1. Introduction
The mature arteriovenous fistula (AVF) remains the preferred vascular access for hemodialysis, offering superior long-term patency and fewer complications compared to alternative modalities [
1]. Unfortunately, no therapeutic interventions currently exist to prevent the failure of approximately 40% of newly created accesses due to postoperative venous stenosis. The low success rate of clinical trials aimed at improving AVF maturation is now thought to stem, at least in part, from the misleading pre-clinical data derived from poorly characterized animal models.
Two mouse AVF models are commonly used in mechanistic studies that investigate the adaptive response of the vein to supra-arterial circulation: the carotid–jugular model, which involves an end-to-side anastomosis of the jugular vein to the carotid artery [
2], and the aortocaval puncture model, in which the aorta is connected side-to-side to the inferior vena cava using a needle puncture [
3]. These models enable the use of genetic knockout and knock-in strains for precise loss- and gain-of-function studies to test the involvement of specific genes in postoperative venous remodeling. Between them, the carotid-jugular AVF is the one that most closely resembles the human AVF in anatomic configuration. While these models have provided important insights into the hemodynamic and molecular alterations after AVF creation [
2,
3,
4,
5,
6,
7,
8,
9], a comprehensive understanding of the cellular dynamics and gene expression programs driving AVF maturation remains incomplete.
In this study, we generated the first temporal transcriptional profiles and single-cell transcriptomic maps of the mouse carotid–jugular AVF. These data provide detailed insights into the roles of myofibroblasts, fibroblasts, and immune cells in the healing of the vascular wall and cellular differentiation after anastomosis. Notably, several key molecular changes identified in mice were also observed in human AVFs [
10], underscoring the translational relevance of this model for investigating the fibrotic mechanisms driving AVF failure.
2. Materials and Methods
2.1. Mice and Surgical Procedures
Male and female C57BL/6 mice (Jackson Laboratory) 20-23 g in weight were randomly allocated to bulk (n=25) and single-cell RNA sequencing (scRNA-seq, n=20). All mice underwent two consecutive surgeries: 5/6 nephrectomy and AVF creation 3-4 weeks later by anastomosing the end of the jugular vein to the side of the carotid artery [
9,
11]. For bulk RNA-seq, animals were euthanized and AVFs were harvested at early (3.6 ± 2.4 days), mid (10.0 ± 0.7 days), and late (20.0 ± 1.8 days) time points after AVF creation. The early (5.5 ± 1.5 days) and late (26.1 ± 3.4 days) time points were also profiled by scRNA-seq. Contralateral jugular veins were sequenced as the control group. At the end of the protocol, animals presented a blood urea nitrogen of 52.8 ± 6.64 mg/dL, confirming the success of the 5/6 nephrectomy. All animal procedures were approved by the University of Miami Institutional Animal Care and Use Committee and adhered to the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
2.2. Bulk RNA Sequencing
Tissues were ground to a fine powder in a Spex/Mill 6770 cryogenic grinder (SPEX SamplePrep, Metuchen, NJ). RNA was extracted using Trizol and treated with DNase as previously described [
12]. Samples were submitted to the University of Miami John P. Hussman Institute for Human Genomics, Center for Genome Technology, for sequencing. Total RNA was quantified and qualified using the Agilent Bioanalyzer, ensuring an RNA integrity score > 5. Ribosomal RNA-depleted sequencing libraries were prepared using the Illumina TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero, starting with 500 ng of total RNA. Libraries were sequenced on the Illumina NextSeq500, generating >40 million raw reads per sample. Raw counts were normalized using the DESeq2 package in R Studio, and genes with a base mean < 100 were filtered out from downstream differential gene expression analyses. Pathway and process enrichment analyses based on differentially expressed genes (DEGs; log2[fold change] > 1, p_adj < 0.05) were performed using the Metascape tool [
13]. Matrisome genes were identified from the curated and proteomics-validated blood vessel collection at the Matrisome Database [
14].
2.3. Single-Cell RNA Sequencing
A pool of 10 AVFs or contralateral veins yielded 30,000 – 50,000 optimal cells for scRNA-seq. The cell suspensions were prepared by finely mincing the vessels and incubating them in 1X Vessel Dissociation Enzyme Solution (containing collagenase types XI and II, hyaluronidase, soybean trypsin inhibitor, DNase I, dispase, elastase, and HEPES) for 90 minutes at 37°C as previously described [
15]. The cell suspension was filtered through a 40-μm strainer, washed twice with Hank’s Balanced Salt Solution (HBSS), and resuspended in 0.1% BSA-PBS. Libraries were prepared using the Chromium Single Cell 3' Library and Gel Bead Kit v3 (10X Genomics) targeting 10,000 cells per group and sequenced on the Illumina NovaSeq 6000 at the John P. Hussman Institute.
Data processing and analysis were performed using the Seurat package (Version 4.0.2) in R. Cells expressing fewer than 200 or more than 6,000 genes, or with >10% mitochondrial gene expression, were excluded. Data were normalized using the “
LogNormalize” method and using a scale factor of 10,000. Variable genes were identified using Seurat’s
FindVariableGenes function. Principal component analysis was performed on variable genes, and the top 12 principal components were selected for further analysis. Uniform Manifold Approximation and Projection (UMAP) and clustering were performed with a resolution of 0.5. Marker genes for each cluster were identified using the Wilcoxon rank-sum test via the
FindAllMarkers function. Pathway enrichment analysis was performed using the webpage-based tool Metascape [
13]. Gene set analysis was conducted using GeneAgent [
16] focusing on genes with an average log2(fold change) > 1 and p_adj < 0.05. Functional scores were added to the integrated Seurat object using the function
AddModuleScore in Seurat 4.0. Scores were based on the “GOBP_Leukocyte_Activation_Involved_In_Inflammatory_Response", “GOBP_Positive_Regulation_Of_Extracellular_Matrix_Organization.v2025”, “COATES_Macrophage_M1_VS_M2_DN.v2025”, and “GAVISH_3CA_Metaprogram_Fibroblasts_Myofibroblasts.v2024” gene signature modules from the Molecular Signatures Database [
17]. Ligand-receptor interactomes were analyzed using CellChat v2 [
18]. For comparison purposes, we downloaded the single cell datasets of human pre-access veins, early AVFs (5-7 days postop), and transposition AVFs (~90 days postop) from the Gene Expression Omnibus repository (accession number GSE305423). Differential gene expression analyses of human mono/macs and myofibroblast/fibroblasts included the same number of cells as the corresponding analyses in mice. Pseudotime trajectory analysis was performed as described in the Monocle3 vignette [
19,
20]. The top 50 transcription factors in fibroblasts were predicted based on the expression of target genes in the DoRothEA database [
21].
3. Results
3.1. The Mouse AVF Remodels Through Waves of Inflammation and ECM Deposition
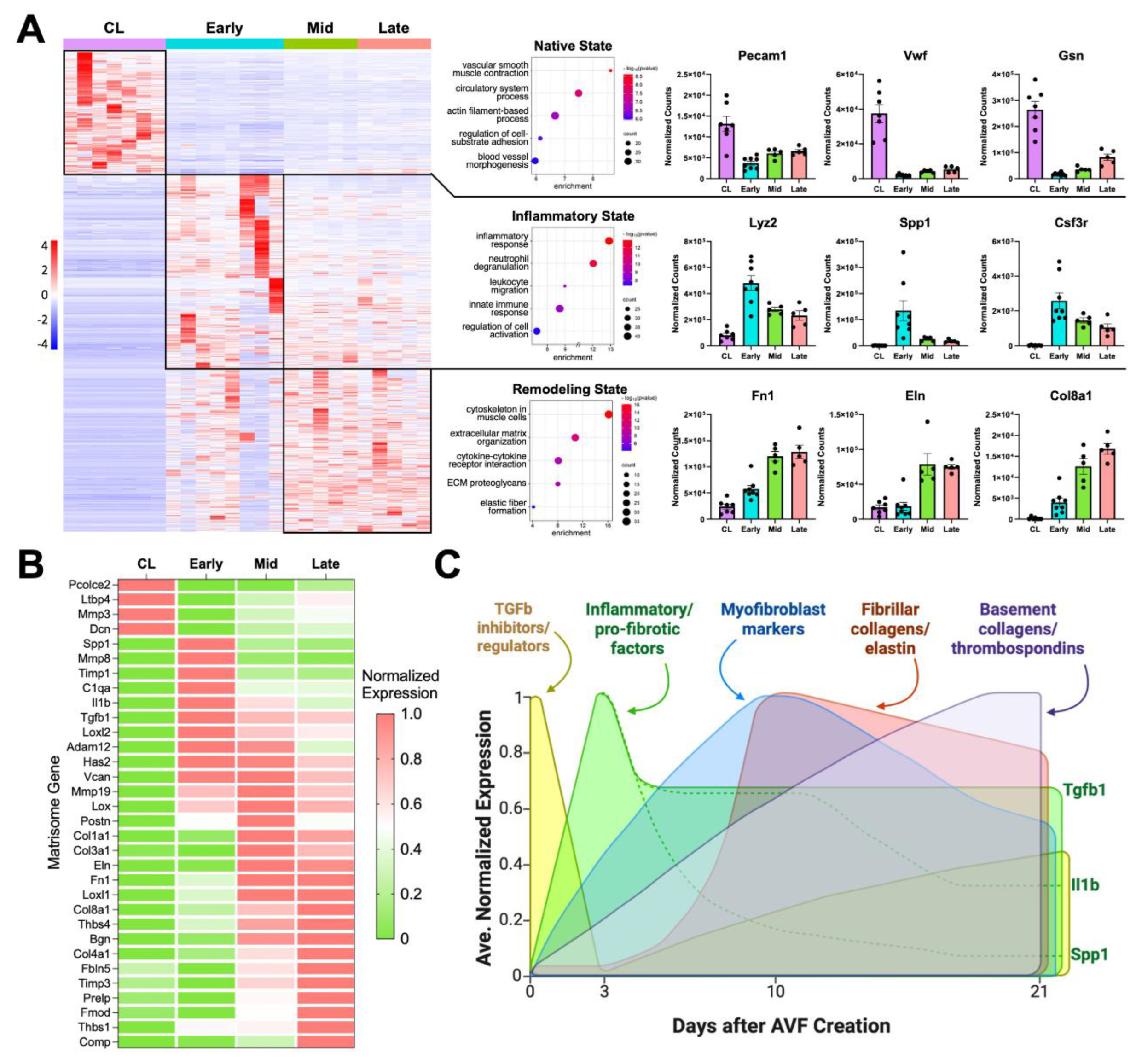
We first performed bulk RNA sequencing of the native contralateral vein and the outflow vein of the AVF collected at early (3.6 ± 2.4 days), mid (10.0 ± 0.7 days), and late (20.0 ± 1.8 days) time points after anastomosis to identify the global transcriptional changes during remodeling of the mouse AVF. The vein, prior to AVF creation, was predominantly characterized by transcriptional programs associated with homeostatic vascular functions, including smooth muscle cell (SMC) contraction, circulatory system regulation, and actin filament–based processes, compared with the postoperative groups (
Figure 1A). Exposure of the vein to supraphysiological conditions significantly altered the expression of a fourth of the transcriptome (4,541 of 20,094 captured genes) (
Figure 1A). Differentially expressed genes (log₂ fold change ≥ 1, adjusted p ≤ 0.01) revealed two distinct transcriptional phases after anastomosis: an early postoperative phase characterized by strong inflammatory activation, and a mid-to-late phase dominated by wall remodeling and healing. The early postoperative phase showed upregulation of myeloid or lymphoid markers (e.g.,
Lyz2, Csf3r, Cx3cr1), complement genes (e.g.,
C1qa, C1qb, C1qc), and cytokines (e.g.,
Spp1, Il1b, Cxcl5) (
Figure 1A-B, Data Set S1). This early response was also accompanied by a marked decline in canonical endothelial (e.g.,
Pecam1, Vwf) and SMC markers (e.g.,
Myh11, Cnn1, Lmod1) that persisted during remodeling.
Inhibitors of TGF-β signaling and pro-collagen processing (e.g.,
Ltbp4, Pcolce2, Dcn) [
22,
23,
24], as well as metalloproteinase activators (
Mmp3) [
25], decreased acutely after anastomosis to set the stage for new ECM deposition (
Figure 1B). By postoperative day 3, there was a balance of metalloproteases and their inhibitors (
Mmp8, Adam12, Timp1), along with the upregulation of pro-fibrotic factors (
Spp1, Tgfb1), ECM cross-linking enzymes (
Loxl2), and genes involved in the production of a provisional ECM (
Has2, Vcan) [
26,
27]. By day 10, gene activity associated with vascular wall healing/remodeling become even more evident. Expression of periostin (
Postn) peaked, indicating maximal myofibroblast activation [
28,
29]. Accordingly, there was significant upregulation of fibrillar collagens (
Col1a1, Col3a1), elastin (
Eln), fibronectin (
Fn1), and additional crosslinking enzymes (
Lox, Loxl1) that remained high in late fistulas. The expression of basement collagens (
Col8a1, Col4a1) and thrombospondins (
Thbs1, Thbs4) peaked at 21 days, likely reflecting the stabilization of cell-ECM interactions and enhanced mechanotransduction. Upregulation of the mechanosensitive machinery also reflects the ability of the vein to continue remodeling. In fact, additional matrisome genes involved in exacerbated ECM deposition (
Prelp, Comp) and inhibition of metalloproteinases (
Timp3) were significantly upregulated during this late remodeling period (
Figure 1B-C).
3.2. Immune Cell Infiltration and Activation of Fibroblasts Explain the Stages of Venous Remodeling
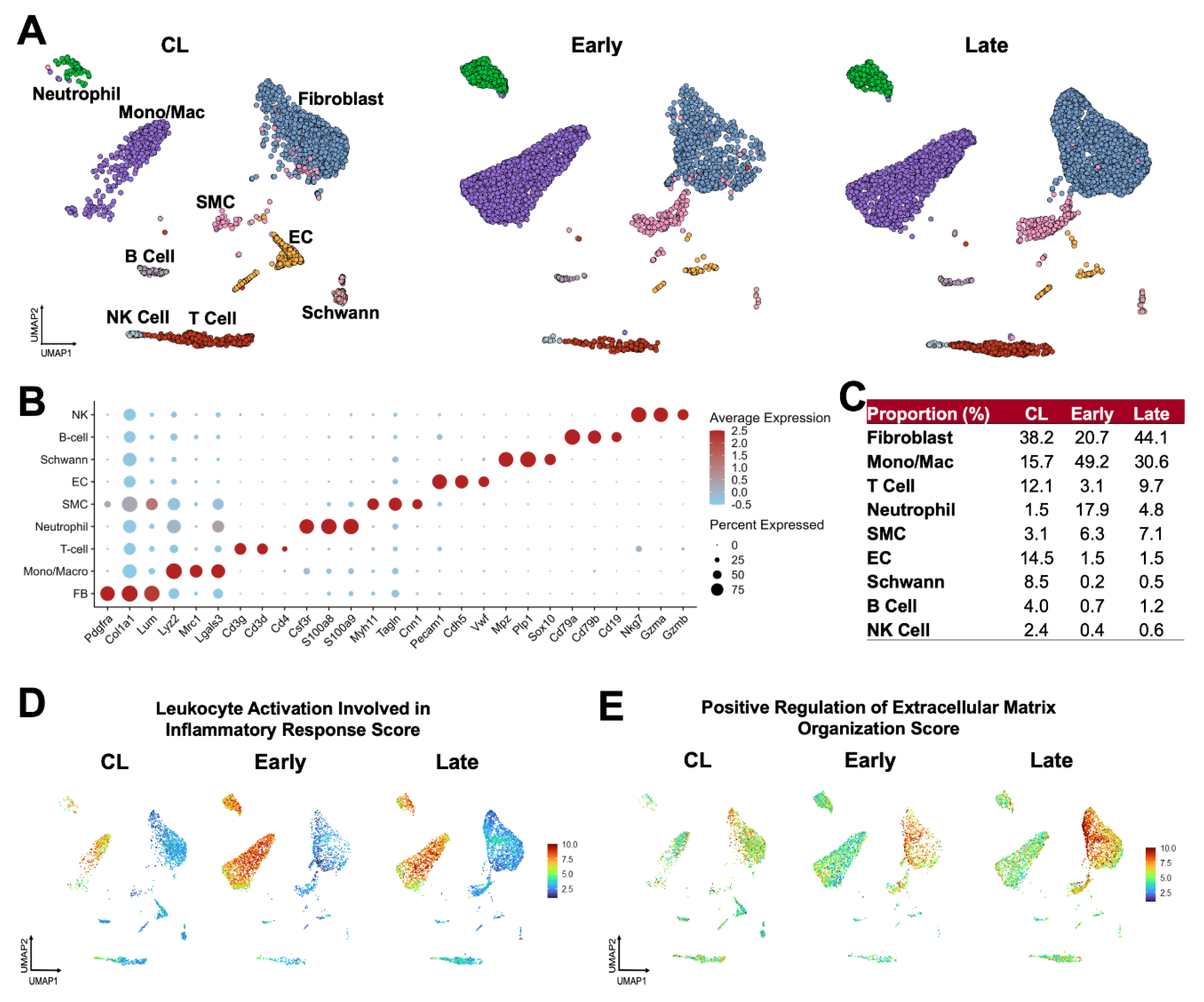
To better define the cell populations contributing to the early and late remodeling of the mouse AVF, we performed scRNA-seq of contralateral jugular veins and the outflow veins of early (5.5 ± 1.5 days postop) and late AVFs (26.1 ± 3.4 days) (
Figure 2A-C). Similar to human veins and AVFs [
10], the most abundant populations in the three experimental groups were fibroblasts (range 20.7 – 44.1%), monocytes/macrophages (15.7 – 49.2%), and T cells (3.1 – 12.1%). We identified a de novo infiltration of mono/macs (3-fold increase) and neutrophils (12-fold) in early AVFs that explains the inflammatory burst inferred by bulk RNA-seq (
Figure 2C). The proportion of fibroblasts increased modestly from 38.2% in contralateral veins to 44.1% in AVFs, while SMCs increased from 3.1 to 7.1%. Postoperatively, the latter were characterized by significant upregulation of stress-response genes (e.g.,
Spp1, S100a4, Atf3), likely as a result of mechanical stretching and high oxygen conditions (
Figure S1A). In contrast, endothelial cells (EC) decreased in proportion after AVF creation (14.5 to 1.5%). In those that remained, a lower expression of canonical EC markers (e.g.,
Vwf, Flt1) and upregulation of contractile genes (e.g.,
Acta2, Tagln) suggested an endothelial-to-mesenchymal transformation (
Figure S1B).
The global changes in cellular functions in the murine jugular vein were characterized using gene signature scores (
Figure 2D-E). The inflammatory leukocyte activation score was elevated in mono/macs of early AVFs and remained high in late AVFs compared with the contralateral veins. Neutrophils were also highly inflammatory but present at lower proportions. Concurrently, the score of ECM organization illustrated the acute activation of fibroblasts after anastomosis and a progressive increment from contralateral veins to late AVFs. There were no significant changes in the DNA synthesis and cell division scores after anastomosis (
Figure S2). These findings indicate that the postoperative upregulation of matrisome genes observed by bulk RNA-seq is due to the phenotypic activation of ECM-producing fibroblasts and mural cells more than cell proliferation. Importantly, the temporal dynamics of inflammation and ECM remodeling scores in the mouse AVF mimic the postoperative profiles of single cell populations from early (5-7 days postop) and late (~90 days) human fistulas [
10].
3.3. Macrophages Undergo a Pro-Fibrotic Reprogramming After AVF Creation
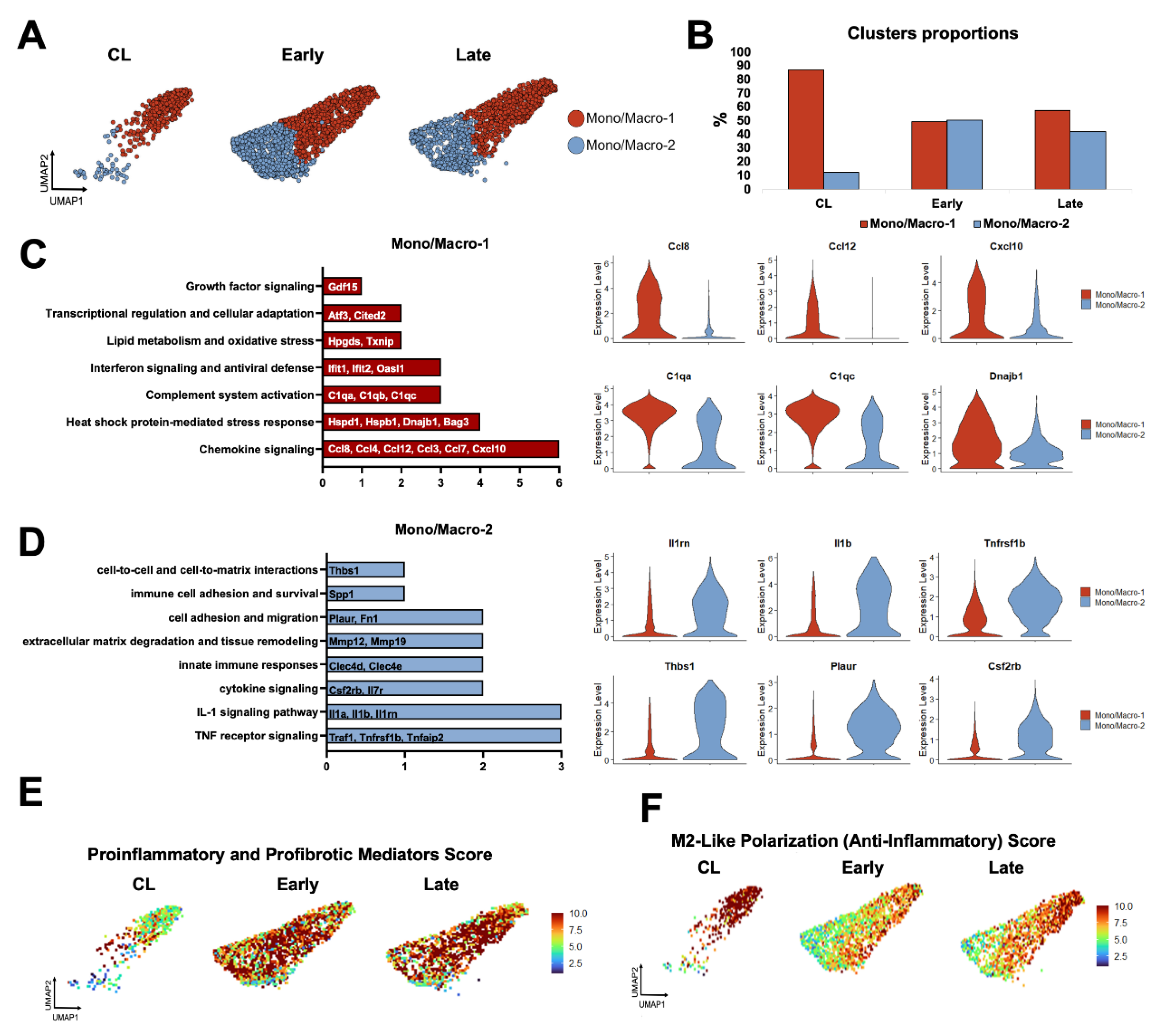
Next, we re-clustered mono/macs at resolution 0.5 to identify two transcriptionally distinct phenotypes orchestrating the remodeling of the murine AVF (
Figure 3A-D). Mono/Mac-1 cells were the predominant type in contralateral veins and expressed a gene program consistent with immune regulation and resolution of inflammation. This subset was defined by upregulation of chemokines (e.g.,
Ccl4, Ccl8, Cxcl10), heat shock proteins (e.g.,
Hspd1, Hspb1, Dnajb1), complement genes (e.g.,
C1qa, C1qb, C1qc), interferon-stimulated genes (e.g.,
Ifit1, Ifit2, Oasl1), and oxidative stress regulators (e.g.,
Hpgds, Txnip). The high expression of the transcription factors
Atf3 and
Cited2 further supports a stress-adaptive, homeostatic phenotype (
Figure 3C) [
30,
31,
32]. Mono/Mac-1 cells decreased significantly after anastomosis but seemed to participate in the resolution of inflammation in late AVFs, as illustrated by the elevated M2-like polarization score and the upregulation of
Mrc1, Klf4, Irf4, Cd36 and
Retnla compared with early fistulas (
Figure 3E-F,
Figure S3A, Data Set S2). The gene repertoire of this late pro-resolving phenotype also has unique antioxidant adaptations compared with the Mono/Mac-1 of contralateral veins, including the higher expression of
Hmox1, Sqstm1, and
Hspa1a (
Figure S3B, Data Set S2). Mono/Mac-2 cells, in turn, increased significantly after anastomosis and displayed a highly pro-inflammatory and tissue-remodeling program (
Figure 3D-E). This included upregulation of TNF and IL-1 signaling mediators (e.g.,
Traf1, Tnfrsf1b, Il1b), cytokine receptors (e.g.,
Csf2rb, Il7r), and pattern-recognition receptors (e.g.,
Clec4d, Clec4e). Elevated expression of
Mmp12 and
Mmp19 reflect an ECM remodeling function; while
Acod1, Txnrd1, and
Prdx1 indicate metabolic reprogramming and redox regulation. Additional upregulated genes, such as
Plaur, Fn1, Ptgs2, Spp1, and
Thbs1, highlight the role of this phenotype in adhesion, fibrosis, and inflammation (
Figure 3D) [
32,
33,
34,
35].
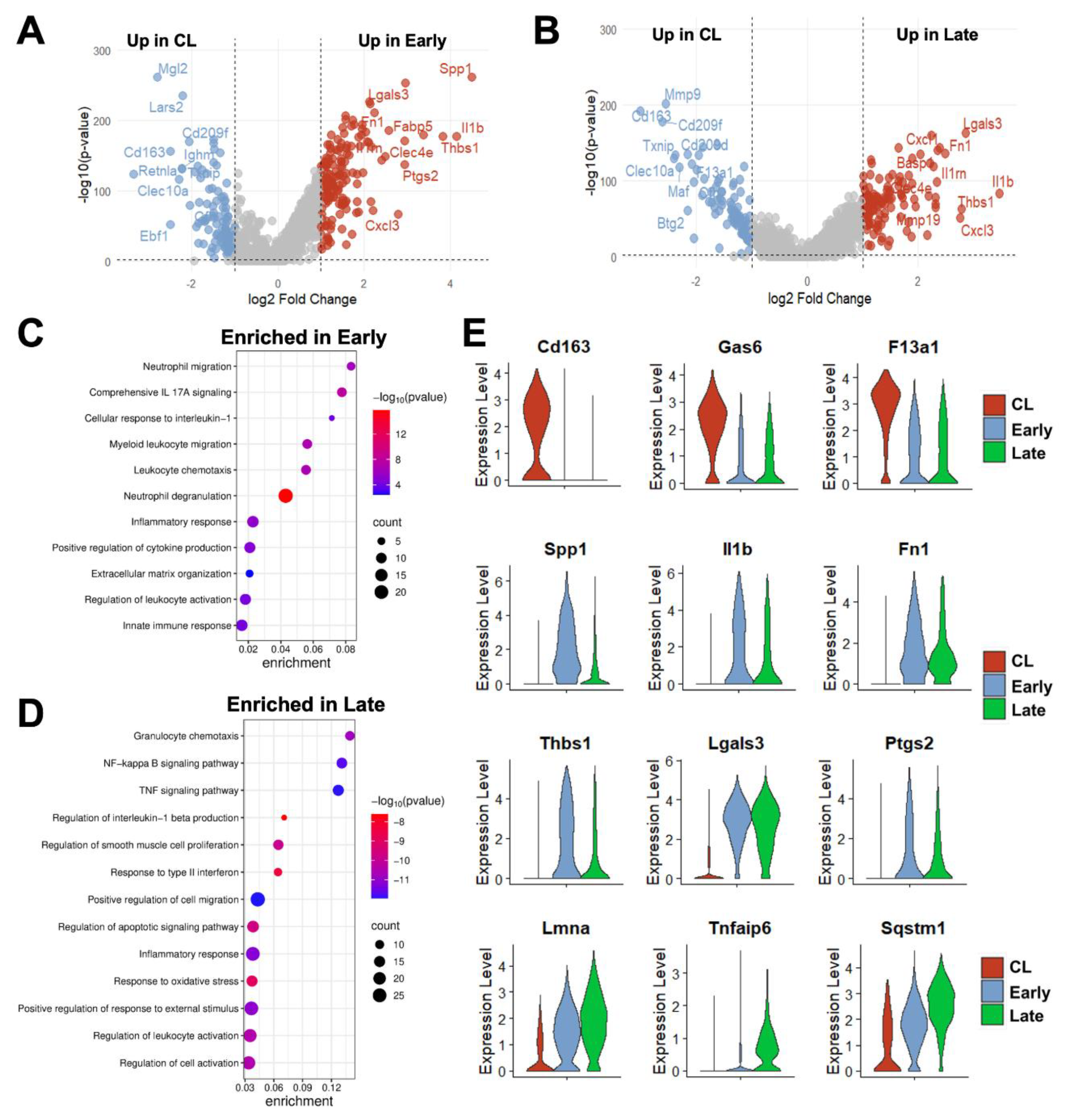
Due to both changes in phenotype proportions and in transcriptional programs of individual cells over time (
Figure 3B, E-F), over 200 DEGs were detected in mono/macs from early (280) and late AVFs (219) compared with those from the contralateral veins (
Figure 4A-B, Data Set S3-S4). Eighty-eight of the upregulated DEGs and 70 of the downregulated ones were shared in common between both time points and were mostly associated with a postoperative enhancement of inflammatory processes (e.g., up in AVFs:
Spp1, Il1b, Lgals3, Ptgs2) and a reduction in homeostatic functions (e.g., down in AVFs:
Cd163, Lyve1, F13a1, Gas6) (
Figure 4C-E). The pathway analysis of the late time point demonstrated an enrichment in some regulatory functions associated with resolution of inflammation (
Figure 4D).
We then compared the DEGs in early and late mono/macs (vs. collateral veins) with those detected in the corresponding populations from early (5-7 days postop) and late (~90 days) human fistulas (
Figure S4, Data Set S5-S6). We focused on genes expressed in >30% of the cells with log2 fold change ≥1 and p_adj ≤ 0.01. Twenty-eight percent of upregulated DEGs in mouse mono/macs at the early time point were also upregulated in early human AVFs compared with pre-access veins. These consisted, for the most part, of inflammatory mediators associated with tissue remodeling (e.g.,
Il1b, Spp1, Thbs1, Ptgs2). Similarly, 13% of upregulated genes in late mono/macs of mice were upregulated in late human fistulas. Many of the upregulated inflammatory and pro-fibrotic mediators from early AVFs (e.g.,
Il1b, Fn1) were included in this set, as well as genes associated with resolution of inflammation (e.g.,
Il10, Il1rn, Trem2, Apoe, Sod2, Prdx1, Fth1, Metrnl). Among the downregulated DEGs in mouse mono/macs, 20% at the early time point and 11% in late AVFs were downregulated in the corresponding groups of human fistulas (
Figure S4). These included several homeostatic genes (e.g.,
Lyve1, F13a1, Gas6), the transcription factor
Maf, which determines the anti-inflammatory polarization of mono/macs [
36,
37,
38], and scavenger receptors (e.g.,
Mrc1, Cd163). These comparisons identified conserved molecular changes in the immunoregulation of venous remodeling in response to surgical trauma and/or supraphysiological flow.
3.4. Fibroblasts Transform from a Quiescent to an Activated State after AVF Creation
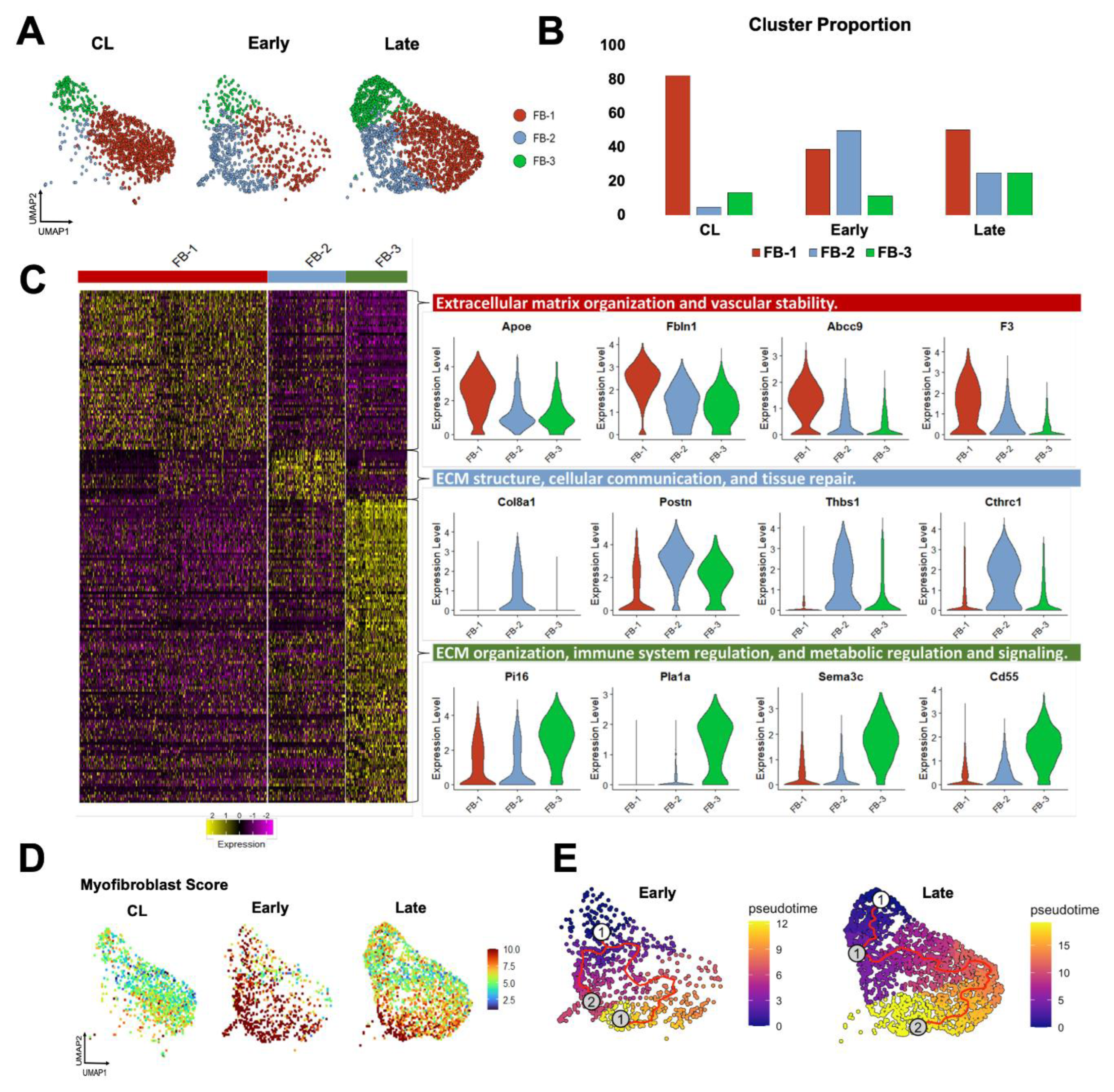
Fibroblasts are more abundant in the vein than mural cells and play a critical role in postoperative hemostasis, wall healing, and reorganization of the ECM. Therefore, we studied the temporal differentiation of fibroblasts in mouse AVFs and their similarities to those from human fistulas. After re-clustering at resolution 0.5, three transcriptionally distinct fibroblast subsets (FB-1 to 3) were identified that changed in abundance and function during AVF remodeling. In contralateral veins, fibroblasts were largely quiescent, with FB-1 representing more than 80% of the population (
Figure 5A-B). FB-1 cells were characterized by genes that maintain ECM integrity (e.g.,
Eln, Dcn, Podn), vascular stability (e.g.,
Gas6, Abcc9, F3), and redox balance (e.g.,
Apoe, Txnip, Inmt) (
Figure 5C, Data Set S7). In early AVFs, FB-2 cells expanded nearly ten-fold, representing ~50% of the overall fibroblast cluster (
Figure 5B). Gene signature scores and trajectory analyses identified this subpopulation as myofibroblasts originating from FB-1 cells and from a minor fibroblast subtype, named FB-3 (
Figure 5D-E). During this differentiation, FB-2 cells gained significant expression of contractile markers (e.g.,
Tagln, Acta2, Tpm2), mechanosensitive genes (e.g.,
Thbs1, Thbs4), and ECM proteins (e.g.,
Postn, Col8a1, Col1a1, Tnc). Upregulated protein chaperones such as
P4hb, Serpinh1, and
Pdia6 facilitate collagen maturation, while metalloproteinases and inhibitors (e.g.,
Mmp14, Timp1) regulate ECM turnover (
Figure 5C, Data Set S8). The differentiation of FB-2 cells continued in late AVFs, with further upregulation of basement collagens, mechanosensitive genes, complement factors, cytokines, and gelsolin (
Gsn) to support cell migration and immune activation (
Figure S5A) [
39,
40]. In contrast to the early postoperative expansion of FB-2 cells, FB-3 cells remained low in proportion in early AVFs but increased in late fistulas. Transcriptionally, they were characterized by upregulation of genes involved in immune regulation and metabolic signaling (
Figure 5C, Data Set S9). These include regulators of inflammation and complement activity (e.g.,
Cd55, Tnfaip6), and enzymes involved in glycosaminoglycan biosynthesis (e.g.,
Ugdh, Uap1, Gfpt2). The relative abundances of fibroblast subtypes over time and their temporal profiles of matrisome gene expression (
Figure S5B) demonstrate that, in terms of magnitude, FB-2 cells are the main population responsible for postoperative ECM deposition.
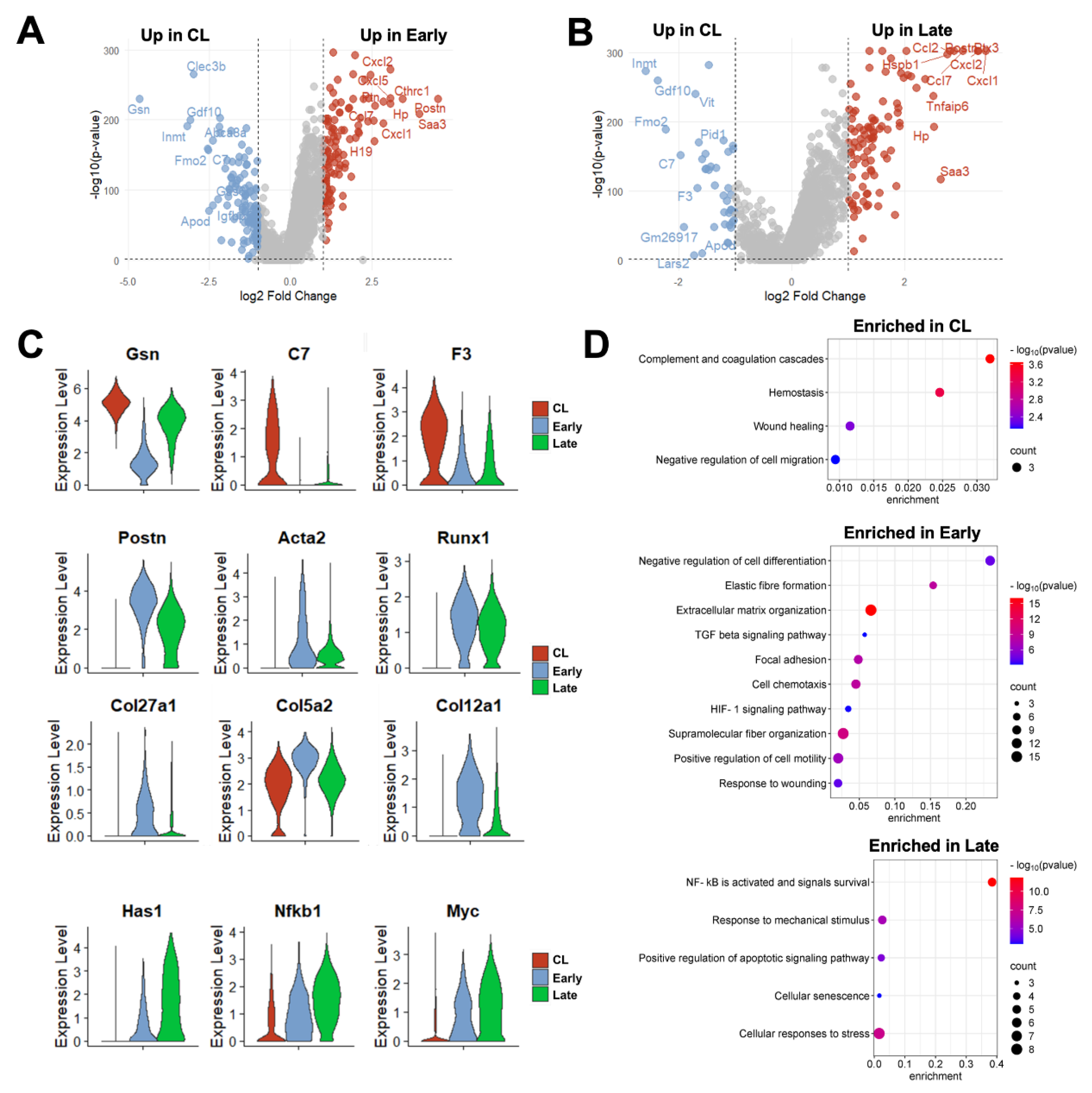
Taking into consideration changes in proportions and in the expression profiles of individual cells, a total of 226 DEGs were detected in fibroblasts from early AVFs and 138 in late fistulas compared with those from contralateral veins (
Figure 6A, Data Set S10-S11). Fifty-one of the upregulated DEGs were found at both time points and included markers of myofibroblast differentiation (e.g.,
Postn, Acta2, Runx1), mechanosensitive genes (e.g.,
Thbs1, Vcam1), and inflammatory transcriptional regulators (e.g.,
Nfkb1, Atf3) (
Figure 6B-C). Thirty-six of the downregulated DEGs were also shared by both early and late fibroblasts. These included regulators of actin assembly (e.g.,
Gsn, Tmsb10), complements and coagulation factors (e.g.,
C7, F3) and inhibitors of TGF-β signaling (e.g.,
Fmo2, Gdf10). The pathway analyses illustrated the shifts in fibroblast functions from the native vein to the late AVF, which concur with the matrisome changes reported by bulk RNA-seq (
Figure 1,
Figure 6D). Fibroblasts of contralateral veins showed activation of hemostasis and the complement cascade, but negative regulation of cell migration. In contrast, processes associated with ECM remodeling, cell motility, and responses to wounding were enriched in early fibroblasts. Late fibroblasts showed enrichment of mechanosensitive pathways and inflammatory activation. Considering that the peak of vascular inflammation occurs during the early time point, the pathways enriched in late fibroblasts suggest a chronic inflammatory activation and/or phenotypic differentiation. An inflammatory basis for either of these processes is supported by the analysis of the top 50 transcription factors activated in early and late fibroblasts based on the expression of target genes (
Figure S5C). This analysis predicted maximum activation of Smad3 and 4 in myofibroblasts (FB-2) of early AVFs and a progressive increase of Nfkb1 and Rela activity in FB-2 and FB-3 cells over time.
Comparison of DEGs in early and late murine fibroblasts (vs. collateral veins) with DEGs in the corresponding populations from early and late human fistulas uncovered significant similarities, particularly in the genes expressed by the myofibroblast (FB-2) subpopulation (
Figure S6, Data Set S12-S13). These similarities were observed in both the upregulated and downregulated directions. Thirty-two percent of upregulated DEGs and 37% of downregulated genes in the overall fibroblast cluster of mice were differentially expressed in the same direction in early human AVFs compared with pre-access veins. Similarly, 21% of upregulated genes and 17% of downregulated DEGs in late fibroblasts of mice were shared in common with late human fistulas. Altogether, these findings demonstrate that, in mice and humans, the differentiation of fibroblasts into myofibroblasts is a dominant process during postoperative AVF remodeling.
3.5. Cell-to-Cell Communication Analyses Single out Macrophages and Fibroblasts as the Drivers of Postoperative Remodeling
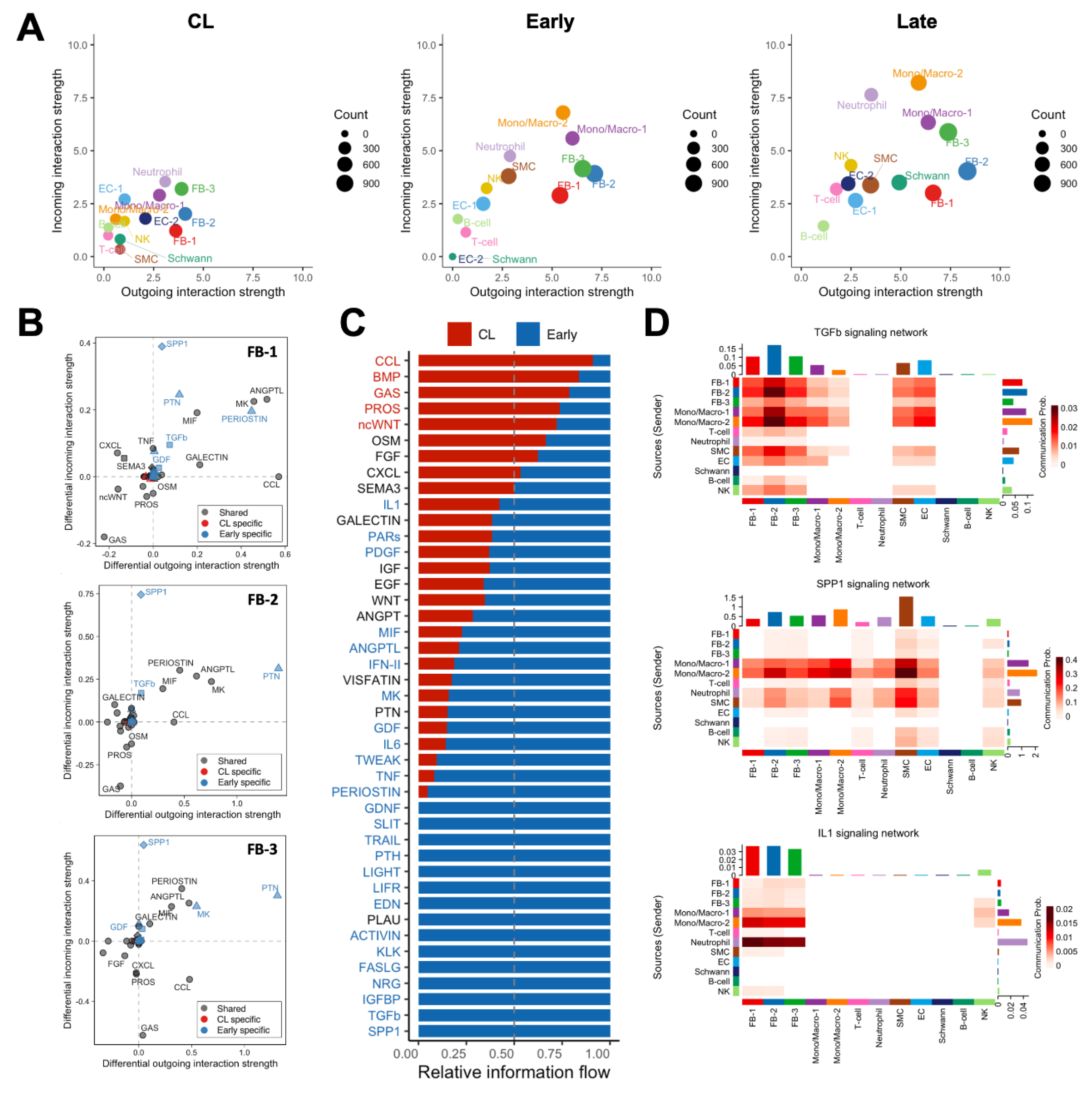
We performed global cell-to-cell communication analyses to infer the strength and direction of signaling interactions contributing to the postoperative remodeling of the mouse AVF. The total number and strength of ligand-receptor interactions increased progressively after anastomosis to reach the highest levels in late AVFs (
Figure 7A,
Figure S7A). Postoperatively, fibroblasts and mono/macs were the top senders of signals (ligand secretion), while mono/macs and neutrophils were the top receivers (receptor binding) (
Figure 7A). The top ligands secreted by fibroblasts of early AVFs compared with those from contralateral veins were periostin, pleiotrophin (PTN), angiopoietin-like proteins (ANGPTLs), midkine (MK), macrophage migration inhibitory factor (MIF), and chemokines (
Figure 7B). These outgoing signals remained enriched in fibroblasts from late fistulas and were the top incoming interactions for macrophages of both postoperative time points (
Figures S7B and S8A). Concurrently, osteopontin (SPP1) originating from mono/macs was the top incoming signal in both early and late AVFs (
Figure 7B-D). Other significant incoming interactions compared with contralateral veins included TGF-β and IL-1 (
Figure 7B-D,
Figure S8B). The former was mostly derived from Mono/Mac-2 cells and myofibroblasts (FB-2), while Mono/Mac-2 and neutrophils were the main sources of IL-1 (
Figure 7D). Altogether, these analyses uncovered a unique macrophage-fibroblast crosstalk in early and late AVFs that, not only provides for the survival and differentiation of macrophages, but also triggers and perpetuates fibroblast activation for the pro-fibrotic remodeling of the AVF wall.
4. Discussion
The murine carotid–jugular AVF model has become increasingly prevalent in mechanistic studies aimed at dissecting the processes by which the vein successfully adapts to supraphysiological conditions after AVF creation. Leveraging novel omics technologies, this study provides a comprehensive transcriptional characterization of venous remodeling in this model in nephrectomized mice, critically evaluating the extent to which it recapitulates clinically relevant molecular pathways of AVF maturation. We have validated the value of the carotid–jugular AVF model to study (1) the early surge of inflammation mediated by monocytes/macrophages and neutrophils, (2) the chronic inflammatory activation of fibroblasts and their differentiation into ECM-producing myofibroblasts, and (3) the establishment of a multifaceted macrophage-fibroblast interaction network that both initiates and regulates ECM deposition.
According to our transcriptomic study, the mouse carotid–jugular AVF represents an excellent model to dissect the mechanisms by which early inflammation after anastomosis drives venous remodeling. As we recently described in humans [
10], neutrophils and mono/macs predominate during the early postoperative phase in the murine AVF, together accounting for nearly 70% of all captured cells in the vessel wall. The abundance of these cells reflects the de novo infiltration of myeloid cells into the injured vasculature to orchestrate the debridement and reconstruction of the vein after hemodynamic and surgical trauma. Accordingly, depletion of macrophages using clodronate-containing liposomes impaired the adaptive thickening of the vascular wall [
41], a process that is essential for vascular resilience.
While most prior studies exploring macrophage involvement in venous remodeling have relied on the reductionist M1/M2 polarization model, which is now considered oversimplified, our data support the existence of distinct inflammatory and reparative macrophage populations in the murine AVF. The reparative subset, designated Mono/Mac-1, expressed stress-adaptive and immunoregulatory genes, including complement components and interferon-stimulated transcripts. These cells were also abundant in the contralateral veins, suggesting a role in maintaining vascular homeostasis. Paradoxically, accumulation of reparative macrophages within the AVF wall correlated with wall thickening and extracellular matrix (ECM) deposition [
10]. On the other hand, the pro-inflammatory macrophage subset Mono/Mac-2 that remained elevated in the murine AVF until the end of the experimental time exhibited a highly inflammatory and pro-fibrotic transcriptional profile characterized by elevated expression of
Il1b, Fn1, Spp1, Thbs1, Thbs4, and
Ptgs2. Notably, the upregulation of mechanosensitive thrombospondin genes in
Spp1⁺ Mono/Mac-2 suggests a mechanistic trigger that integrates ECM-derived mechanical cues with inflammatory signaling, enhancing immune cell recruitment, promoting IL-1β activation, and facilitating TGF-β1 release [
42,
43]. Higher proportion of pro-inflammatory mono/macs was associated with AVF failure in human AVFs [
10], underscoring their potential contribution to venous stenosis. Spp1⁺ macrophages, in particular, are a source of pro-fibrotic factors in the heart [
44] and were recently linked to human AVF failure [
10]. The functional role of Spp1+ macrophages, should be further validated using genetic and pharmacologic approaches. Collectively, our findings support the use of the carotid–jugular AVF model to further investigate the pro-fibrotic macrophage crosstalk with fibroblasts and other stromal cells after AVF creation, particularly involving SPP1, TGF-β, and IL-1 signaling.
Our study also demonstrates the utility of the carotid–jugular AVF model to investigate the role of fibroblasts in venous remodeling. We observed striking phenotypic transitions of venous fibroblasts after anastomosis. The homeostatic FB-1 fibroblasts, which were abundant in the pre-operative mouse vein, likely transitioned into
Postn⁺ myofibroblasts (aka FB-2) in response to biomechanical stress and inflammatory cues. This postoperative fibroblast population expands markedly and adopts a fibrotic and inflammatory phenotype characterized by the upregulation of collagens, matricellular proteins, and chemokines during postoperative venous remodeling. Like postoperative macrophages,
Postn⁺ myofibroblasts constitute an important source of TGF-β within the fistula wall. Notably, one of the defining markers of
Postn⁺ myofibroblasts is
Col8a1, whose upregulation has been associated with maturation failure of human AVFs [
12]. Importantly, our analysis does not exclude a potential contribution of SMCs to this population, although a definitive validation will require single-cell lineage tracing coupled with histological analyses. In the heart, Postn⁺ myofibroblasts have likewise been found enriched within infarcted and fibrotic regions, and targeted ablation of this population reduces fibrotic remodeling without impairing tissue healing [
45].
While our study provides valuable insights about the cellular and molecular mechanisms driving AVF remodeling in mice, several limitations must be acknowledged. First, the mouse model lacks a clear definition of stenosis, limiting its ability to accurately simulate human maturation failure. Second, there are profound differences between human and murine AVFs in the cellular heterogeneity of fibroblasts, mural cells, ECs, and mono/mac populations, reflecting the greater histological complexity of human veins. Despite these limitations, our study demonstrates that — as in humans — remodeling of the carotid–jugular AVF in mice is orchestrated by early immune infiltration, fibroblast activation, and loss or reprogramming of ECs. Comparisons with human data indicate that the murine model recapitulates the core inflammatory-fibrotic axis but lacks the overall complexity of human remodeling. Therefore, integrating insights from both biological systems will be instrumental for designing mechanistic studies of ECM remodeling and inflammatory signaling within a clinically relevant framework, minimizing the risk of misleading interpretations.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figure S1: Transcriptional characteristics of smooth muscle cells (SMC) and endothelial cells (EC) after creation of the mouse arteriovenous fistula (AVF); Figure S2: Proliferation scores in cell populations of the mouse arteriovenous fistula (AVF) by scRNA-seq; Figure S3: Differentially expressed genes (DEG) in monocyte/macrophage phenotypes after creation of the mouse arteriovenous fistula (AVF); Figure S4: Common differentially expressed genes (DEG) in monocyte/macrophages from mouse and human arteriovenous fistulas (AVF); Figure S5: Differentiation of venous fibroblast after creation of the mouse arteriovenous fistula (AVF); Figure S6: Common differentially expressed genes (DEG) in fibroblasts (FB) from mouse and human arteriovenous fistulas (AVF); Figure S7: Comparative cell-to-cell communication analyses in the mouse arteriovenous fistula (AVF) using CellChat; Figure S8: Cell-to-cell communication analyses in the mouse arteriovenous fistula (AVF); Data Sets S1-S13: Single cell markers and differential gene expression analyses.
Author Contributions
Conceptualization, F.F.S.C., R.I.V.P., and L.M.; methodology, F.F.S.C., A.K., M.G.R., Y.W., R.I.V.P., and L.M.; software, F.F.S.C., M.G.R., and L.M.; validation, F.F.S.C., A.K., Y.W., and L.M.; formal analysis, F.F.S.C., M.G.R., and L.M.; investigation, F.F.S.C., A.K., M.G.R., Y.W., M.S.S., J.S.L.M., M.T., X.Y., R.I.V.P., and L.M.; resources, M.S.S., M.T., X.Y., R.I.V.P., and L.M.; data curation, F.F.S.C., M.G.R., R.I.V.P., and L.M.; writing—original draft preparation, F.F.S.C. and L.M.; writing—review and editing, F.F.S.C., A.K., M.G.R., Y.W., M.S.S., J.S.L.M., M.T., X.Y., R.I.V.P., and L.M.; visualization, F.F.S.C. and L.M.; supervision, R.I.V.P. and L.M.; project administration, R.I.V.P. and L.M.; funding acquisition, F.F.S.C., R.I.V.P. and L.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Institutes of Health grant numbers R01-DK132888 and R01-DK136297 to R.I.V.P., R01-DK144860 to L.M., and F32-HL158216 to F.F.S.C., and the Department of Veterans Affairs grant numbers IK6-BX006823 and I01-BX006080 to R.I.V.P.
Institutional Review Board Statement
The animal study protocol was approved by the University of Miami Institutional Animal Care and Use Committee.
Data Availability Statement
Sequencing data were deposited in the Gene Expression Omnibus, accession number GSEXXXXXX.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Lok, C.E.; Huber, T.S.; Lee, T.; Shenoy, S.; Yevzlin, A.S.; Abreo, K.; Allon, M.; Asif, A.; Astor, B.C.; Glickman, M.H.; et al. KDOQI Clinical Practice Guideline for Vascular Access: 2019 Update. Am J Kidney Dis 2020, 75, S1-S164. [CrossRef]
- Yang, B.; Shergill, U.; Fu, A.A.; Knudsen, B.; Misra, S. The mouse arteriovenous fistula model. Journal of vascular and interventional radiology : JVIR 2009, 20, 946-950. [CrossRef]
- Yamamoto, K.; Protack, C.D.; Tsuneki, M.; Hall, M.R.; Wong, D.J.; Lu, D.Y.; Assi, R.; Williams, W.T.; Sadaghianloo, N.; Bai, H.; et al. The mouse aortocaval fistula recapitulates human arteriovenous fistula maturation. Am J Physiol Heart Circ Physiol 2013, 305, H1718-1725. [CrossRef]
- Applewhite, B.; Gupta, A.; Wei, Y.; Yang, X.; Martinez, L.; Rojas, M.G.; Andreopoulos, F.; Vazquez-Padron, R.I. Periadventitial beta-aminopropionitrile-loaded nanofibers reduce fibrosis and improve arteriovenous fistula remodeling in rats. Front Cardiovasc Med 2023, 10, 1124106. [CrossRef]
- Duque, J.C.; Martinez, L.; Mesa, A.; Wei, Y.; Tabbara, M.; Salman, L.H.; Vazquez-Padron, R.I. CD4(+) lymphocytes improve venous blood flow in experimental arteriovenous fistulae. Surgery 2015, 158, 529-536. [CrossRef]
- Hernandez, D.R.; Applewhite, B.; Martinez, L.; Laurito, T.; Tabbara, M.; Rojas, M.G.; Wei, Y.; Selman, G.; Knysheva, M.; Velazquez, O.C.; et al. Inhibition of Lysyl Oxidase with beta-aminopropionitrile Improves Venous Adaptation after Arteriovenous Fistula Creation. Kidney360 2021, 2, 270-278. [CrossRef]
- Krishnamoorthy, M.K.; Banerjee, R.K.; Wang, Y.; Zhang, J.; Sinha Roy, A.; Khoury, S.F.; Arend, L.J.; Rudich, S.; Roy-Chaudhury, P. Hemodynamic wall shear stress profiles influence the magnitude and pattern of stenosis in a pig AV fistula. Kidney Int 2008, 74, 1410-1419. [CrossRef]
- Yassari, R.; Sayama, T.; Jahromi, B.S.; Aihara, Y.; Stoodley, M.; Macdonald, R.L. Angiographic, hemodynamic and histological characterization of an arteriovenous fistula in rats. Acta Neurochir (Wien) 2004, 146, 495-504. [CrossRef]
- Pike, D.; Shiu, Y.T.; Cho, Y.F.; Le, H.; Somarathna, M.; Isayeva, T.; Guo, L.; Symons, J.D.; Kevil, C.G.; Totenhagen, J.; et al. The effect of endothelial nitric oxide synthase on the hemodynamics and wall mechanics in murine arteriovenous fistulas. Sci Rep 2019, 9, 4299. [CrossRef]
- Martinez, L.; Stoyell-Conti, F.F.; Tabbara, M.; Rojas, M.G.; Pereira-Simon, S.; Falcon, N.S.; Lopez, R.H.; Galtes, D.; Kosanovic, C.; Griswold, A.J.; et al. The single-cell landscape of the human vein after arteriovenous fistula creation and implications for maturation failure. Kidney Int 2025. [CrossRef]
- Pike, D.; Shiu, Y.T.; Somarathna, M.; Guo, L.; Isayeva, T.; Totenhagen, J.; Lee, T. High resolution hemodynamic profiling of murine arteriovenous fistula using magnetic resonance imaging and computational fluid dynamics. Theor Biol Med Model 2017, 14, 5. [CrossRef]
- Martinez, L.; Rojas, M.G.; Tabbara, M.; Pereira-Simon, S.; Santos Falcon, N.; Rauf, M.A.; Challa, A.; Zigmond, Z.M.; Griswold, A.J.; Duque, J.C.; et al. The Transcriptomics of the Human Vein Transformation After Arteriovenous Fistula Anastomosis Uncovers Layer-Specific Remodeling and Hallmarks of Maturation Failure. Kidney Int Rep 2023, 8, 837-850. [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019, 10, 1523. [CrossRef]
- Shao, X.; Taha, I.N.; Clauser, K.R.; Gao, Y.T.; Naba, A. MatrisomeDB: the ECM-protein knowledge database. Nucleic Acids Res 2020, 48, D1136-D1144. [CrossRef]
- Stoyell-Conti, F.F.; Suresh Kumar, M.; Zigmond, Z.M.; Rojas, M.G.; Santos Falcon, N.; Martinez, L.; Vazquez-Padron, R.I. Gene inactivation of lysyl oxidase in smooth muscle cells reduces atherosclerosis burden and plaque calcification in hyperlipidemic mice. Atherosclerosis 2024, 397, 118582. [CrossRef]
- Wang, Z.; Jin, Q.; Wei, C.H.; Tian, S.; Lai, P.T.; Zhu, Q.; Day, C.P.; Ross, C.; Leaman, R.; Lu, Z. GeneAgent: self-verification language agent for gene-set analysis using domain databases. Nat Methods 2025, 22, 1677-1685. [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015, 1, 417-425. [CrossRef]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using CellChat. Nat Commun 2021, 12, 1088. [CrossRef]
- Cao, J.; Spielmann, M.; Qiu, X.; Huang, X.; Ibrahim, D.M.; Hill, A.J.; Zhang, F.; Mundlos, S.; Christiansen, L.; Steemers, F.J.; et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 2019, 566, 496-502. [CrossRef]
- Qiu, X.; Mao, Q.; Tang, Y.; Wang, L.; Chawla, R.; Pliner, H.A.; Trapnell, C. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 2017, 14, 979-982. [CrossRef]
- Garcia-Alonso, L.; Holland, C.H.; Ibrahim, M.M.; Turei, D.; Saez-Rodriguez, J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res 2019, 29, 1363-1375. [CrossRef]
- Jahanyar, J.; Joyce, D.L.; Southard, R.E.; Loebe, M.; Noon, G.P.; Koerner, M.M.; Torre-Amione, G.; Youker, K.A. Decorin-mediated transforming growth factor-beta inhibition ameliorates adverse cardiac remodeling. J Heart Lung Transplant 2007, 26, 34-40. [CrossRef]
- Xu, H.; Thomas, M.J.; Kaul, S.; Kallinger, R.; Ouweneel, A.B.; Maruko, E.; Oussaada, S.M.; Jongejan, A.; Cense, H.A.; Nieuwdorp, M.; et al. Pcpe2, a Novel Extracellular Matrix Protein, Regulates Adipocyte SR-BI-Mediated High-Density Lipoprotein Uptake. Arterioscler Thromb Vasc Biol 2021, 41, 2708-2725. [CrossRef]
- Noda, K.; Dabovic, B.; Takagi, K.; Inoue, T.; Horiguchi, M.; Hirai, M.; Fujikawa, Y.; Akama, T.O.; Kusumoto, K.; Zilberberg, L.; et al. Latent TGF-beta binding protein 4 promotes elastic fiber assembly by interacting with fibulin-5. Proc Natl Acad Sci U S A 2013, 110, 2852-2857. [CrossRef]
- Wan, J.; Zhang, G.; Li, X.; Qiu, X.; Ouyang, J.; Dai, J.; Min, S. Matrix Metalloproteinase 3: A Promoting and Destabilizing Factor in the Pathogenesis of Disease and Cell Differentiation. Front Physiol 2021, 12, 663978. [CrossRef]
- Tammi, M.I.; Day, A.J.; Turley, E.A. Hyaluronan and homeostasis: a balancing act. J Biol Chem 2002, 277, 4581-4584. [CrossRef]
- Wight, T.N.; Kang, I.; Evanko, S.P.; Harten, I.A.; Chang, M.Y.; Pearce, O.M.T.; Allen, C.E.; Frevert, C.W. Versican-A Critical Extracellular Matrix Regulator of Immunity and Inflammation. Front Immunol 2020, 11, 512. [CrossRef]
- Lindner, V.; Wang, Q.; Conley, B.A.; Friesel, R.E.; Vary, C.P. Vascular injury induces expression of periostin: implications for vascular cell differentiation and migration. Arterioscler Thromb Vasc Biol 2005, 25, 77-83. [CrossRef]
- Kanisicak, O.; Khalil, H.; Ivey, M.J.; Karch, J.; Maliken, B.D.; Correll, R.N.; Brody, M.J.; SC, J.L.; Aronow, B.J.; Tallquist, M.D.; et al. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nat Commun 2016, 7, 12260. [CrossRef]
- Hai, T.; Wolfgang, C.D.; Marsee, D.K.; Allen, A.E.; Sivaprasad, U. ATF3 and stress responses. Gene Expr 1999, 7, 321-335.
- Mattes, K.; Geugien, M.; Korthuis, P.M.; Brouwers-Vos, A.Z.; Fehrmann, R.S.N.; Todorova, T.I.; Steidl, U.; Vellenga, E.; Schepers, H. Transcriptional regulators CITED2 and PU.1 cooperate in maintaining hematopoietic stem cells. Exp Hematol 2019, 73, 38-49 e37. [CrossRef]
- Behrendt, N.; Ronne, E.; Dano, K. The structure and function of the urokinase receptor, a membrane protein governing plasminogen activation on the cell surface. Biol Chem Hoppe Seyler 1995, 376, 269-279.
- Zhao, Y.; Huang, Z.; Gao, L.; Ma, H.; Chang, R. Osteopontin/SPP1: a potential mediator between immune cells and vascular calcification. Front Immunol 2024, 15, 1395596. [CrossRef]
- Gao, Q.; Gao, S.; Li, H.; Chen, Z.; Zhang, R.; Li, Y.; Zhang, H. Multi-Omics Exploration of the Role of PTGS2 as a Hub Gene in Ferroptosis Within the Artery of Takayasu Arteritis. J Inflamm Res 2024, 17, 9135-9146. [CrossRef]
- Sweetwyne, M.T.; Murphy-Ullrich, J.E. Thrombospondin1 in tissue repair and fibrosis: TGF-beta-dependent and independent mechanisms. Matrix Biol 2012, 31, 178-186. [CrossRef]
- Liu, M.; Tong, Z.; Ding, C.; Luo, F.; Wu, S.; Wu, C.; Albeituni, S.; He, L.; Hu, X.; Tieri, D.; et al. Transcription factor c-Maf is a checkpoint that programs macrophages in lung cancer. J Clin Invest 2020, 130, 2081-2096. [CrossRef]
- Nassar, M.; Tabib, Y.; Capucha, T.; Mizraji, G.; Nir, T.; Pevsner-Fischer, M.; Zilberman-Schapira, G.; Heyman, O.; Nussbaum, G.; Bercovier, H.; et al. GAS6 is a key homeostatic immunological regulator of host-commensal interactions in the oral mucosa. Proc Natl Acad Sci U S A 2017, 114, E337-E346. [CrossRef]
- Gemmati, D.; Vigliano, M.; Burini, F.; Mari, R.; El Mohsein, H.H.; Parmeggiani, F.; Serino, M.L. Coagulation Factor XIIIA (F13A1): Novel Perspectives in Treatment and Pharmacogenetics. Curr Pharm Des 2016, 22, 1449-1459. [CrossRef]
- Feldt, J.; Welss, J.; Schropp, V.; Gelse, K.; Tsokos, M.; Paulsen, F. Recombinant human gelsolin promotes the migration of human articular cartilage chondrocytes by regulating gene expression in vitro. Osteoarthr Cartil Open 2020, 2, 100124. [CrossRef]
- Li, G.H.; Shi, Y.; Chen, Y.; Sun, M.; Sader, S.; Maekawa, Y.; Arab, S.; Dawood, F.; Chen, M.; De Couto, G.; et al. Gelsolin regulates cardiac remodeling after myocardial infarction through DNase I-mediated apoptosis. Circ Res 2009, 104, 896-904. [CrossRef]
- Danenberg, H.D.; Fishbein, I.; Gao, J.; Monkkonen, J.; Reich, R.; Gati, I.; Moerman, E.; Golomb, G. Macrophage depletion by clodronate-containing liposomes reduces neointimal formation after balloon injury in rats and rabbits. Circulation 2002, 106, 599-605. [CrossRef]
- Murphy-Ullrich, J.E.; Suto, M.J. Thrombospondin-1 regulation of latent TGF-beta activation: A therapeutic target for fibrotic disease. Matrix Biol 2018, 68-69, 28-43. [CrossRef]
- Stein, E.V.; Miller, T.W.; Ivins-O'Keefe, K.; Kaur, S.; Roberts, D.D. Secreted Thrombospondin-1 Regulates Macrophage Interleukin-1beta Production and Activation through CD47. Sci Rep 2016, 6, 19684. [CrossRef]
- Uhlig, M.; Billig, S.; Wienhold, J.; Schumacher, D. Pro-Fibrotic Macrophage Subtypes: SPP1+ Macrophages as a Key Player and Therapeutic Target in Cardiac Fibrosis? Cells 2025, 14. [CrossRef]
- Kaur, H.; Takefuji, M.; Ngai, C.Y.; Carvalho, J.; Bayer, J.; Wietelmann, A.; Poetsch, A.; Hoelper, S.; Conway, S.J.; Mollmann, H.; et al. Targeted Ablation of Periostin-Expressing Activated Fibroblasts Prevents Adverse Cardiac Remodeling in Mice. Circ Res 2016, 118, 1906-1917. [CrossRef]
Figure 1.
Transcriptional regulation of postoperative venous remodeling in the mouse carotid-jugular arteriovenous fistula (AVF) per bulk RNA sequencing analyses: (A) Heatmap of 4,541 differentially expressed genes among contralateral jugular veins (CL, n=7), and outflow veins from early (~3 days post-op, n=8), mid (~10 days, n=5), and late AVFs (~21 days, n=5). Pathway enrichment analyses and representative upregulated genes of the three transcriptional states identified in the heatmap. (B) Heatmap of differentially expressed matrisome genes illustrating the transcriptional stages of extracellular matrix (ECM) remodeling in the AVF. The average gene expression per experimental group was normalized so that the lowest level across the groups was set at 0 and the highest level at 1. (C) Temporal diagram of the main stages of ECM remodeling in the AVF based on the differential expression of matrisome genes.
Figure 1.
Transcriptional regulation of postoperative venous remodeling in the mouse carotid-jugular arteriovenous fistula (AVF) per bulk RNA sequencing analyses: (A) Heatmap of 4,541 differentially expressed genes among contralateral jugular veins (CL, n=7), and outflow veins from early (~3 days post-op, n=8), mid (~10 days, n=5), and late AVFs (~21 days, n=5). Pathway enrichment analyses and representative upregulated genes of the three transcriptional states identified in the heatmap. (B) Heatmap of differentially expressed matrisome genes illustrating the transcriptional stages of extracellular matrix (ECM) remodeling in the AVF. The average gene expression per experimental group was normalized so that the lowest level across the groups was set at 0 and the highest level at 1. (C) Temporal diagram of the main stages of ECM remodeling in the AVF based on the differential expression of matrisome genes.
Figure 2.
Postoperative venous remodeling in the mouse carotid-jugular arteriovenous fistula (AVF) per single-cell RNA sequencing: (A) Uniform manifold approximation and projection (UMAP) plot of 10,500 cells isolated from contralateral veins (CL, n=10) and the outflow veins from early (~5 days post-op, n=10) and late AVFs (~26 days, n=10). (B) Markers used for cell cluster identification. The color of the dots indicates the average gene expression per cluster, while their size represents the percentage of cells in the cluster expressing the gene. (C) Relative proportions of cell clusters per experimental group. (D-E) Functional profiling of single cell clusters in the experimental groups according to gene signature scores.
Figure 2.
Postoperative venous remodeling in the mouse carotid-jugular arteriovenous fistula (AVF) per single-cell RNA sequencing: (A) Uniform manifold approximation and projection (UMAP) plot of 10,500 cells isolated from contralateral veins (CL, n=10) and the outflow veins from early (~5 days post-op, n=10) and late AVFs (~26 days, n=10). (B) Markers used for cell cluster identification. The color of the dots indicates the average gene expression per cluster, while their size represents the percentage of cells in the cluster expressing the gene. (C) Relative proportions of cell clusters per experimental group. (D-E) Functional profiling of single cell clusters in the experimental groups according to gene signature scores.
Figure 3.
Monocyte/macrophage phenotypes during remodeling of the mouse arteriovenous fistula (AVF): (A-B) Focused UMAPs and relative proportions of monocyte/macrophage phenotypes in contralateral veins (CL) and the outflow veins from early and late AVFs. (C-D) Pathway enrichment analyses and representative differentially expressed genes between the two mono/mac phenotypes. (E-F) Functional profiling of mono/macs before and after AVF creation according to gene signature scores.
Figure 3.
Monocyte/macrophage phenotypes during remodeling of the mouse arteriovenous fistula (AVF): (A-B) Focused UMAPs and relative proportions of monocyte/macrophage phenotypes in contralateral veins (CL) and the outflow veins from early and late AVFs. (C-D) Pathway enrichment analyses and representative differentially expressed genes between the two mono/mac phenotypes. (E-F) Functional profiling of mono/macs before and after AVF creation according to gene signature scores.
Figure 4.
Time-dependent transcriptional changes in murine monocyte/macrophages after arteriovenous fistula (AVF) creation: (A-B) Volcano plots of differentially expressed genes between mono/macs from early or late AVFs compared with those from contralateral veins (CL). (C-D) Pathway enrichment analyses of mono/macs from early and late AVFs. (E) Time-dependent changes in RNA expression of selected genes in mono/macs after AVF creation.
Figure 4.
Time-dependent transcriptional changes in murine monocyte/macrophages after arteriovenous fistula (AVF) creation: (A-B) Volcano plots of differentially expressed genes between mono/macs from early or late AVFs compared with those from contralateral veins (CL). (C-D) Pathway enrichment analyses of mono/macs from early and late AVFs. (E) Time-dependent changes in RNA expression of selected genes in mono/macs after AVF creation.
Figure 5.
Fibroblast phenotypes during remodeling of the mouse arteriovenous fistula (AVF): (A-B) Focused UMAPs and relative proportions of fibroblast phenotypes in contralateral veins (CL) and the outflow veins from early and late AVFs. (C) Heatmap and representative examples of differentially expressed genes among the three fibroblast phenotypes. (D) Functional profiling of fibroblasts before and after AVF creation according to the myofibroblast gene signature score. (E) Pseudotime trajectory analysis of fibroblasts from early and late AVFs. The white dot indicates the root of the trajectory, while the gray dots are the main leaves.
Figure 5.
Fibroblast phenotypes during remodeling of the mouse arteriovenous fistula (AVF): (A-B) Focused UMAPs and relative proportions of fibroblast phenotypes in contralateral veins (CL) and the outflow veins from early and late AVFs. (C) Heatmap and representative examples of differentially expressed genes among the three fibroblast phenotypes. (D) Functional profiling of fibroblasts before and after AVF creation according to the myofibroblast gene signature score. (E) Pseudotime trajectory analysis of fibroblasts from early and late AVFs. The white dot indicates the root of the trajectory, while the gray dots are the main leaves.
Figure 6.
Time-dependent transcriptional changes in murine fibroblasts after arteriovenous fistula (AVF) creation: (A-B) Volcano plots of differentially expressed genes between fibroblasts from early or late AVFs compared with those from contralateral veins (CL). (C) Time-dependent changes in RNA expression of selected genes in fibroblasts after AVF creation. (D) Pathway enrichment analyses of fibroblasts from the three experimental groups.
Figure 6.
Time-dependent transcriptional changes in murine fibroblasts after arteriovenous fistula (AVF) creation: (A-B) Volcano plots of differentially expressed genes between fibroblasts from early or late AVFs compared with those from contralateral veins (CL). (C) Time-dependent changes in RNA expression of selected genes in fibroblasts after AVF creation. (D) Pathway enrichment analyses of fibroblasts from the three experimental groups.
Figure 7.
Macrophage-fibroblast crosstalk during remodeling of the mouse arteriovenous fistula (AVF): (A) Global cell-to-cell communication analyses in contralateral veins (CL) and outflow veins from early and late AVFs. The x- and y- axes indicate the strengths of outgoing and incoming interactions, respectively, while the sizes of circles represent the number of predicted interactions. (B) Differential analysis of interactions in the three fibroblasts phenotypes from early AVFs compared with those from contralateral veins. Positive values in the x- and y- axes indicate outgoing and incoming interactions, respectively, that are significantly enhanced in early AVFs. Ligands in blue denote pathways that are specific in early AVFs; those in red are specific in contralateral veins; and ligands in gray indicate pathways that occur in both. Triangles indicate outgoing specific; squares, incoming specific; diamonds, incoming and outgoing specific; circles, occur in both. (C) Ranking of cell-cell interactions originating from all cell populations in early AVFs and received by fibroblasts. The red and blue fonts in the ligand names indicate interactions that are statistically significant in contralateral veins and early AVFs, respectively. (D) Heatmap of TGF-b, SPP1, and IL-1 mediated interactions in early AVFs. Senders of ligands are shown on the left, while the receivers are shown at the bottom of each heatmap.
Figure 7.
Macrophage-fibroblast crosstalk during remodeling of the mouse arteriovenous fistula (AVF): (A) Global cell-to-cell communication analyses in contralateral veins (CL) and outflow veins from early and late AVFs. The x- and y- axes indicate the strengths of outgoing and incoming interactions, respectively, while the sizes of circles represent the number of predicted interactions. (B) Differential analysis of interactions in the three fibroblasts phenotypes from early AVFs compared with those from contralateral veins. Positive values in the x- and y- axes indicate outgoing and incoming interactions, respectively, that are significantly enhanced in early AVFs. Ligands in blue denote pathways that are specific in early AVFs; those in red are specific in contralateral veins; and ligands in gray indicate pathways that occur in both. Triangles indicate outgoing specific; squares, incoming specific; diamonds, incoming and outgoing specific; circles, occur in both. (C) Ranking of cell-cell interactions originating from all cell populations in early AVFs and received by fibroblasts. The red and blue fonts in the ligand names indicate interactions that are statistically significant in contralateral veins and early AVFs, respectively. (D) Heatmap of TGF-b, SPP1, and IL-1 mediated interactions in early AVFs. Senders of ligands are shown on the left, while the receivers are shown at the bottom of each heatmap.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).