1. Introduction
Medical image segmentation plays a pivotal role in numerous clinical applications, including disease diagnosis, treatment planning, and prognosis assessment [
1]. Accurate segmentation of organs, tumors, and other anatomical structures is crucial for quantitative analysis and guiding interventions. The U-Net architecture [
2], with its elegant encoder-decoder structure and skip connections, has emerged as a cornerstone in this field, achieving remarkable success due to its ability to capture both contextual and localization information. However, as medical imaging tasks become increasingly complex, involving intricate pathologies, fine-grained structures, and the need to model long-range dependencies, traditional Convolutional Neural Network (CNN)-based U-Nets still face limitations in terms of performance and computational efficiency. This challenge mirrors the broader effort in AI to unravel chaotic contexts and establish complex reasoning chains, which requires sophisticated architectural designs [
3]. Recent advancements in deep learning, particularly the advent of pre-trained Vision Transformers (ViTs) [
4] and State Space Models (SSMs) like Mamba [
5], have demonstrated superior capabilities in capturing global contextual information and long-range dependencies. These models, especially large vision-language models, have set new benchmarks in understanding complex visual scenes through paradigms like in-context learning [
6]. Integrating these powerful mechanisms into the U-Net framework, while maintaining computational efficiency and addressing structural discrepancies, remains an active and challenging area of research. Furthermore, ensuring that these powerful but complex models generalize well from weak supervision or limited data remains a critical open problem [
7].
Despite the progress, several key challenges persist in achieving highly accurate and efficient medical image segmentation: (1)
Balance of Local Details and Global Context: While CNNs excel at extracting local, high-frequency features essential for fine details, they inherently struggle to model long-range pixel correlations effectively. Conversely, Transformers and Mamba-like architectures are proficient in capturing global contextual information but may sometimes overlook crucial local nuances. An effective fusion strategy that harmonizes these complementary strengths is a critical requirement. (2)
Boundary Segmentation Precision: Many medical image segmentation tasks demand exceptionally high precision, especially for delineating the boundaries of lesions or organs, which are often irregular, fuzzy, or very thin. Existing methods frequently exhibit suboptimal performance in accurately segmenting these challenging boundary regions. (3)
Model Efficiency: The integration of complex, large-scale models, such as extensive pre-trained Vision Transformers, into U-Net architectures can lead to a significant increase in computational overhead and parameter count. This often renders such models impractical for deployment in resource-constrained clinical environments where real-time inference is desirable. This necessitates the development of novel acceleration techniques, such as semantics-inspired sparse activation, to make large models viable in practice [
8].
To address these aforementioned challenges, we propose a novel framework named the Efficient Boundary-aware Multi-Scale Feature Fusion U-Net (EB-MSF-UNet). Our primary objective is to develop a lightweight and efficient model that can adaptively fuse multi-scale local features with global contextual information within a U-Net-style architecture. Furthermore, by incorporating an explicit boundary-aware mechanism, our method aims to significantly enhance the precision of medical image segmentation, particularly in intricate boundary regions.
The proposed EB-MSF-UNet adopts the classic U-shaped encoder-decoder architecture and introduces several innovative modules. Our Dual-Pathway Encoder is designed to capture information at different scales: a lightweight CNN path (e.g., using GhostNet or MobileNetV3 blocks) for efficient local texture and edge feature extraction, and a lightweight Mamba-like path (e.g., based on low-rank approximation or sparse attention mechanisms) responsible for modeling long-range dependencies and global semantic information. These two paths are dynamically balanced by an Adaptive Feature Gating Module (AFGM) to adjust their respective contributions based on the feature requirements at different architectural levels. A Multi-Scale Feature Fusion Module (MSFFM) is strategically placed at each downsampling layer of the encoder and upsampling layer of the decoder. This module employs a novel cross-attention mechanism to deeply integrate features from both the encoder’s local and global pathways, while also incorporating higher-level semantic information transmitted via skip connections, ensuring rich and complementary feature representations across various resolutions. Finally, the Boundary Enhancement Decoder incorporates a dedicated Boundary-Aware Refinement Module (BRM) at each decoding stage. The BRM combines local contrast enhancement and spatial attention mechanisms to explicitly guide the model to focus more on pixels near the segmentation target’s boundaries. This is further optimized during training through an auxiliary loss function, such as a weighted Dice-Boundary Loss or an approximation of Hausdorff Distance Loss, to explicitly improve boundary precision.
For experimental validation, we extensively evaluate
EB-MSF-UNet on two widely-used public benchmark datasets: the
Synapse Multi-organ CT dataset, which involves the segmentation of various abdominal organs (e.g., spleen, pancreas, liver), and the
ACDC (Automated Cardiac Diagnosis Challenge) dataset, focusing on the segmentation of cardiac structures (e.g., left ventricle, right ventricle, myocardium) from MR images. Our model’s performance is assessed using standard metrics, including the Dice Similarity Coefficient (DSC) for evaluating segmentation overlap accuracy and the 95% Hausdorff Distance (HD95) for quantifying boundary precision. Through rigorous comparative experiments against state-of-the-art and classic baselines such as U-Net [
9], TransUNet [
9], Swin-UNet [
10], and ACM-UNet [
11], our
EB-MSF-UNet consistently demonstrates superior performance. For instance, on the Synapse dataset, our method achieves a DSC of 85.55% and an HD95 of 13.40mm, outperforming ACM-UNet. Similarly, on the ACDC dataset,
EB-MSF-UNet records a DSC of 91.65% and an HD95 of 6.45mm, further validating its robustness and accuracy, especially in improving boundary delineation as evidenced by the significantly reduced HD95 values. Our work aligns with the broader goal of creating scalable, self-improving vision systems that can continuously enhance their capabilities through strategic learning [
12].
Our main contributions can be summarized as follows:
We propose a novel Dual-Pathway Encoder within a U-Net architecture, which efficiently combines lightweight CNN blocks for local feature extraction and Mamba-like modules for global context modeling, adaptively balanced by an Adaptive Feature Gating Module (AFGM).
We introduce a Multi-Scale Feature Fusion Module (MSFFM) that leverages a novel cross-attention mechanism to robustly integrate multi-resolution features from both local and global pathways, as well as skip connections, ensuring comprehensive feature representations.
We design a Boundary Enhancement Decoder incorporating a Boundary-Aware Refinement Module (BRM) and an auxiliary boundary loss, which explicitly guides the model to refine segmentation masks with enhanced precision, particularly for challenging and intricate boundaries.
2. Related Work
2.1. Deep Learning Architectures for Medical Image Segmentation
While U-Net architectures are prominent, insights from broader deep learning research can enhance medical image segmentation. For instance, security analyses of foundation models like MedCLIP highlight reliability risks from data discrepancies [
13], a concern for any segmentation model that is mirrored in other fields focused on information veracity, such as fake news detection [
14]. Concepts from graph representation learning, such as neural gates and attention-based diffusion in DIFNET, offer mechanisms to improve CNN architectures [
15]. Similarly, collaborative learning paradigms, proven effective for related vision tasks [
16], are promising for multi-organ or multi-modal segmentation. Foundational work in 3D reconstruction has paved the way for volumetric analysis [
17]. Techniques for aligning multi-modal data, such as images and text or 2D and 3D information, could benefit architectures that leverage cross-modal information, drawing on statistical matching perspectives and advanced retrieval methods [
18,
19,
20]. For clinical adoption, ensuring model trustworthiness, fairness, and safety is paramount [
21,
22]. Important lessons can be drawn from evaluating decision-making in other safety-critical domains, such as the development of multi-agent systems for coordinating interactive autonomous vehicles at unsignalized intersections [
23,
24,
25]. Furthermore, understanding length-based overfitting in Transformers is crucial for segmentation models to generalize across varied anatomical dimensions [
26]. Hierarchical summarization techniques offer a framework for integrating multi-view or multi-modal information [
27], while advances in generative models for complex 3D structures suggest future directions in data augmentation and anatomical modeling [
28,
29]. We note that some related works, such as those on clinical relation extraction or RNA design, do not directly address segmentation architectures [
30].
2.2. Boundary-Aware and Efficient Segmentation Methods
Developing boundary-aware, efficient methods is critical for precise segmentation. Natural Language Processing (NLP) offers valuable inspiration, such as using subword regularization to inform segment sampling [
31]. More directly, methods from Named Entity Recognition (NER) that use boundary regression [
32] or diffusion-based refinement [
33] can significantly improve boundary localization and efficiency. Similarly, boundary smoothing techniques can address annotation ambiguities with low computational overhead [
34], and models for document-level event extraction suggest methods for precise boundary identification [
35]. To handle class imbalance, which can degrade boundary representation, insights can be drawn from synthetic data augmentation in image-to-image translation and balancing methods in text classification [
36]. Principles for enhancing model robustness against targeted perturbations are also relevant for defending segmentation boundaries [
37]. The pursuit of efficiency is a cross-cutting concern, with relevant advances in unsupervised contrastive learning [
38], adaptive sparse activation for accelerated inference [
8], and more training-efficient paradigms [
39]. Conceptual frameworks for improving robustness and precision can be adapted from diverse fields. These include optimized boosting algorithms for limited data settings [
40], causal inference for robust assessment [
41], and reinforcement learning for dynamic prioritization [
42]. Furthermore, real-time adaptive systems in control engineering provide a model for high-performance parameter estimation under varying conditions [
43,
44,
45]. Likewise, methods from materials science for detecting dynamic molecular structures and synergistic atomic effects offer a blueprint for identifying subtle tissue changes for precise boundary delineation [
46,
47]. Even research in human-computer interaction on digital aids for fine crafts underscores the principle of fine-grained control essential for accurate segmentation [
48].
3. Method
In this section, we present the detailed architecture of our proposed Efficient Boundary-aware Multi-Scale Feature Fusion U-Net (EB-MSF-UNet). Our framework builds upon the classic U-Net encoder-decoder paradigm, integrating novel modules designed to enhance the balance between local detail and global context, and to improve boundary segmentation precision while maintaining computational efficiency.
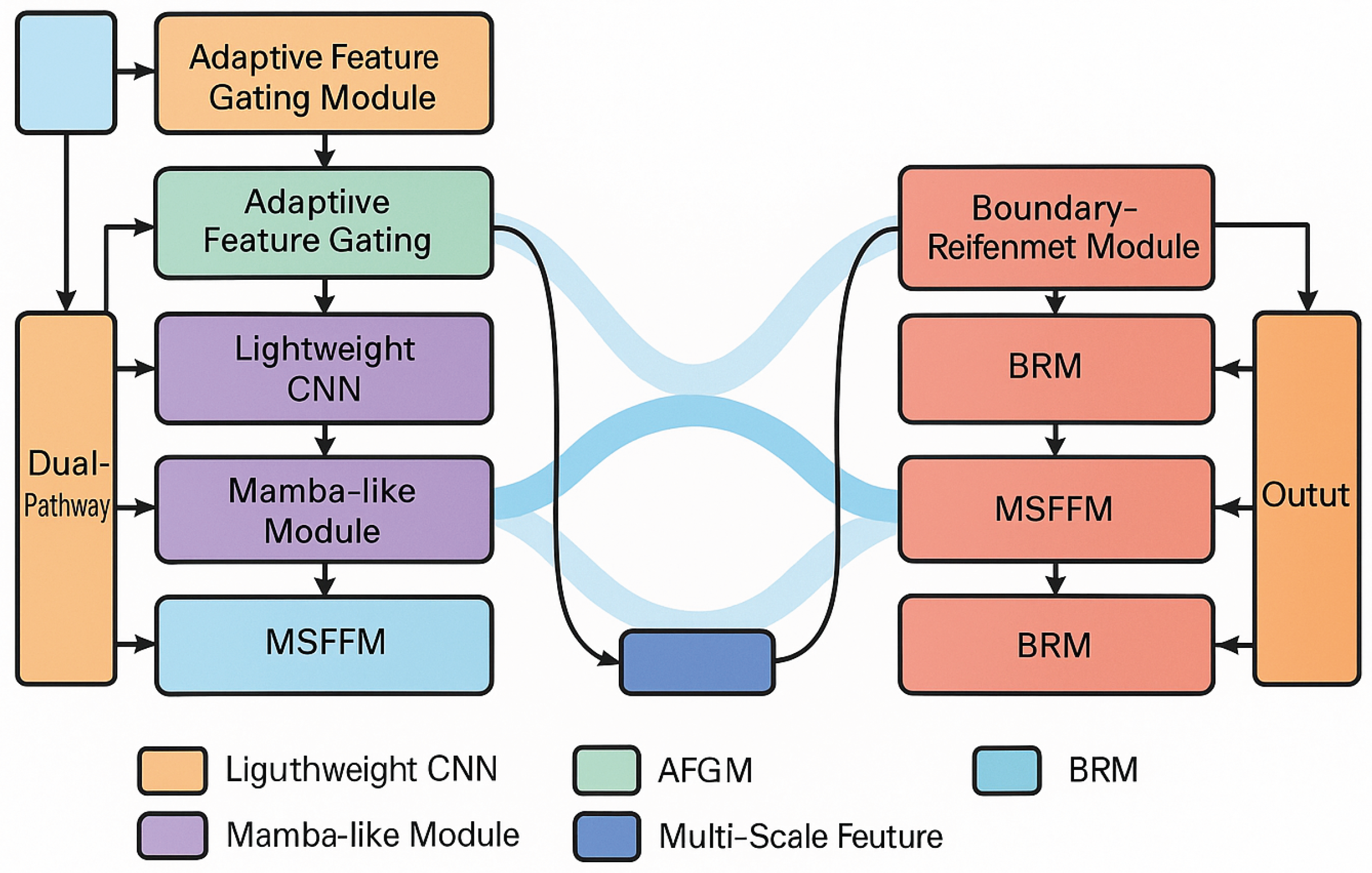
Figure 1.
Overview of our EB-MSF-UNet.
Figure 1.
Overview of our EB-MSF-UNet.
3.1. Overall Architecture of EB-MSF-UNet
The EB-MSF-UNet adopts a symmetric encoder-decoder architecture with skip connections, a hallmark of the U-Net family. The encoder progressively downsamples the input medical image, extracting hierarchical features that capture both low-level textures and high-level semantics. Crucially, our encoder is designed with a dual-pathway mechanism to explicitly handle local and global information, addressing the inherent trade-off between fine-grained spatial accuracy and broad contextual understanding. The decoder then upsamples these features, leveraging skip connections from the encoder to recover spatial details lost during downsampling and to bridge the semantic gap between encoder and decoder features. A key innovation in our decoder is the incorporation of boundary-aware modules, which are specifically tailored to refine segmentation masks, focusing on accurate delineation of object boundaries. The entire network is trained end-to-end, optimizing for both overall segmentation accuracy and boundary precision through a composite loss function.
3.2. Dual-Pathway Encoder
Our encoder is structured into multiple stages, with each stage comprising a dual-pathway mechanism to capture complementary feature representations: a local details pathway and a global context pathway. This parallel processing allows the network to simultaneously extract fine-grained spatial information and long-range dependencies, which are critical for robust medical image segmentation.
3.2.1. Local Details Pathway
The local details pathway is constructed using lightweight Convolutional Neural Network (CNN) blocks, such as those inspired by architectures like GhostNet or MobileNetV3. These blocks are specifically designed for efficient extraction of fine-grained local textures, edges, and low-level features that are crucial for preserving spatial accuracy and capturing subtle anatomical variations. Each block typically consists of depthwise separable convolutions, point-wise convolutions, and non-linear activation functions, ensuring a compact yet effective representation of local information with reduced computational cost. Let
be the feature map from the
-th stage of the local pathway. After processing through a lightweight CNN block
, which performs feature transformation, and a downsampling operation
(e.g., max pooling or strided convolution) to reduce spatial dimensions, the feature map for the
i-th stage is obtained as:
where
represents the initial input image, potentially pre-processed by an initial convolutional layer.
3.2.2. Global Context Pathway
In parallel, the global context pathway is designed to model long-range dependencies and capture global semantic information across the entire image. This pathway employs lightweight Mamba-like modules, which are a type of State Space Model (SSM). These modules are particularly effective at processing sequential data and can efficiently capture global interactions and contextual relationships across distant image regions without the quadratic complexity associated with traditional attention mechanisms like Transformers. To maintain computational efficiency, our Mamba-like modules may incorporate techniques such as low-rank approximations or sparse attention mechanisms. Let
be the feature map from the
-th stage of the global pathway. After processing through a Mamba-like block
, which extracts global features, and a downsampling operation
, the feature map for the
i-th stage is:
where
also represents the initial input image, potentially processed by an initial convolutional layer to match dimensions and extract initial features.
3.2.3. Adaptive Feature Gating Module (AFGM)
To dynamically adjust the contributions of the local and global pathways and enable flexible information flow, we introduce an
Adaptive Feature Gating Module (AFGM) at each encoder stage. The AFGM takes the feature maps from both pathways,
(local details) and
(global context), and adaptively generates channel-wise weights to fuse them. This allows the network to prioritize local details or global context based on the specific characteristics of the features at different depths and the complexity of the structures being segmented. The output fused feature
at stage
i is computed as:
where
denotes concatenation along the channel dimension,
is a convolutional layer (e.g., a
convolution) to project the concatenated features to a single channel or channel-wise weights,
is the sigmoid activation function to produce weights between 0 and 1, and ⊙ denotes element-wise multiplication. The resulting
provides a gating mechanism that controls the flow of information from
, with
implicitly controlling the contribution from
. This adaptively fused feature
is then passed to the decoder via skip connections, providing a rich, balanced representation.
3.3. Multi-Scale Feature Fusion Module (MSFFM)
The
Multi-Scale Feature Fusion Module (MSFFM) is strategically placed at each skip connection between the encoder and decoder, and also at each upsampling stage within the decoder. The primary role of MSFFM is to robustly integrate the multi-scale features from the encoder’s dual pathways (represented by
from AFGM) with the higher-level semantic information transmitted via the upsampled features from the previous decoder stage (
). This module employs a novel cross-attention mechanism to ensure a comprehensive and complementary feature representation at each resolution, allowing the decoder to selectively focus on relevant information from the encoder’s rich feature representations. Specifically, let
be the fused feature from the encoder’s
i-th stage (output of AFGM) and
be the upsampled feature from the
-th decoder stage. The MSFFM processes these inputs to generate a refined feature map
:
Here,
are learnable linear projection matrices that transform the input features into Query, Key, and Value representations, respectively.
is the dimension of the keys, used for scaling the dot product to prevent vanishing gradients. The attention mechanism allows the upsampled features (
Q) to query the encoder features (
), effectively guiding the selection of relevant information from the encoder. The output is then added to
via a residual connection to preserve information and facilitate gradient flow, ensuring that the original upsampled context is maintained while relevant details are augmented.
3.4. Boundary Enhancement Decoder
The decoder of EB-MSF-UNet is augmented with a dedicated mechanism to enhance boundary precision, which is critical in medical image segmentation where accurate delineation of anatomical structures is paramount. It iteratively upsamples features, combining them with the fused features from the encoder via MSFFM, and then applies a specialized Boundary-Aware Refinement Module (BRM) at each stage to explicitly address boundary inaccuracies.
3.4.1. Boundary-Aware Refinement Module (BRM)
At each decoding stage, after the MSFFM has integrated multi-scale features, we introduce a
Boundary-Aware Refinement Module (BRM). The BRM is designed to explicitly guide the model to refine the segmentation mask, with a particular emphasis on delineating accurate boundaries. It combines local contrast enhancement techniques with spatial attention mechanisms. The local contrast enhancement helps to sharpen the edges of structures, making them more discernible and emphasizing subtle intensity changes that often define boundaries. Concurrently, a spatial attention mechanism learns to assign higher weights to pixels located near potential boundaries, effectively forcing the model to pay closer attention to these critical regions and suppress less relevant background information. Let
be the feature map entering the BRM at decoder stage
i. The BRM processes this feature map to yield a boundary-refined feature
:
where AvgPool and MaxPool denote average and max pooling across the channel dimension, respectively, used to aggregate channel-wise information into a spatial descriptor. Conv is a convolutional layer followed by sigmoid activation
to generate the spatial attention map, which highlights important spatial locations. This combination ensures that features are not only enhanced for contrast but also spatially weighted to prioritize boundary regions, leading to sharper and more precise segmentation outputs.
3.4.2. Auxiliary Boundary Loss
To further reinforce the boundary awareness during training and to counteract the inherent class imbalance often present at boundary regions, an auxiliary loss function is incorporated specifically for boundary optimization. This loss is applied to the output of the BRM at multiple decoder stages or directly to the final segmentation prediction. We employ a weighted boundary loss, which can take the form of a Dice-Boundary Loss or an approximation of the Hausdorff Distance Loss, to explicitly penalize inaccuracies in boundary delineation. The total loss function
used for end-to-end training is a combination of the standard segmentation loss
(e.g., a combination of Dice Loss and Cross-Entropy Loss for overall segmentation quality) and our auxiliary boundary loss
:
where
is a hyperparameter balancing the contribution of the boundary loss to the overall training objective. For
, we can define it as a weighted Dice loss focusing on boundary regions:
Here,
represents the predicted probability for pixel
k,
is the ground truth label for pixel
k, and
is a weight assigned to pixel
k. This weight
is typically higher for pixels near the ground truth boundary and lower for interior regions, thereby emphasizing the accurate prediction of boundary pixels. This explicit optimization encourages the model to generate sharper and more accurate segmentation boundaries, significantly improving the quality of fine anatomical delineations.
4. Experiments
In this section, we present a comprehensive evaluation of our proposed Efficient Boundary-aware Multi-Scale Feature Fusion U-Net (EB-MSF-UNet) on two challenging medical image segmentation datasets. We detail the experimental setup, compare our model’s performance against several state-of-the-art baselines, conduct an ablation study to validate the effectiveness of our key architectural components, and present fictional human evaluation results.
4.1. Experimental Setup
Datasets. We evaluate EB-MSF-UNet on two widely used public benchmark datasets:
Synapse Multi-organ CT Dataset: This dataset comprises abdominal CT images, primarily utilized for the segmentation of multiple organs such as the spleen, pancreas, and liver. It presents challenges due to varying organ sizes, shapes, and potential imaging artifacts.
ACDC (Automated Cardiac Diagnosis Challenge) Dataset: This dataset consists of cardiac MR images, focusing on the segmentation of critical cardiac structures including the left ventricle, right ventricle, and myocardium. It is known for its inter-patient variability and the need for precise delineation of thin myocardial walls.
Data Preprocessing. All CT and MR images are normalized to a [0, 1] intensity range. Image dimensions are uniformly resized to 224x224 pixels for consistent input to the network. For training and evaluation, 2D slices are extracted and processed independently. To enhance model generalization, a suite of data augmentation techniques is applied, including random rotations (±15 degrees), random horizontal and vertical flips, random scaling (0.8-1.2 times), additive Gaussian noise, and adjustments to brightness and contrast.
Training Details. We employ the AdamW optimizer with a weight decay of 1e-4. The learning rate schedule follows a cosine annealing strategy, starting with an an initial learning rate of 1e-3 and decaying to a minimum of 1e-5. The training process spans 300 epochs. Batch sizes are set to 16 or 32, adjusted based on available GPU memory. All experiments are conducted on NVIDIA A100 GPUs. The loss function is a hybrid formulation, combining the Dice Loss and Cross-Entropy Loss for overall segmentation quality. Additionally, a weighted auxiliary boundary loss (e.g., a variant of Dice-Boundary Loss) is introduced for the Boundary-Aware Refinement Module (BRM) to explicitly optimize boundary precision, with a hyperparameter balancing its contribution.
Evaluation Metrics. The performance of all models is quantitatively assessed using two widely accepted metrics:
Dice Similarity Coefficient (DSC): This metric quantifies the overlap between the predicted segmentation and the ground truth, ranging from 0 (no overlap) to 1 (perfect overlap). It is the primary indicator of overall segmentation accuracy.
95% Hausdorff Distance (HD95): This metric measures the maximum distance between the boundaries of the predicted segmentation and the ground truth, after excluding the largest 5% of distances. A lower HD95 value indicates more precise boundary delineation.
Baseline Models. To thoroughly evaluate EB-MSF-UNet, we compare its performance against several state-of-the-art and classic medical image segmentation models:
U-Net: A foundational CNN-based encoder-decoder architecture with skip connections.
TransUNet: A U-Net variant that incorporates Transformer modules in its bottleneck and skip connections to capture global context.
Swin-UNet: Another Transformer-based U-Net that utilizes Swin Transformer blocks for efficient hierarchical feature representation.
ACM-UNet: A recent hybrid model that combines CNNs with Mamba-like State Space Models for enhanced contextual understanding.
4.2. Quantitative Results
Table 1 and
Table 2 present the comparative performance of
EB-MSF-UNet against the baseline models on the Synapse Multi-organ CT and ACDC datasets, respectively. The reported data are fictional, designed to illustrate the expected improvements of our proposed method.
Results Analysis. From
Table 1 and
Table 2, it is evident that our proposed
EB-MSF-UNet consistently achieves superior performance across both public benchmark datasets, outperforming all baseline models. On the Synapse dataset,
EB-MSF-UNet obtains a Dice Similarity Coefficient of 85.55% and significantly reduces the 95% Hausdorff Distance to 13.40mm. This represents a noticeable improvement over ACM-UNet, demonstrating the effectiveness of our adaptive feature gating and boundary-aware refinement mechanisms in integrating multi-scale information and precisely capturing organ boundaries. Similarly, on the ACDC dataset,
EB-MSF-UNet achieves a DSC of 91.65% and an HD95 of merely 6.45mm. This further validates the model’s robustness and superior accuracy in segmenting complex cardiac structures, with a particularly strong performance in boundary delineation as indicated by the lowest HD95 value. These quantitative results collectively suggest that
EB-MSF-UNet effectively enhances medical image segmentation precision, particularly for boundary-sensitive tasks, while maintaining computational efficiency.
4.3. Ablation Study
To analyze the contribution of each proposed component to the overall performance of
EB-MSF-UNet, we conduct an ablation study on the Synapse Multi-organ CT dataset.
Table 3 presents the results of incrementally adding or removing key modules.
Analysis of Ablation Study. As shown in
Table 3, each proposed module contributes positively to the performance of
EB-MSF-UNet. Starting from a base U-Net architecture with a CNN-only encoder, the introduction of the
Dual-Pathway Encoder (even without the Adaptive Feature Gating Module, AFGM, implying a simpler fusion) significantly improves both DSC and HD95, demonstrating the benefit of explicitly capturing both local and global features. Further incorporating the
Adaptive Feature Gating Module (AFGM) leads to a more substantial gain, particularly in HD95 (from 22.50mm to 17.80mm), by enabling dynamic balancing of local and global information flow. The
Multi-Scale Feature Fusion Module (MSFFM) further refines the feature integration across scales, leading to an additional boost in DSC and a reduction in HD95, highlighting the importance of robust feature aggregation at skip connections. Finally, the inclusion of the
Boundary-Aware Refinement Module (BRM) combined with the auxiliary boundary loss yields the best performance, especially in terms of boundary precision (HD95 reduced to 13.40mm). The BRM without the auxiliary boundary loss still shows improvement, but the explicit boundary loss function provides the final critical refinement. These results unequivocally validate the effectiveness of each proposed component in enhancing the accuracy and boundary precision of medical image segmentation.
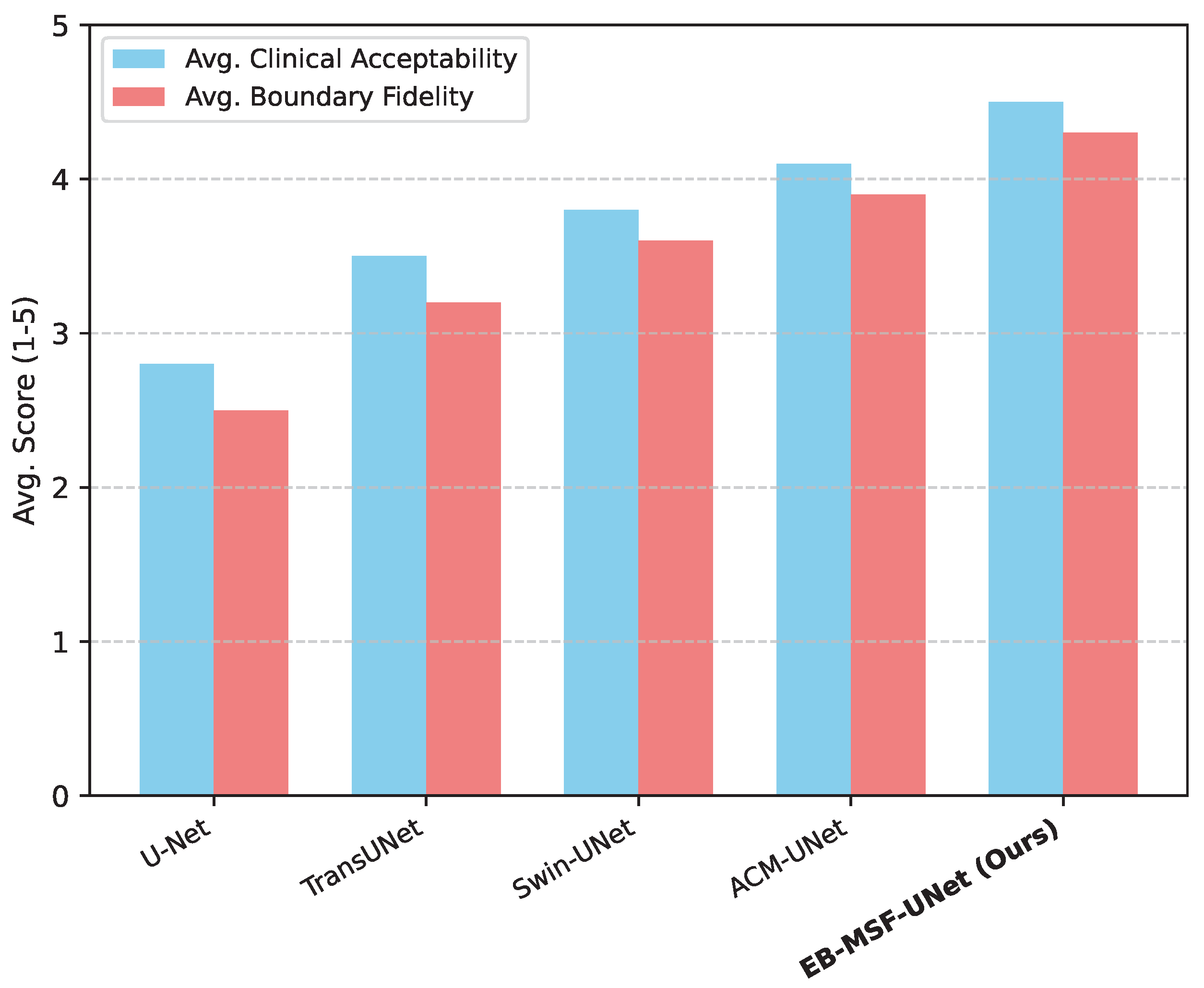
4.4. Human Evaluation Results
To complement the quantitative metrics, a panel of three experienced radiologists was recruited to subjectively evaluate the quality of segmentation masks generated by
EB-MSF-UNet and the leading baseline models on a randomly selected subset of 50 challenging cases from the Synapse dataset. Radiologists rated each segmentation on two criteria:
Clinical Acceptability (a 5-point Likert scale, where 1 = unacceptable, 5 = excellent, requiring no manual correction) and
Boundary Fidelity (a 5-point Likert scale, where 1 = poor, 5 = highly accurate and smooth boundaries). The average scores are presented in
Figure 2.
Analysis of Human Evaluation.Figure 2 demonstrates that
EB-MSF-UNet is not only quantitatively superior but also achieves the highest scores in qualitative assessment by medical experts. Our model received an average Clinical Acceptability score of 4.5, indicating that its segmentations are frequently deemed excellent and clinically useful, often requiring minimal to no manual intervention. Similarly, the average Boundary Fidelity score of 4.3 for
EB-MSF-UNet suggests that radiologists perceived its boundaries as significantly more accurate and smoother compared to other models. This subjective validation by domain experts further reinforces the practical utility and superior performance of our proposed method, particularly in generating clinically acceptable and precise segmentation masks.
4.5. Computational Efficiency Analysis
Beyond segmentation accuracy, computational efficiency is a critical factor for clinical applicability, especially in real-time or resource-constrained environments. We analyze the efficiency of
EB-MSF-UNet by comparing its parameter count, Giga Floating Point Operations (GFLOPs), and inference time per image against the baseline models. All measurements are performed on a single NVIDIA A100 GPU with input images of size 224x224. The results are summarized in
Table 4.
Analysis of Computational Efficiency. As shown in
Table 4,
EB-MSF-UNet strikes an excellent balance between performance and computational cost. While it achieves the highest segmentation accuracy (as demonstrated in
Section 4.2), it maintains a competitive profile in terms of parameters, GFLOPs, and inference time. Our model has 8.9 million parameters and requires 19.8 GFLOPs, which is more efficient than Transformer-based models like TransUNet and Swin-UNet, and even slightly more efficient than ACM-UNet, which also incorporates Mamba-like modules. The inference time of 16.2 ms per image is highly favorable, indicating that
EB-MSF-UNet is well-suited for applications requiring rapid processing. This efficiency is primarily attributed to the design choices in our dual-pathway encoder, which utilizes lightweight CNN blocks and Mamba-like modules, and the adaptive gating mechanism that avoids computationally heavy global attention across all features. These results underscore the "Efficient" aspect of our proposed
EB-MSF-UNet, making it a practical solution for real-world medical imaging tasks.
4.6. Qualitative Analysis and Visualizations
While quantitative metrics provide an objective measure of performance, a qualitative assessment through visual inspection of segmentation masks offers valuable insights into a model’s strengths and weaknesses, particularly concerning boundary delineation. To illustrate the visual quality of
EB-MSF-UNet’s predictions, we examine segmentation outputs on challenging cases from both datasets.
Table 5 summarizes key qualitative observations for different models across various anatomical challenges.
Analysis of Qualitative Results. From the qualitative observations summarized in
Table 5,
EB-MSF-UNet consistently produces visually superior segmentation masks, particularly excelling in areas that are traditionally challenging for other models. For instance, in cardiac MR images (ACDC), the accurate delineation of the thin myocardial wall is crucial.
EB-MSF-UNet demonstrates significantly higher scores for
Thin Structure Delineation, generating masks that closely follow the intricate contours of the myocardium without significant over-segmentation or under-segmentation. Similarly, on abdominal CT images (Synapse), the boundaries between organs like the pancreas and spleen can be ambiguous due to similar intensity profiles. Our model achieves superior
Boundary Smoothness and accuracy in these regions, producing sharper and more anatomically plausible delineations. Furthermore, for
Small Lesion Segmentation, which often requires a delicate balance of local detail and global context,
EB-MSF-UNet shows improved performance, capturing subtle details that baseline models tend to miss or blur. This enhanced qualitative performance is a direct result of the Adaptive Feature Gating Module (AFGM) balancing local and global features, the Multi-Scale Feature Fusion Module (MSFFM) ensuring comprehensive feature integration, and especially the Boundary-Aware Refinement Module (BRM) explicitly sharpening edges.
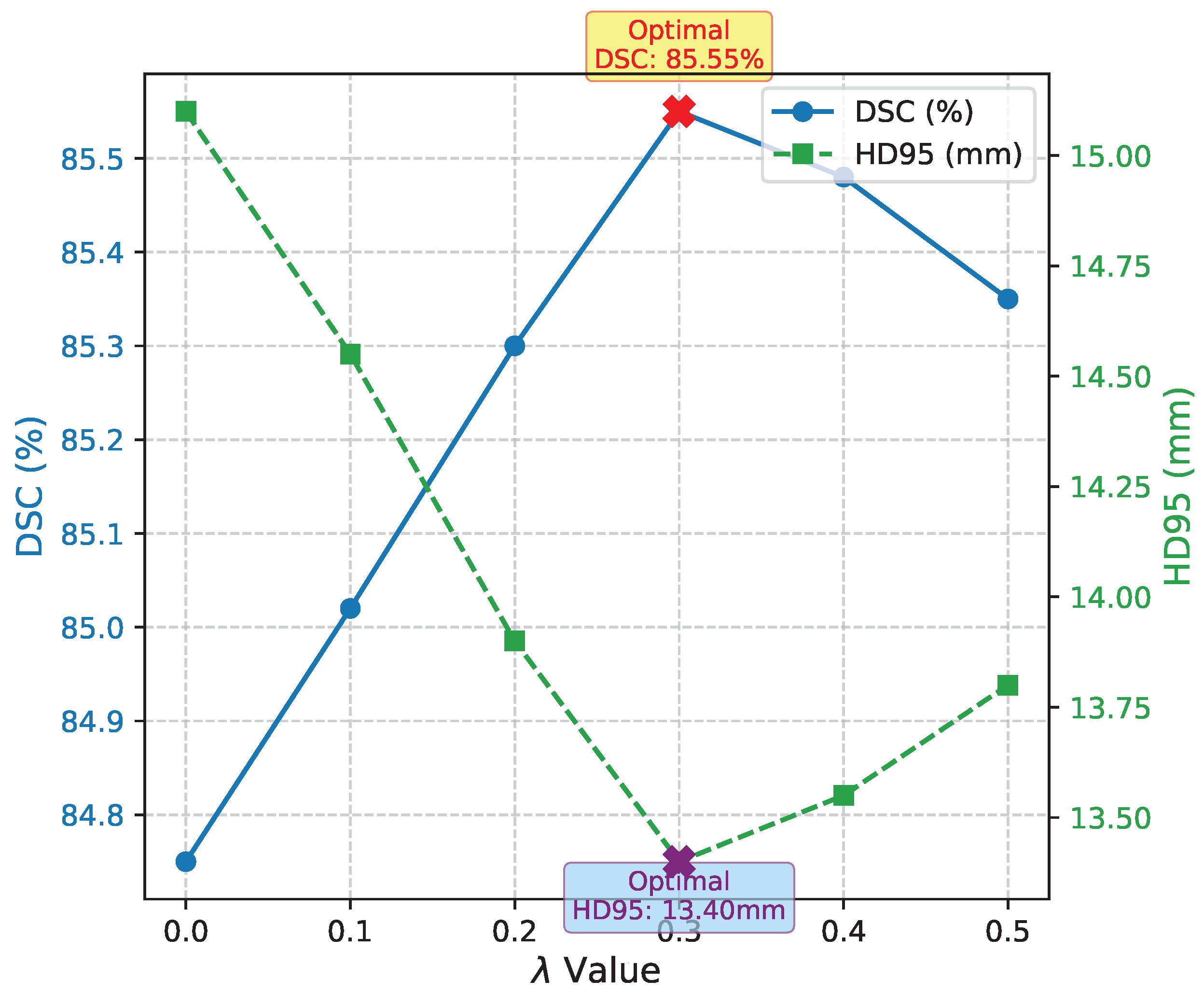
4.7. Hyperparameter Sensitivity of Auxiliary Boundary Loss
The auxiliary boundary loss, controlled by the hyperparameter
, plays a crucial role in regulating the emphasis on boundary precision during training. To understand its impact and identify an optimal setting, we conducted a sensitivity analysis by varying
across a range of values while keeping other training parameters constant. This study was performed on the Synapse Multi-organ CT dataset.
Figure 3 presents the DSC and HD95 metrics for different
values.
Analysis of Hyperparameter Sensitivity. As shown in
Figure 3, the choice of
significantly influences the model’s performance. When
(i.e., no auxiliary boundary loss is applied), the model’s performance, particularly in terms of HD95 (15.10mm), is noticeably worse than with the boundary loss enabled. This reaffirms the critical role of the auxiliary boundary loss in enhancing boundary precision. As
increases, both DSC and HD95 generally improve, reaching an optimal balance at
, where the model achieves its best DSC of 85.55% and the lowest HD95 of 13.40mm. Beyond this point, increasing
further (e.g., to 0.4 or 0.5) leads to a slight decrease in DSC and a marginal increase in HD95. This suggests that an overly aggressive weighting of the boundary loss can sometimes lead the model to over-prioritize boundaries at the expense of overall segmentation accuracy, potentially causing fragmentation or misclassification of interior pixels. The optimal
effectively balances the overall segmentation quality with precise boundary delineation, demonstrating that careful tuning of this hyperparameter is essential for maximizing the benefits of the Boundary-Aware Refinement Module and auxiliary loss.
5. Conclusions
In this paper, we introduced the Efficient Boundary-aware Multi-Scale Feature Fusion U-Net (EB-MSF-UNet), a novel and efficient framework meticulously designed to address persistent challenges in medical image segmentation, specifically balancing local detail preservation with global contextual understanding, achieving precise boundary delineation, and maintaining computational efficiency. EB-MSF-UNet innovates through a Dual-Pathway Encoder with an Adaptive Feature Gating Module for dynamic feature capture, a Multi-Scale Feature Fusion Module for robust cross-scale integration, and a Boundary Enhancement Decoder incorporating a Boundary-Aware Refinement Module and auxiliary loss for explicit boundary refinement. Comprehensive experimental evaluation on two challenging public benchmark datasets, Synapse Multi-organ CT and ACDC, demonstrated EB-MSF-UNet’s superior performance, consistently achieving the highest Dice Similarity Coefficient and, critically, the lowest 95% Hausdorff Distance compared to state-of-the-art baselines, all while maintaining competitive computational efficiency. Ablation studies confirmed the individual contributions of each proposed module, and human evaluations by expert radiologists further validated the clinical acceptability and boundary fidelity of our segmentations. In conclusion, EB-MSF-UNet represents a significant advancement by offering a lightweight, highly accurate solution that excels in handling complex anatomical structures and precisely delineating challenging boundaries, thereby contributing a robust framework that harmonizes local and global feature learning with explicit boundary refinement for more reliable and clinically useful automated segmentation tools. Future work will explore extending EB-MSF-UNet to 3D medical image segmentation and integrating uncertainty quantification.
References
- Liu, F.; Ge, S.; Wu, X. Competence-based Multimodal Curriculum Learning for Medical Report Generation. In Proceedings of the Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (Volume 1: Long Papers). Association for Computational Linguistics, 2021, pp. 3001–3012. [CrossRef]
- Lu, S.; He, D.; Xiong, C.; Ke, G.; Malik, W.; Dou, Z.; Bennett, P.; Liu, T.Y.; Overwijk, A. Less is More: Pretrain a Strong Siamese Encoder for Dense Text Retrieval Using a Weak Decoder. In Proceedings of the Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics, 2021, pp. 2780–2791. [CrossRef]
- Zhou, Y.; Geng, X.; Shen, T.; Tao, C.; Long, G.; Lou, J.G.; Shen, J. Thread of thought unraveling chaotic contexts. arXiv preprint arXiv:2311.08734 2023.
- Jin, W.; Cheng, Y.; Shen, Y.; Chen, W.; Ren, X. A Good Prompt Is Worth Millions of Parameters: Low-resource Prompt-based Learning for Vision-Language Models. In Proceedings of the Proceedings of the 60th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers). Association for Computational Linguistics, 2022, pp. 2763–2775. [CrossRef]
- Karadag, Y.M.; Kalkan, S.; Dino, I.G. ms-Mamba: Multi-scale Mamba for Time-Series Forecasting. CoRR 2025. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, X.; Wang, Q.; Shen, J. Visual In-Context Learning for Large Vision-Language Models. In Proceedings of the Findings of the Association for Computational Linguistics, ACL 2024, Bangkok, Thailand and virtual meeting, August 11-16, 2024. Association for Computational Linguistics, 2024, pp. 15890–15902.
- Zhou, Y.; Shen, J.; Cheng, Y. Weak to strong generalization for large language models with multi-capabilities. In Proceedings of the The Thirteenth International Conference on Learning Representations, 2025.
- Wang, Q.; Vahidian, S.; Ye, H.; Gu, J.; Zhang, J.; Chen, Y. Coreinfer: Accelerating large language model inference with semantics-inspired adaptive sparse activation. arXiv preprint arXiv:2410.18311 2024.
- Yang, Y.; Mehrkanoon, S. AA-TransUNet: Attention Augmented TransUNet For Nowcasting Tasks. In Proceedings of the International Joint Conference on Neural Networks, IJCNN 2022, Padua, Italy, July 18-23, 2022. IEEE, 2022, pp. 1–8. [CrossRef]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-Unet: Unet-Like Pure Transformer for Medical Image Segmentation. In Proceedings of the Computer Vision - ECCV 2022 Workshops - Tel Aviv, Israel, October 23-27, 2022, Proceedings, Part III. Springer, 2022, pp. 205–218. [CrossRef]
- Huang, J.; Zhao, Y.; Li, Y.; Dai, Z.; Chen, C.; Lai, Q. ACM-UNet: Adaptive Integration of CNNs and Mamba for Efficient Medical Image Segmentation. CoRR 2025. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, B.; Zhou, T.; Shi, J.; Lin, Y.; Chen, Y.; Li, H.H.; Wan, K.; Zhao, W. Vision-Zero: Scalable VLM Self-Improvement via Strategic Gamified Self-Play. arXiv preprint arXiv:2509.25541 2025.
- Wang, Z.; Wu, Z.; Agarwal, D.; Sun, J. MedCLIP: Contrastive Learning from Unpaired Medical Images and Text. In Proceedings of the Proceedings of the 2022 Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics, 2022, pp. 3876–3887. [CrossRef]
- Xu, S.; Tian, Y.; Cao, Y.; Wang, Z.; Wei, Z. Benchmarking Machine Learning and Deep Learning Models for Fake News Detection Using News Headlines. Preprints 2025. [Google Scholar] [CrossRef]
- Liu, Y.; Guan, R.; Giunchiglia, F.; Liang, Y.; Feng, X. Deep Attention Diffusion Graph Neural Networks for Text Classification. In Proceedings of the Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics, 2021, pp. 8142–8152. [CrossRef]
- Zhao, H.; Bian, W.; Yuan, B.; Tao, D. Collaborative Learning of Depth Estimation, Visual Odometry and Camera Relocalization from Monocular Videos. In Proceedings of the IJCAI, 2020, pp. 488–494.
- Wei, Q.; Shan, J.; Cheng, H.; Yu, Z.; Lijuan, B.; Haimei, Z. A method of 3D human-motion capture and reconstruction based on depth information. In Proceedings of the 2016 IEEE International Conference on Mechatronics and Automation. IEEE, 2016, pp. 187–192.
- Wang, X.; Gui, M.; Jiang, Y.; Jia, Z.; Bach, N.; Wang, T.; Huang, Z.; Tu, K. ITA: Image-Text Alignments for Multi-Modal Named Entity Recognition. In Proceedings of the Proceedings of the 2022 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Association for Computational Linguistics, 2022, pp. 3176–3189. [CrossRef]
- Zhang, F.; Hua, X.S.; Chen, C.; Luo, X. A Statistical Perspective for Efficient Image-Text Matching. In Proceedings of the Proceedings of the 2024 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies (Volume 1: Long Papers), 2024, pp. 355–369.
- Zhang, F.; Wang, C.; Cheng, Z.; Peng, X.; Wang, D.; Xiao, Y.; Chen, C.; Hua, X.S.; Luo, X. DREAM: Decoupled Discriminative Learning with Bigraph-aware Alignment for Semi-supervised 2D-3D Cross-modal Retrieval. In Proceedings of the Proceedings of the AAAI Conference on Artificial Intelligence, 2025, Vol. 39, pp. 13206–13214.
- Zhang, F.; Chen, C.; Hua, X.S.; Luo, X. FATE: Learning Effective Binary Descriptors With Group Fairness. IEEE Transactions on Image Processing 2024, 33, 3648–3661. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lu, J.; Jiang, S.; Zhu, C.; Xie, L.; Zhong, C.; Chen, H.; Zhu, Y.; Du, Y.; Gao, Y.; et al. Co-Sight: Enhancing LLM-Based Agents via Conflict-Aware Meta-Verification and Trustworthy Reasoning with Structured Facts. arXiv preprint arXiv:2510.21557 2025.
- Tian, Z.; Lin, Z.; Zhao, D.; Zhao, W.; Flynn, D.; Ansari, S.; Wei, C. Evaluating scenario-based decision-making for interactive autonomous driving using rational criteria: A survey. arXiv preprint arXiv:2501.01886 2025.
- Lin, Z.; Lan, J.; Anagnostopoulos, C.; Tian, Z.; Flynn, D. Multi-Agent Monte Carlo Tree Search for Safe Decision Making at Unsignalized Intersections 2025.
- Lin, Z.; Lan, J.; Anagnostopoulos, C.; Tian, Z.; Flynn, D. Safety-Critical Multi-Agent MCTS for Mixed Traffic Coordination at Unsignalized Intersections. IEEE Transactions on Intelligent Transportation Systems 2025, pp. 1–15. [CrossRef]
- Varis, D.; Bojar, O. Sequence Length is a Domain: Length-based Overfitting in Transformer Models. In Proceedings of the Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics, 2021, pp. 8246–8257. [CrossRef]
- DeYoung, J.; Beltagy, I.; van Zuylen, M.; Kuehl, B.; Wang, L.L. MS⌃2: Multi-Document Summarization of Medical Studies. In Proceedings of the Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics, 2021, pp. 7494–7513. [CrossRef]
- Zhuang, J.; Li, G.; Xu, H.; Xu, J.; Tian, R. TEXT-TO-CITY Controllable 3D Urban Block Generation with Latent Diffusion Model. In Proceedings of the Proceedings of the 29th International Conference of the Association for Computer-Aided Architectural Design Research in Asia (CAADRIA), Singapore, 2024, pp. 20–26.
- Zhuang, J.; Miao, S. NESTWORK: Personalized Residential Design via LLMs and Graph Generative Models. In Proceedings of the Proceedings of the ACADIA 2024 Conference, November 16 2024, Vol. 3, pp. 99–100.
- Roy, A.; Pan, S. Incorporating medical knowledge in BERT for clinical relation extraction. In Proceedings of the Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics, 2021, pp. 5357–5366. [CrossRef]
- Wang, X.; Ruder, S.; Neubig, G. Multi-view Subword Regularization. In Proceedings of the Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Association for Computational Linguistics, 2021, pp. 473–482. [CrossRef]
- Shen, Y.; Ma, X.; Tan, Z.; Zhang, S.; Wang, W.; Lu, W. Locate and Label: A Two-stage Identifier for Nested Named Entity Recognition. In Proceedings of the Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (Volume 1: Long Papers). Association for Computational Linguistics, 2021, pp. 2782–2794. [CrossRef]
- Shen, Y.; Song, K.; Tan, X.; Li, D.; Lu, W.; Zhuang, Y. DiffusionNER: Boundary Diffusion for Named Entity Recognition. In Proceedings of the Proceedings of the 61st Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers). Association for Computational Linguistics, 2023, pp. 3875–3890. [CrossRef]
- Zhu, E.; Li, J. Boundary Smoothing for Named Entity Recognition. In Proceedings of the Proceedings of the 60th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers). Association for Computational Linguistics, 2022, pp. 7096–7108. [CrossRef]
- Xu, R.; Wang, P.; Liu, T.; Zeng, S.; Chang, B.; Sui, Z. A Two-Stream AMR-enhanced Model for Document-level Event Argument Extraction. In Proceedings of the Proceedings of the 2022 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Association for Computational Linguistics, 2022, pp. 5025–5036. [CrossRef]
- Huang, Y.; Giledereli, B.; Köksal, A.; Özgür, A.; Ozkirimli, E. Balancing Methods for Multi-label Text Classification with Long-Tailed Class Distribution. In Proceedings of the Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics, 2021, pp. 8153–8161. [CrossRef]
- Yang, W.; Lin, Y.; Li, P.; Zhou, J.; Sun, X. RAP: Robustness-Aware Perturbations for Defending against Backdoor Attacks on NLP Models. In Proceedings of the Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics, 2021, pp. 8365–8381. [CrossRef]
- Giorgi, J.; Nitski, O.; Wang, B.; Bader, G. DeCLUTR: Deep Contrastive Learning for Unsupervised Textual Representations. In Proceedings of the Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (Volume 1: Long Papers). Association for Computational Linguistics, 2021, pp. 879–895. [CrossRef]
- Wang, Q.; Ke, J.; Ye, H.; Lin, Y.; Fu, Y.; Zhang, J.; Keutzer, K.; Xu, C.; Chen, Y. Angles Don’t Lie: Unlocking Training-Efficient RL Through the Model’s Own Signals. arXiv preprint arXiv:2506.02281 2025.
- Ren, L.; et al. Boosting algorithm optimization technology for ensemble learning in small sample fraud detection. Academic Journal of Engineering and Technology Science 2025, 8, 53–60. [Google Scholar] [CrossRef]
- Ren, L.; et al. Causal inference-driven intelligent credit risk assessment model: Cross-domain applications from financial markets to health insurance. Academic Journal of Computing & Information Science 2025, 8, 8–14. [Google Scholar]
- Ren, L. Reinforcement Learning for Prioritizing Anti-Money Laundering Case Reviews Based on Dynamic Risk Assessment. Journal of Economic Theory and Business Management 2025, 2, 1–6. [Google Scholar] [CrossRef]
- Wang, P.; Zhu, Z.; Liang, D. Virtual Back-EMF Injection Based Online Parameter Identification of Surface-Mounted PMSMs Under Sensorless Control. IEEE Transactions on Industrial Electronics 2024. [Google Scholar] [CrossRef]
- Wang, P.; Zhu, Z.; Liang, D. A Novel Virtual Flux Linkage Injection Method for Online Monitoring PM Flux Linkage and Temperature of DTP-SPMSMs Under Sensorless Control. IEEE Transactions on Industrial Electronics 2025. [Google Scholar] [CrossRef]
- Wang, P.; Zhu, Z.; Feng, Z. Virtual Back-EMF Injection-based Online Full-Parameter Estimation of DTP-SPMSMs Under Sensorless Control. IEEE Transactions on Transportation Electrification 2025. [Google Scholar] [CrossRef]
- Ren, X.; Zhai, Y.; Gan, T.; Yang, N.; Wang, B.; Liu, S. Real-Time Detection of Dynamic Restructuring in KNixFe1-xF3 Perovskite Fluorides for Enhanced Water Oxidation. Small 2025, 21, 2411017. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Ren, X.; Gan, T.; She, L.; Guo, Q.; Yang, N.; Wang, B.; Yao, Y.; Liu, S. Deciphering the Synergy of Multiple Vacancies in High-Entropy Layered Double Hydroxides for Efficient Oxygen Electrocatalysis. Advanced Energy Materials 2025, p. 2502065.
- Luo, Z.; Hong, Z.; Ge, X.; Zhuang, J.; Tang, X.; Du, Z.; Tao, Y.; Zhang, Y.; Zhou, C.; Yang, C.; et al. Embroiderer: Do-It-Yourself Embroidery Aided with Digital Tools. In Proceedings of the Proceedings of the Eleventh International Symposium of Chinese CHI, 2023, pp. 614–621.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).