successful complex formation for both the original serum sample (
Figure 5a) and isolated antibody fractions (
Figure 5 b-d), verifying preserved bioactivity in the purified eluates. Crucially, the absence of cross-reactivity with non-cognate antigens on the microarray indicates that the SLG-based elution process does not induce polyreactivity in the isolated antibodies.
Despite the limited binding capacity of individual microarray gel elements, the platform offers several compelling advantages. Its primary strength lies in exceptional economic efficiency: the system requires only 6 µg of antigen protein to analyze up to 1,000 serum samples, enabling antibody isolation even against rare or expensive targets. Furthermore, antigen immobilization within a polyacrylamide hydrogel matrix stabilizes the protein’s tertiary structure, preserving epitopes in a native conformation for at least one year[
22]. For applications demanding higher antibody yields, such as subsequent ELISA, the platform can be scaled by fabricating capture microarrays with increased numbers or larger dimensions of gel elements. In summary, this approach represents an inexpensive, rapid, and versatile platform for parallel isolation and analysis of specific antibodies from multiple samples.
The mild-elution-based antibody isolation method was successfully adapted also to a magnetic particle platform to demonstrate its versatility.
Figure 6 presents the analysis of the isolated serum antibodies against CMV, Tg, and BSA captured with magnetic beads and 2% SLG elution. While the original sample contained antibodies against all three antigens (
Figure 6a), each isolated fraction demonstrated high specificity: anti-CMV (
Figure 6c), anti-Tg (
Figure 6d), and anti-BSA antibodies (
Figure 6e). The multiple interactions observed for anti-BSA antibodies (
Figure 6f) are attributed to assay interference, as BSA is commonly used as a stabilizer in commercial protein preparations [
15]. This was confirmed by direct analysis with fluorescently labeled anti-BSA antibodies (
Figure 6f). The specificity of the detecting antibodies used was also validated in a separate control (
Figure 6b). Multiplex analysis confirmed the highly selective extraction of target antibodies, with the isolated antibodies retaining functionality and showing no cross-reactivity against unrelated array antigens. These results demonstrate the reversible nature of the SLG-antibody interaction and underscore the method’s high selectivity.
The method’s specificity was demonstrated using a complex serum sample containing multiple distinct antibodies, including anti-CMV (viral), anti-BSA (dietary), anti-Tg (autoimmune) targets (
Figure 5 and
Figure 6). This successful parallel isolation confirms the platform’s capability for comparative studies of antibody populations across different categories—enabling direct investigation of interaction specificity between autoantibodies, pathogen-specific, and food-specific antibodies within the same experimental framework.
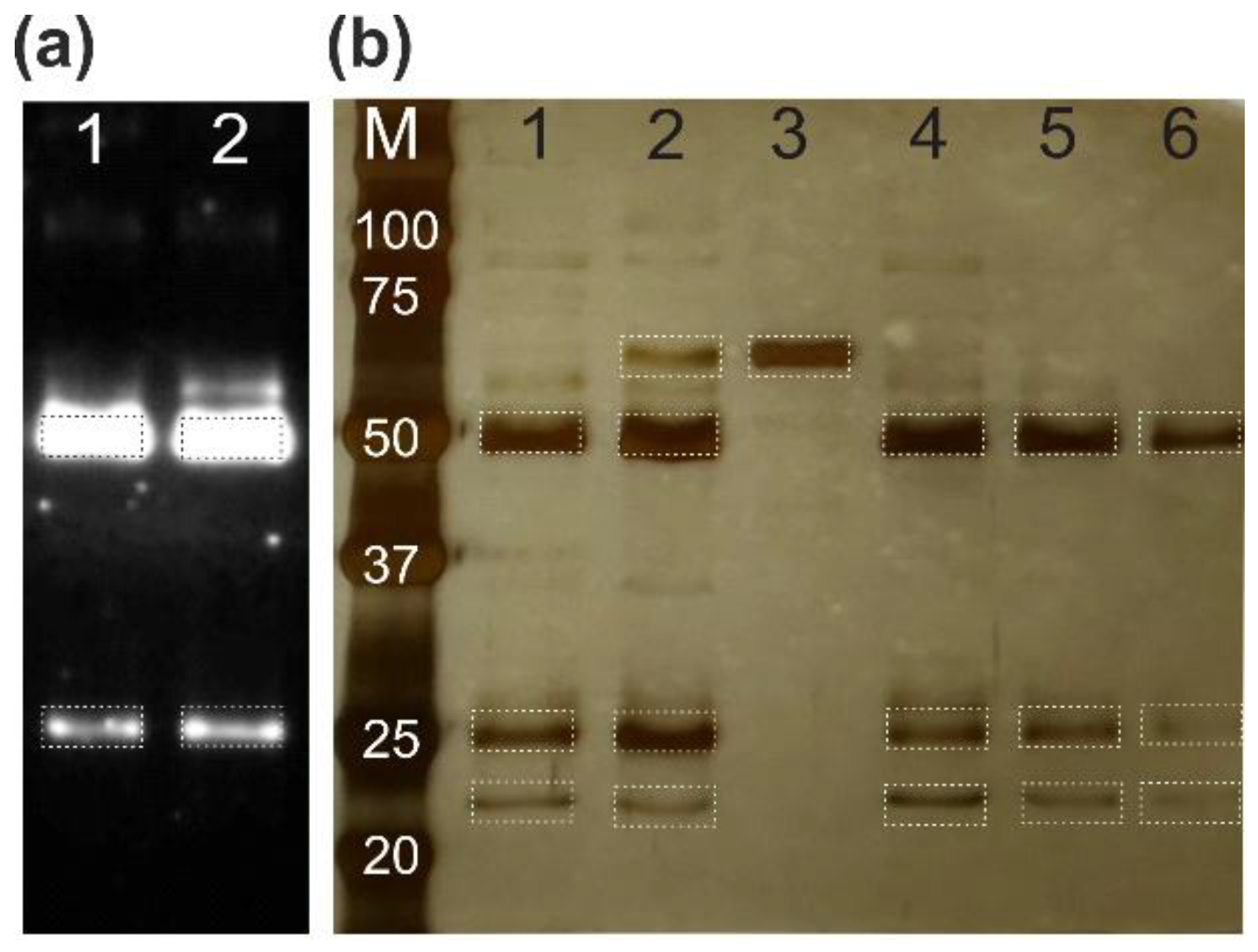
The mild anionic detergent elution method was further validated for isolating total IgG from human serum using Protein G-conjugated magnetic beads. A direct comparison was made between standard acidic elution (Gly-HCl, pH 2.5) and mild elution with 2% SLG. Western blot analysis confirmed successful IgG isolation with both methods (
Figure 7a), while silver-stained gels demonstrated comparable purity with minimal co-elution of serum proteins such as albumin (
Figure 7b). The consistent band patterns and high purity across multiple serum samples establish SLG elution as a robust alternative to conventional acidic conditions.
Consequently, this strategy enables the selective isolation of target antibodies through mild anionic detergents and versatile immunosorbents, yielding high-purity preparations with preserved structural integrity.