1. Introduction
For over five decades, small nucleolar RNAs have occupied a seemingly specialized niche in cell biology. These molecules of 60 to 300 nucleotides, initially discovered in the 1960s as components of small nucleolar ribonucleoprotein complexes, were long considered secondary players in the context of gene regulation. Their primary function appeared clear and limited: guiding chemical modifications of ribosomal RNA within the nucleolus, where ribosomes are assembled. However, as frequently occurs in molecular biology, this simplistic view has given way to a much more nuanced and fascinating understanding of these small yet powerful molecules.
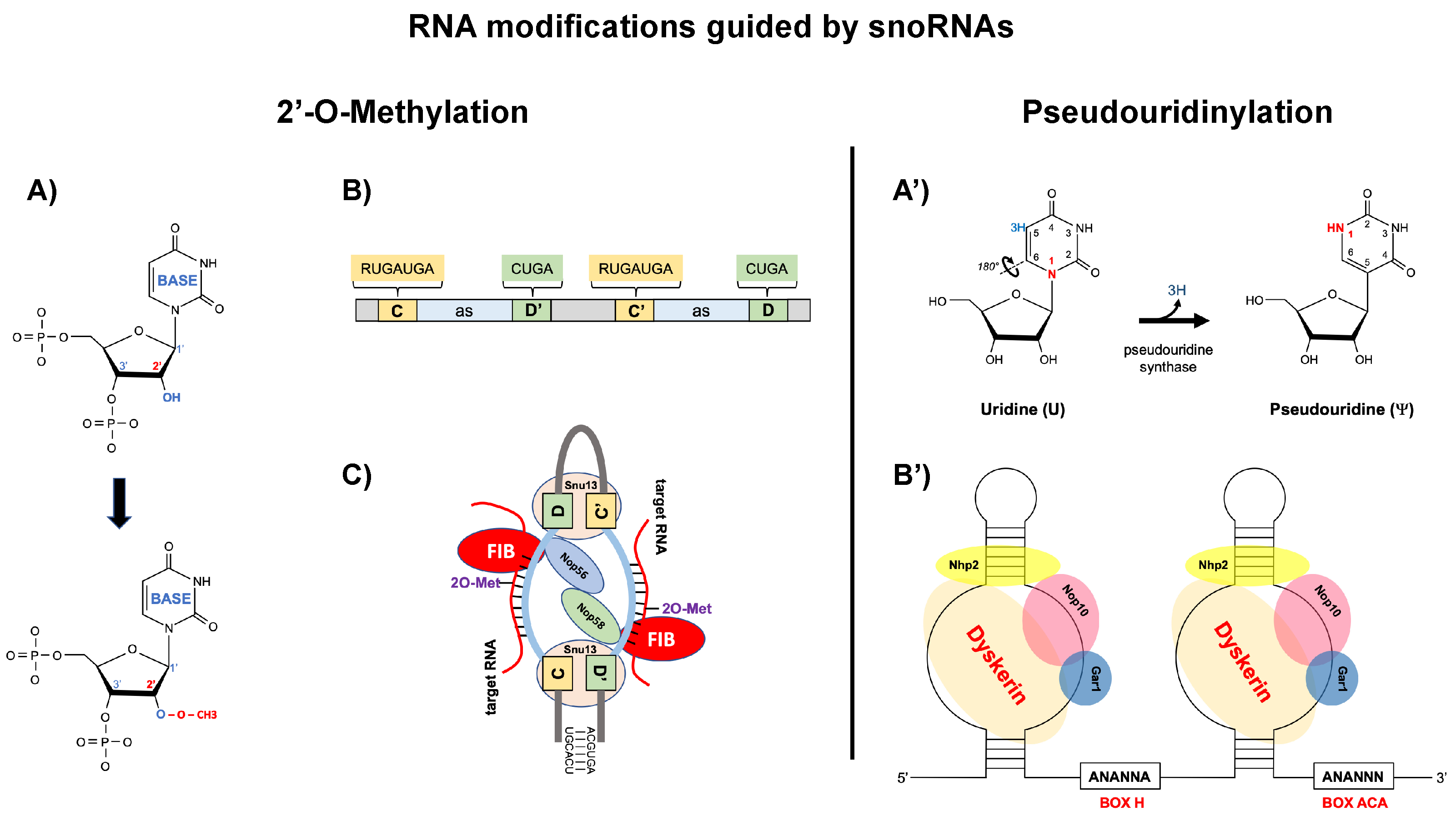
As shown in
Figure 1, snoRNAs are classified into two main families based on their conserved sequence motifs and the type of chemical modification they guide (Kiss, 2002; Matera et al., 2007) [
1,
2]. Box C/D snoRNAs (also designated as SNORDs) contain characteristic C (RUGAUGA) and D (CUGA) box motifs near their 5’ and 3’ ends, respectively, with internal C’ and D’ boxes often present. These snoRNAs guide 2’-O-methylation, a modification where a methyl group (CH₃) is added to the 2’ hydroxyl position of the ribose sugar, converting the 2’-OH to 2’-O-CH₃. This methylation occurs precisely 5 nucleotides upstream from the D or D’ box, with substrate specificity determined by 10-21 nucleotide antisense elements that form Watson-Crick base pairs with the target RNA. The box C/D snoRNP complex assembles with four essential proteins: fibrillarin (the methyltransferase enzyme), Nop56, Nop58, and Snu13 (15.5K in humans), which together position the catalytic site for precise modification (
Figure 1) [
3].
Box H/ACA snoRNAs (designated as SNORAs) possess a distinct bipartite structure consisting of two hairpins connected by a single-stranded hinge region containing the conserved H box (ANANNA), terminating with an ACA box precisely 3 nucleotides before the 3’ end. These snoRNAs guide pseudouridylation, an isomerization reaction that converts uridine to pseudouridine (Ψ) by breaking the N-glycosidic bond and rotating the uracil base 180° to form a C-glycosidic bond. This modification creates an additional hydrogen bond donor, enhancing RNA stability and affecting RNA structure. The target uridine is identified through complementary sequences in internal loops called pseudouridylation pockets, positioned exactly 14-16 nucleotides from either the H or ACA box. The H/ACA snoRNP machinery comprises four core proteins: dyskerin (the pseudouridine synthase), Gar1, Nop10, and Nhp2, which form a stable complex that catalyzes this isomerization (
Figure 1) [
1,
2].
While most snoRNAs localize to the nucleolus where ribosome biogenesis occurs, a subset called small Cajal body-specific RNAs (scaRNAs) concentrate in Cajal bodies, nuclear structures involved in snRNP maturation. scaRNAs guide modifications of spliceosomal snRNAs and can contain both C/D and H/ACA domains within a single molecule, enabling them to catalyze both types of modifications. The trafficking of snoRNAs between the nucleolus and nucleoplasm is mediated by specific localization signals and protein interactions, with some snoRNAs showing dynamic localization patterns in response to cellular conditions.
The human genome harbors approximately 400 snoRNA genes, a number that continues to expand as next-generation sequencing techniques reveal previously unnoticed transcripts. The snoDB database has systematically cataloged these genes and their tissue expression patterns, revealing unexpected complexity [
4]. Many snoRNAs show highly specific expression profiles that do not always correlate with those of their host genes, suggesting sophisticated post-transcriptional regulatory mechanisms. This tissue specificity acquires particular relevance in the cancer context, where altered snoRNA expression patterns emerge as distinctive molecular signatures of different tumor types.
The revolution in our understanding of snoRNAs began with the discovery of their non-canonical functions. Pioneering studies by Kishore and colleagues (2010) demonstrated that certain snoRNAs can be processed into smaller fragments capable of regulating alternative splicing [
5]. Subsequently, it was discovered that some snoRNAs participate in mRNA stabilization, modulate chromatin structure, and can even function analogously to microRNAs, regulating gene expression at the post-transcriptional level [
6,
7,
8]. This functional versatility has transformed our perception of snoRNAs from simple modification guides to multifaceted regulators capable of influencing numerous aspects of cellular physiology.
The link between snoRNAs and cancer has strengthened considerably in recent years. The accumulated evidence indicates that dysregulation of specific snoRNAs is not merely a passive consequence of malignant transformation, but an active participant in oncogenesis. Different snoRNAs can function as oncogenes or tumor suppressors, and their altered expression levels correlate with important clinical features such as tumor stage, metastatic capacity, and treatment response [
9,
10,
11]. This association has opened new perspectives both for understanding the fundamental mechanisms of cancer and for developing innovative diagnostic and therapeutic tools.
2. Dysregulation of snoRNAs in Cancer
The alteration in snoRNA expression patterns constitutes a recurring feature in virtually all cancer types studied to date. However, this dysregulation is far from being a random or chaotic phenomenon. On the contrary, consistent patterns emerge that suggest specific functional roles for different snoRNAs in tumor biology. The complexity of these patterns reflects the inherent heterogeneity of cancer and underscores the need to understand snoRNAs not as isolated entities, but as components integrated into broader regulatory networks.
2.1. Oncogenic snoRNAs
Among the snoRNAs that actively promote malignant transformation and progression, several have been characterized in sufficient detail to understand their mechanisms of action. Some examples are examined here. For example, SNORA21 illustrates how snoRNAs can function as molecular oncogenes. In colorectal cancer, SNORA21 exhibits significantly elevated expression levels and actively participates in the modulation of multiple cancer-related signaling pathways, including the Hippo, Wnt, and Axon guidance pathways, which collectively regulate cell cycle progression, proliferation, and invasion [
12]. Importantly, this oncogenic activity appears to extend beyond canonical rRNA modification mechanisms, suggesting that SNORA21 influences these signaling cascades through non-traditional functional mechanisms, thereby illustrating the expanding repertoire of snoRNA functions in cancer biology.
Other case is SNORD78 that provides interesting insights into how alteration of rRNA modifications can contribute to oncogenesis. This snoRNA, consistently overexpressed in colorectal tumors, directs hypermethylation of specific sites in 28S rRNA [
13]. These aberrant modifications create ribosomes with altered translational properties that favor the synthesis of proteins encoded by mRNAs with particular structural features in their 5’ untranslated regions. Many oncogenes and growth factors possess precisely these characteristics, resulting in their preferential translation by these modified “oncoribosomes” [
14,
15]. In a recent work Lan and colleagues have elucidated additional aspects of this mechanism, demonstrating that cancer cells can actively remodel their translational machinery to favor pro-tumoral gene expression programs [
13].
The case of SNORD16 emerged as a particularly relevant prognostic biomarker in digestive tract cancers [
16]. Several studies established that SNORD16 overexpression not only correlates with adverse clinical features such as increased tumor size and presence of lymph node metastases but functions as an independent prognostic factor [
16]. At the molecular level, SNORD16 promotes cell proliferation and inhibits apoptosis through mechanisms involving both its canonical function in rRNA modification and non-canonical effects on specific mRNA stability [
16]. The situation is further complicated by the fact that SNORD16 resides within the lncRNA SNHG16, which also possesses independent oncogenic functions. This nested genomic architecture creates complex regulatory circuits where the host lncRNA and embedded snoRNA can act synergistically to promote the malignant phenotype [
17].
A recent discovery of particular clinical relevance is the role of SNORA47 in Luminal A breast cancer. SNORA47 is not only overexpressed in this breast cancer subtype but actively contributes to two critical cancer characteristics: maintenance of tumor stemness and chemotherapy resistance. The underlying mechanism involves a previously uncharacterized regulatory axis including transcription factor EBF3, ribosomal protein RPL11, and the oncogene c-Myc [
18]. This molecular cascade directly connects snoRNA expression with regulation of one of the most important oncogenes in cancer biology, providing a clear example of how snoRNAs can influence critical nodes of tumor signaling networks.
2.2. Tumor Suppressor snoRNAs
At the other extreme of the functional spectrum, several snoRNAs have been demonstrated to act as molecular guardians against malignant transformation. SNORA24 exemplifies this protective role through its essential function in maintaining translational fidelity. This snoRNA guides pseudouridylation at U609 and U863 within 18S rRNA, modifications that are critical for accurate ribosome decoding. When SNORA24 expression is reduced or lost, ribosomes lacking these modifications exhibit compromised translational accuracy, resulting in increased coding errors that accumulate progressively and contribute to proteomic instability. In a hepatocellular carcinoma model driven by oncogenic RAS, McMahon and colleagues demonstrated that this loss of translational fidelity enables cells to bypass oncogene-induced senescence, thereby allowing tumor development and validating SNORA24 as tumor suppressor [
19].
SNORD44 represents another molecular guardian whose loss facilitates tumor progression. This C/D box snoRNA, encoded within the GAS5 lncRNA, exhibits downregulated expression in colorectal cancer and is subject to p53-dependent regulation. Yuan and colleagues demonstrated that therapeutic restoration of SNORD44 expression via an oncolytic adenovirus effectively suppressed colorectal tumor growth both in vitro and in vivo, with enhanced efficacy when combined with rapamycin treatment. These findings established SNORD44 as a tumor suppressor whose loss compromises cellular growth arrest mechanisms [
20].
Another important feature to note is the potential tissue specificity of snoRNAs. An example of this can be found in SNORD113-1. This snoRNA shows markedly reduced expression specifically in hepatocellular carcinoma, where it normally functions by inhibiting the MAPK/ERK and STAT3 signaling pathways [
21]. In this regard, Xu and colleagues (2014) demonstrated that the loss of SNORD113-1 directly contributes to hepatocarcinogenesis by allowing the uncontrolled activation of these proliferative pathways. Furthermore, experimental restoration of SNORD113-1 in hepatocarcinoma cell lines suppresses their growth and reduces their tumorigenic capacity, confirming its role as a liver-specific tumor suppressor [
21].
Other additional snoRNAs have also emerged as critical tumor suppressors across different cancer types. SNORD50A and SNORD50B, located at chromosome 6q14.3, represent particularly compelling examples, as they are recurrently deleted in 10-40% of twelve common cancer types, with their loss associated with reduced patient survival [
22]. These snoRNAs directly bind to K-Ras protein and inhibit its oncogenic activity by preventing farnesyltransferase-mediated activation and reducing GTP-bound active K-Ras levels, thereby suppressing hyperactivation of the Ras-ERK signaling pathway [
22]. Functional studies have demonstrated that SNORD50A/B undergo frequent genomic deletions and transcriptional downregulation in both prostate and breast cancers, with specific mutations showing significant associations with clinically aggressive disease [
23,
24]. Similarly, SNORD47 demonstrates tumor suppressive properties in glioma by suppressing cell proliferation, inducing G2 phase arrest, and inhibiting invasion and epithelial-mesenchymal transition, with low expression correlating with advanced tumor stage and poor overall survival [
25]. Together, these snoRNAs exemplify the diverse mechanisms through which small nucleolar RNAs can act as molecular guardians against malignant transformation.
2.3. Context-Dependent Functional Plasticity
Perhaps one of the most intriguing aspects of snoRNA biology in cancer is the observation that some of these RNAs can function as oncogenes or tumor suppressors depending on cellular context. SNORD76 exemplifies this remarkable functional duality. In hepatocellular carcinoma, SNORD76 functions as an oncogene, with its expression significantly upregulated in tumor tissues and correlating with poorer patient survival [
26,
27]. It promotes tumorigenesis through activation of the Wnt/β-catenin pathway, enhanced cell proliferation, and induction of epithelial-mesenchymal transition [
26]. In contrast, the same SNORD76 molecule exhibits tumor suppressor properties in glioblastoma, where it is selectively downregulated in WHO grade IV tumors and its enforced expression inhibits proliferation by inducing cell cycle arrest at S phase [
25,
28]. Several mechanisms may underlie this plasticity, including tissue-specific availability of binding partners, differential activation of signaling pathways across cancer types, and distinct epigenetic landscapes. This functional duality has critical therapeutic implications: strategies targeting snoRNAs must be carefully tailored to specific cancer types and molecular contexts, as depleting a snoRNA acting as an oncogene in one cancer could paradoxically promote tumor growth in another malignancy where it functions as a tumor suppressor [
29,
30]. Additionally, SNORA13 acts as a tumor suppressor by inducing cellular senescence through direct interaction with ribosomal protein RPL23, which impairs 60S ribosomal subunit assembly and triggers p53 activation [
31]. This mechanism links ribosome biogenesis surveillance to tumor suppression through senescence induction.
Table 1.
Example of snoRNAs with Validated Roles in Cancer.
Table 1.
Example of snoRNAs with Validated Roles in Cancer.
| snoRNA |
Type |
Function |
Cancer Type |
Primary Mechanism |
Validation |
Reference |
| SNORA21 |
H/ACA |
Oncogene |
Colorectal |
Hippo/Wnt pathways |
In vitro/in vivo |
[12] |
| SNORD78 |
C/D |
Oncogene |
Colorectal |
Oncoribosome formation |
Patient samples |
[13] |
| SNORA47 |
H/ACA |
Oncogene |
Breast |
EBF3/RPL11/c-Myc axis |
Xenografts |
[18] |
| SNORA24 |
H/ACA |
Tumor suppressor |
HCC |
Translational fidelity |
RAS model |
[19] |
| SNORD44 |
C/D |
Tumor suppressor |
Colorectal |
p53 pathway |
Oncolytic virus |
[20] |
| SNORD113-1 |
C/D |
Tumor suppressor |
HCC |
MAPK/STAT3 inhibition |
In vitro/in vivo |
[21] |
| SNORA13 |
H/ACA |
Tumor suppressor |
Multiple |
Senescence via RPL23/p53 |
Cell models |
[31] |
| SNORD50A/B |
C/D |
Context-dependent |
Multiple |
K-Ras/TRIM21 |
Xenografts |
[22,23,24] |
| SNORD76 |
C/D |
Context-dependent |
HCC/Glioblastoma |
Wnt/Cell cycle |
Patient tissues |
[26,27,28] |
3. Molecular Mechanisms of snoRNA-Mediated Effects in Cancer
SnoRNAs drive cancer progression through multifaceted molecular mechanisms spanning ribosomal dysfunction, post-transcriptional gene regulation, epigenetic control, microRNA-like functions, and integration with cellular signaling networks through dynamic protein interactions.
3.1. Ribosomal Dysfunction and Translational Control
The concept of specialized ribosomes or “onco-ribosomes” represents an emerging paradigm in contemporary cancer biology. For decades, ribosomes were considered uniform translation machines. However, experimental evidence now demonstrates that cells generate ribosomes with distinct modification patterns that exhibit selective translational preferences [
32]. In vivo CRISPR screens and ribosome profiling have revealed that differential expression of ribosomal proteins like RPL15 creates specialized ribosomes that selectively translate specific mRNA subsets, promoting metastatic progression [
33].
SnoRNAs play a central role in this specialization by directing site-specific 2’-O-methylations and pseudouridylations that alter ribosomal structure and function, influencing which mRNAs are preferentially translated [
15]. Cancer cells actively exploit this mechanism to remodel their proteome without need for additional genetic changes. For instance, SNORD78 overexpression in colorectal cancer does not simply result in “more” rRNA methylation, but in specific methylation patterns that create preferential binding sites for certain mRNAs. These mRNAs, which incidentally encode many proteins related to proliferation, survival, and metastasis, are more efficiently translated by these modified ribosomes [
34,
35]. Nait Slimane and colleagues (2020) has extensively documented this phenomenon, demonstrating that oncoribosomes can selectively increase translation of oncogenes while maintaining or even reducing synthesis of tumor suppressor proteins [
14]. Ribosomal heterogeneity extends beyond simple changes in translational efficiency. snoRNA-directed modifications can affect translation fidelity, reading frame selection, and even the ribosome’s capacity for translational reinitiation. These effects have profound consequences for cellular homeostasis and can contribute to multiple cancer hallmarks, from apoptosis evasion to metabolic reprogramming [
14].
3.2. Alternative Splicing Regulation
Pre-mRNA alternative splicing is a fundamental process that allows cells to generate multiple proteins from a single gene. In cancer, splicing patterns are frequently altered, producing protein isoforms that promote tumor growth, invasion, and therapy resistance [
36]. snoRNAs have emerged as important regulators of this process, acting through mechanisms that extend beyond their canonical functions.
The capacity of snoRNAs to modulate alternative splicing was first demonstrated by Kishore and Stamm (2006) in the context of Prader-Willi syndrome, where they showed that the orphan snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C [
37]. Subsequently, Bazeley and colleagues (2008) used computational approaches to demonstrate that many orphan snoRNAs localize near alternative splicing sites across the genome, suggesting a broader regulatory role for this class of molecules [
38]. This observation was later confirmed by detailed mechanistic studies showing that snoRNAs can be processed into smaller fragments that directly interact with the spliceosome and modulate splicing site selection [
5].
The loss of SNORD44 in various cancer types illustrates the pathological consequences of snoRNA-mediated splicing dysregulation. Under normal conditions, SNORD44 promotes inclusion of exons encoding important functional domains in tumor suppressor proteins, while suppressing exon inclusion in oncogenes that would result in more active isoforms [
20]. Its absence in cancer cells reverses these patterns, creating a protein repertoire that favors tumor proliferation and survival. This mechanism affects not only individual genes but can remodel entire splicing networks, thus amplifying its impact on the cellular phenotype.
3.3. Chromatin Remodeling and Epigenetic Regulation
SnoRNA participation in epigenetic regulation represents a significant expansion beyond their canonical nucleolar functions, establishing new links between RNA metabolism and genome organization. Several snoRNAs demonstrate the capacity to interact with chromatin and modulate its accessibility through distinct mechanisms, thereby influencing gene expression at transcriptional and post-transcriptional levels. The orphan snoRNA SNORA73 exemplifies non-canonical chromatin interaction in acute myeloid leukemia (AML). This snoRNA localizes to chromatin where it forms a non-canonical ribonucleoprotein complex with PARP1 and the canonical H/ACA proteins DKC1/NHP2 at DNA damage sites. SNORA73 inhibits PARP1 auto-PARylation, thereby blocking DNA damage repair and maintaining the genome instability characteristic of hematopoietic malignancies [
39].
SnoRNA-derived RNAs (sdRNA) further expand this epigenetic repertoire. The sdnRNA3, expressed in melanoma tumor-associated macrophages (TAMs), represses Nos2 gene transcription by recruiting the chromatin-remodeling regulator Mi-2β and depositing the repressive histone mark H3K27me3 at the gene promoter, thereby reducing chromatin accessibility and promoting an immunosuppressive tumor microenvironment [
40]. Complementing this direct chromatin-remodeling mechanism, SNORD104 modulates epigenetic regulation in endometrial cancer through 2’-O-methylation of PARP1 mRNA, increasing transcript stability and protein levels of this polymerase involved in DNA repair and chromatin structure modification [
41].
These mechanisms illustrate that snoRNAs and their derivatives transcend their canonical nucleolar roles to function as epigenetic regulators through diverse strategies: from direct recruitment of chromatin-remodeling complexes and histone modification to indirect modulation of key enzymes governing chromosomal architecture.
3.4. MicroRNA-like Functions
The discovery that some snoRNAs can function analogously to microRNAs has revealed a completely new dimension in the biology of these RNAs. This phenomenon involves the regulated processing of snoRNAs into smaller fragments, termed sdRNAs (snoRNA-derived RNAs), which incorporate into the RISC complex and regulate gene expression post-transcriptionally [
42]. Like canonical microRNAs, sdRNAs operate through 6-8 nucleotide “seed” regions that mediate specific binding to the 3’UTR regions of target mRNAs [
43]. However, sdRNAs exhibit distinctive properties in terms of molecular stability, subcellular localization, and diversity of target genes. Recent studies suggest that this processing is not accidental, but can be induced by specific conditions such as cellular stress or oncogenic signals [
44].
Several sdRNAs have demonstrated significant functional roles in different cancer types. SNORA42 generates sdRNAs that target multiple oncogene mRNAs, functioning as endogenous tumor suppressors [
45]. Conversely, sdRNA-93 promotes invasion in breast cancer, particularly in Luminal B HER2+ tumors, by directly regulating PIPOX [
46]. Similarly, SNORD78 and its derived sdRNAs are significantly elevated in patients with metastatic disease in lung and prostate cancer [
47].
Some snoRNAs exhibit dual functionality, simultaneously fulfilling their canonical roles in rRNA modification and generating functional sdRNAs in a context-dependent manner. Pan-cancer analyses of TCGA data have identified hundreds of snoRNAs associated with clinical stage and patient survival [
9], and sdRNAs show more pronounced expression changes than microRNAs in certain tumor contexts [
47]. Notably, many microRNA discovery algorithms have erroneously discarded small RNAs that align with snoRNAs, suggesting that the actual repertoire of post-transcriptional regulators relevant to cancer is substantially larger than currently appreciated. Recent investigations by Huo et al. (2024) have initiated a systematic cataloging of sdRNAs and their roles in cancer, revealing that these RNAs represent an emerging class of regulators with potential as biomarkers and therapeutic targets [
8].
3.5. Integration with Signaling Networks
snoRNAs do not operate in isolation but integrate deeply into cellular signaling networks. This integration occurs at multiple levels, from modulation of key signaling pathway components to regulation of pathway gene expression. For example, SNORA21 has been identified as a key oncogenic snoRNA in colorectal cancer, where its inhibition results in decreased cell proliferation and invasion through modulation of multiple cancer-related pathways, including the Hippo and Wnt signaling cascades [
12]. Similarly, other snoRNAs directly impact critical oncogenic pathways: SNORD60 regulates the PI3K/AKT/mTOR pathway by enhancing the stability and expression of PIK3CA mRNA through 2’-O-methylation, thereby promoting cancer progression [
48]. This multilayered integration allows snoRNAs to influence not only the activity of signaling pathways but also the expression levels and stability of their key components, positioning them as important modulators of cellular signaling networks in cancer. Likewise, the recent discovery of the SNORA47/EBF3/RPL11/c-Myc axis in breast cancer illustrates how snoRNAs can connect different levels of cellular regulation [
18]. SNORA47 influences expression of transcription factor EBF3, which in turn regulates ribosomal protein RPL11. RPL11, beyond its structural function in the ribosome, can sequester MDM2 and stabilize p53, but also affects c-Myc stability and function. This cascade connects ribosomal biogenesis with two of the most important regulators of cell growth and apoptosis, providing multiple potential therapeutic intervention points [
18].
3.6. snoRNA-Protein Interactions and Regulatory Complexes
Beyond their canonical associations with snoRNP proteins, snoRNAs engage in diverse protein interactions that expand their functional repertoire in cancer [
49]. These partnerships create regulatory complexes that influence multiple aspects of tumor biology, from RNA processing to signal transduction. Both aberrant canonical modifications and novel non-canonical protein interactions contribute to snoRNA-mediated oncogenesis.
Dysregulated snoRNAs drive cancer progression through altered protein partnerships that manifest in diverse molecular contexts [
35]. These snoRNAs can exert oncogenic effects through their canonical modification functions—where aberrant expression of modification-guiding snoRNAs alters ribosome composition and selectively enhances translation of oncogenic transcripts—and through non-canonical interactions with proteins outside the typical rRNA/snRNA modification machinery. For example, SNORD50A and SNORD50B directly bind to K-Ras protein, and their loss increases GTP-bound active K-Ras and hyperactivates Ras-ERK1/ERK2 signaling, with deletion of these snoRNAs occurring in 10-40% of common cancers [
22]. Additionally, SNORD50A and SNORD50B enhance the interaction between E3 ubiquitin ligase TRIM21 and its substrate GMPS by forming a complex with them, which promotes tumor progression specifically in p53 wild-type breast cancers by facilitating p53 degradation [
50]. This oncogenic role contrasts with their tumor suppressor function reported in other cancer contexts, where SNORD50A/B inhibit K-Ras pathway activity, highlighting the context-dependent and tissue-specific nature of snoRNA functions in tumorigenesis [
51].
Beyond these cancer-associated interactions, snoRNAs also form complexes with proteins involved in cellular stress responses, a phenomenon that may be co-opted during tumorigenesis. Under various stress conditions, specific snoRNAs are released from their usual nucleolar localization. The rpL13a-encoded snoRNAs U32a, U33, and U35a accumulate in the cytoplasm in response to metabolic stress [
44], while transcriptional inhibition induces the formation of nucleolar caps containing relocated snoRNPs [
52]. These stress-induced relocalization events may contribute to cancer cell adaptation to the tumor microenvironment. More recent evidence suggests that snoRNAs interact with various RNA-binding proteins beyond their canonical partners, forming ribonucleoprotein complexes with distinct functional specificities, as revealed by advanced chimeric eCLIP studies. These studies showed that snoRNA ribonucleoprotein complexes containing accessory proteins like WDR43 and NOLC1 are enriched for specific subsets of snoRNA-target RNA interactions with distinct roles in ribosome and spliceosome biogenesis [
53]. Notably, this work discovered that SNORD89 guides 2′-O-methylation at two neighboring sites in U2 snRNA that fine-tune splice site recognition. Dysregulation of SNORD89 has been implicated in cancer progression promoting, for example; endometrial cancer development through 2′-O-methylation modification of Bim, affecting apoptotic pathways [
54]. The specificity of these interactions and their regulation by cellular conditions in both normal physiology and cancer pathogenesis remain active areas of investigation.
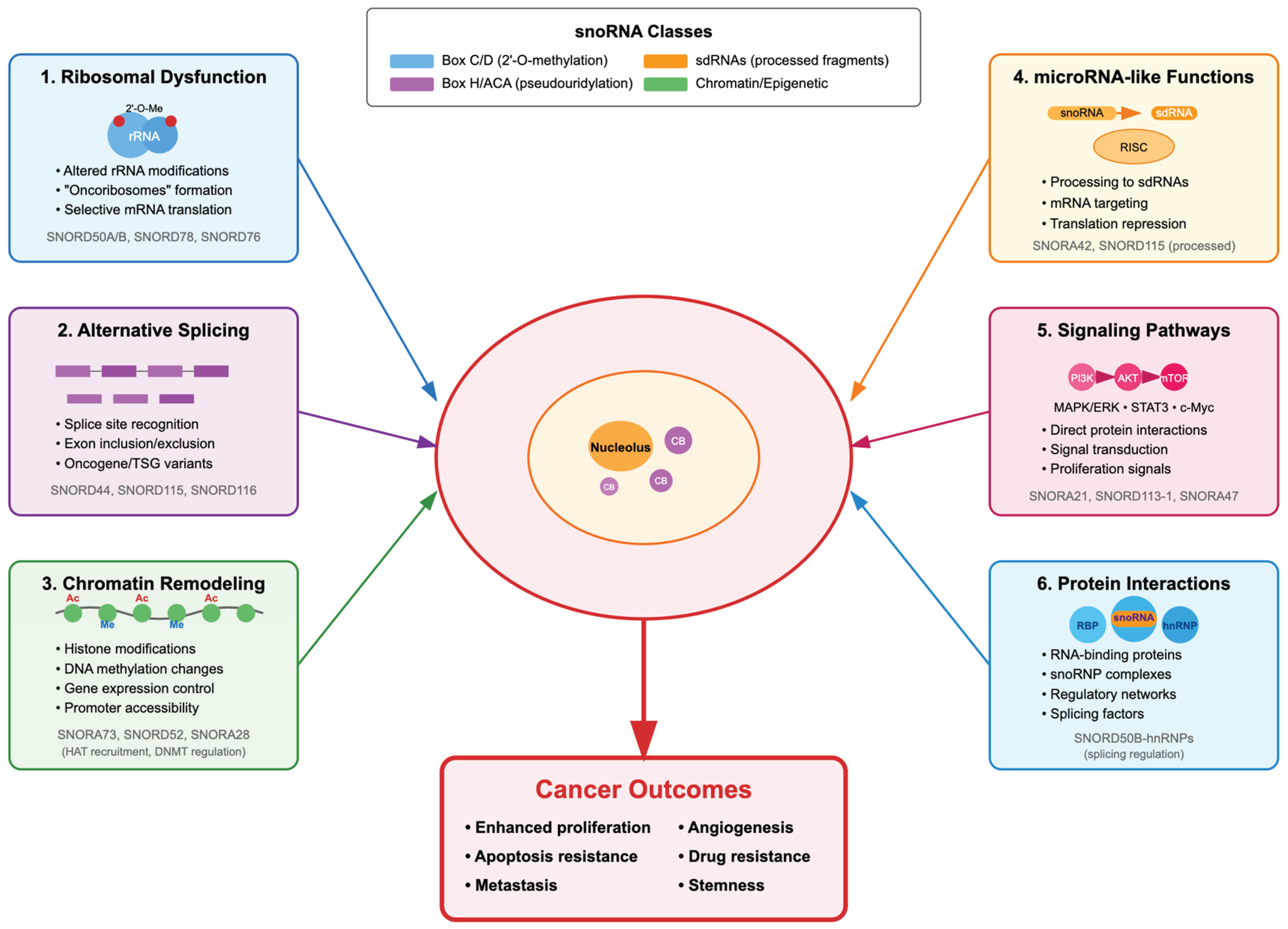
Figure 2.
Integration of snoRNA-mediated mechanisms in cancer biology. The diagram illustrates the convergence of multiple pathways through which dysregulated snoRNAs contribute to the malignant phenotype. Mechanisms include: (1) Generation of oncoribosomes with selective translational capacity, (2) Alteration of alternative splicing patterns, (3) Epigenetic remodeling of the genome, (4) Production of sdRNAs with regulatory functions, (5) Direct modulation of oncogenic signaling pathways, and (6) Formation of regulatory complexes with RNA-binding proteins. These mechanisms act synergistically to promote cancer hallmarks including uncontrolled proliferation, apoptosis evasion, invasive capacity, angiogenesis, and therapeutic resistance. CB: Cajal Bodies; Ac: Acetylation; Me: Methylation.
Figure 2.
Integration of snoRNA-mediated mechanisms in cancer biology. The diagram illustrates the convergence of multiple pathways through which dysregulated snoRNAs contribute to the malignant phenotype. Mechanisms include: (1) Generation of oncoribosomes with selective translational capacity, (2) Alteration of alternative splicing patterns, (3) Epigenetic remodeling of the genome, (4) Production of sdRNAs with regulatory functions, (5) Direct modulation of oncogenic signaling pathways, and (6) Formation of regulatory complexes with RNA-binding proteins. These mechanisms act synergistically to promote cancer hallmarks including uncontrolled proliferation, apoptosis evasion, invasive capacity, angiogenesis, and therapeutic resistance. CB: Cajal Bodies; Ac: Acetylation; Me: Methylation.
4. snoRNAs as Cancer Biomarkers
Translation of fundamental knowledge about snoRNAs toward clinical applications represents one of the most promising developments in this field. snoRNAs possess several characteristics that make them ideal biomarkers: they are stable in biological fluids, show tissue- and tumor-type specific expression patterns, and their levels can be measured with high sensitivity and specificity using established technologies.
4.1. Diagnostic Applications
The diagnostic potential of snoRNAs has been demonstrated across multiple cancer types through diverse biological specimens, each offering distinct advantages for clinical implementation. Circulating biomarkers represent one of the most accessible approaches for widespread screening. Li and colleagues (2024) identified a four-snoRNA panel (SNORD16, SNORA73B, SCARNA4, and SNORD49B) for breast cancer detection in plasma, achieving 74.38% specificity and 66.51% sensitivity. These snoRNAs exhibited remarkable stability, resisting RNase A treatment, and were predominantly found in vesicle-free plasma fractions, simplifying extraction procedures compared to exosome-dependent biomarkers [
55]. Complementing this plasma-based approach, exosomal snoRNAs have proven particularly valuable for addressing specific diagnostic challenges. In lung cancer, Cao and colleagues (2024) validated that serum exosomal SNORD78 and SNORD37 can accurately distinguish malignant from benign pulmonary nodules, a critical clinical need that could reduce unnecessary biopsies while improving early detection of malignancy [
56]. The differential stability and compartmentalization of snoRNAs in various blood fractions thus provides multiple strategies for biomarker development depending on the clinical context.
Beyond blood-based approaches, snoRNA detection in other biological fluids has significantly expanded non-invasive screening capabilities. In colorectal cancer, Gómez-Matas and colleagues (2024) demonstrated that fecal SNORA51, combined with hemoglobin concentration, significantly improved diagnostic accuracy among individuals with positive fecal immunochemical tests, enhancing the specificity of population screening strategies [
57]. This refinement of screening protocols addresses a major challenge in colorectal cancer detection: reducing false-positive rates that lead to unnecessary colonoscopies. Similarly, urinary biomarkers offer another accessible non-invasive avenue. Zhou and colleagues (2023) identified a three-snoRNA signature (SNORD15A, SNORD35B, and SNORD60) in renal cell carcinoma, demonstrating significant upregulation in tumor tissues alongside stable detection in urinary sediment, thus providing a practical monitoring tool for kidney malignancies [
58]. Tissue-based snoRNA profiling has complemented these fluid biomarker studies, with SCARNA12 emerging as a promising diagnostic marker in colorectal cancer tissue samples [
59] and comprehensive profiling revealing distinctive signatures in head and neck squamous cell carcinomas [
60]. Together, these findings across diverse cancer types and specimen sources demonstrate the remarkable versatility of snoRNAs as diagnostic and prognostic biomarkers, establishing them as promising tools for cancer detection and patient stratification in clinical oncology.
Table 2.
Circulating snoRNA Biomarker Panels for Cancer Diagnosis.
Table 2.
Circulating snoRNA Biomarker Panels for Cancer Diagnosis.
| Cancer Type |
snoRNA Panel |
Sample Type |
Sensitivity |
Specificity |
AUC |
Reference |
| Breast |
SNORD16, SNORA73B, SCARNA4, SNORD49B |
Plasma |
66.5% |
74.4% |
N/A |
[54] |
| Lung |
SNORD78, SNORD37 |
Serum exosomes |
N/A |
N/A |
0.85 |
[55] |
| Colorectal |
SNORA51 |
Fecal |
82% |
89% |
0.91 |
[56] |
| Renal cell |
SNORD15A, SNORD35B, SNORD60 |
Urine sediment |
78% |
85% |
0.88 |
[57] |
| HCC |
9-snoRNA signature |
Tissue |
N/A |
N/A |
0.92 |
[60] |
4.2. Prognostic Value and Disease Monitoring
Beyond their diagnostic applications, snoRNAs provide valuable prognostic information that can guide therapeutic decisions and predict clinical outcomes in cancer patients. Specific snoRNA expression patterns have been consistently associated with adverse clinical features including advanced tumor stage, metastatic spread, and reduced overall survival across diverse malignancy types. The prognostic utility of these molecules has been particularly well-demonstrated through the development of multi-snoRNA signatures that can stratify patients into distinct risk groups with superior predictive performance compared to conventional clinicopathological staging systems. In hepatocellular carcinoma, for instance, a 9-snoRNA signature developed through multivariate Cox regression analysis serves as an independent prognostic factor with enhanced predictive accuracy [
61]. Similarly, in breast cancer, the integration of snoRNA profiles with other molecular biomarkers has yielded promising results, with SNORA47 correlating with advanced tumor stage and unfavorable survival outcomes in luminal A subtype patients [
18], while a comprehensive 13-snoRNA signature has demonstrated independent prognostic value in multivariate analysis, effectively stratifying patients for both overall survival and recurrence-free survival [
62]. This stratification capability proves particularly critical for therapeutic decision-making in borderline cases where the benefit of adjuvant chemotherapy remains uncertain.
The clinical relevance of snoRNAs as prognostic biomarkers extends prominently to hematological malignancies, where distinct expression profiles have been characterized across multiple disease subtypes. Comprehensive analyses of acute leukemias have revealed distinctive snoRNA signatures in both myeloid and lymphoblastic variants, with acute promyelocytic leukemia exhibiting a particularly specific expression pattern [
63]. In chronic lymphocytic leukemia, the analysis of 211 patients established a two-snoRNA prognostic model incorporating SNORA70F and SNORD116-18 that successfully distinguished distinct prognostic groups independent of traditional markers such as IGHV mutational status [
64]. The prognostic significance of snoRNAs in lymphoid malignancies extends further to peripheral T-cell lymphoma, where HBII-239 expression correlates significantly with prolonged progression-free and overall survival in patient cohorts [
65].
In solid tumors, snoRNAs have emerged as equally valuable prognostic markers with clinical applicability across multiple cancer types. Non-small cell lung cancer studies have identified several snoRNAs—including SNORA47, SNORA68, and SNORA78—as independent predictors of poor prognosis after adjustment for clinical parameters [
66], while the expression of SNORA3 and SNORA42 in tumor-initiating cells shows inverse correlation with patient survival [
67]. In the colorectal cancer setting, SNORA42 has been established as an independent prognostic factor for both overall survival and disease-free survival, with particular utility in identifying high-risk patients for recurrence among those with stage II disease [
68]. The widespread clinical relevance of snoRNAs across malignancies has been further validated through pan-cancer analyses examining more than 10,000 samples from 31 cancer types, which identified 355 snoRNAs significantly associated with patient survival [
9].
The integration of computational approaches has enhanced the discovery and validation of prognostic snoRNA signatures. Machine learning algorithms applied to snoRNA expression patterns across eight major cancer types have successfully identified discriminative molecules including HBII-52-14, HBII-336, and SNORD123 with demonstrated prognostic value, illustrating the potential of computational methods to systematically extract clinically relevant information from complex expression datasets [
69].
Table 3.
Molecular Mechanisms of snoRNA-Mediated Oncogenesis.
Table 3.
Molecular Mechanisms of snoRNA-Mediated Oncogenesis.
| Mechanism |
snoRNA Examples |
Cancer Effect |
Therapeutic Potential |
References |
| Ribosomal Dysfunction |
|
|
|
|
| Aberrant 2’-O-methylation |
SNORD78, SNORD60 |
Selective oncogene translation |
ASO targeting |
[13,47] |
| Loss of pseudouridylation |
SNORA24 |
Reduced translational fidelity |
Expression restoration |
[19] |
| Oncoribosomes |
SNORD16 |
IRES-mediated translation |
Ribosome inhibitors |
[14,15] |
| Post-transcriptional Regulation |
|
|
|
|
| MicroRNA-like functions |
sdRNA-93, SNORA42 |
Target mRNA regulation |
sdRNA inhibitors |
[44,45] |
| Alternative splicing |
SNORD44, SNORD115 (HBII-52) |
Pro-tumoral isoforms |
Splicing modulators |
[5,20,36] |
| mRNA stability |
SNORD104 |
Enhanced PARP1 expression |
PARP inhibitors |
[40] |
| Chromatin Remodeling |
|
|
|
|
| PARP1 interaction |
SNORA73 |
Genomic instability |
PARP inhibitors |
[38] |
| Histone modification |
sdnRNA3 |
TAM immunosuppression |
Epigenetic therapy |
[39] |
| Signaling Networks |
|
|
|
|
| Oncogenic pathways |
SNORA21, SNORD113-1 |
Proliferation/survival |
Combination therapy |
[12,21] |
| Protein interactions |
SNORD50A/B |
K-Ras activation |
Targeted inhibitors |
[22,48] |
5. snoRNAs as Therapeutic Targets
The increasingly deep understanding of snoRNA functional roles in cancer has opened new opportunities for therapeutic development. Although RNA targeting presents technical challenges, recent advances in oligonucleotide chemistry and delivery systems have made snoRNA-based therapies increasingly viable.
Antisense oligonucleotides (ASOs) represent the most direct approach for modulating snoRNA levels. Designing effective ASOs against snoRNAs requires overcoming substantial obstacles: the highly stable secondary structures characteristic of many snoRNAs can impede efficient ASO hybridization, while their incorporation into ribonucleoprotein complexes may shield them from targeting. Next-generation ASOs with chemical modifications that improve stability and cellular penetration have been developed to overcome these barriers, showing promising efficacy in preclinical models. The therapeutic potential of ASO-based targeting depends critically on understanding the context-dependent biological roles of individual snoRNAs. SNORD50A/B exemplifies this complexity: in p53 wild-type breast cancer, where these snoRNAs promote tumor progression through p53 degradation, ASO-mediated targeting significantly reduces tumor growth and inhibits metastatic potential in murine xenograft models [
50]. Hu and colleagues (2024) have comprehensively reviewed ASO-based therapeutic strategies targeting snoRNAs in cancer, discussing both preclinical successes and challenges remaining for clinical translation [
49]. Sophisticated designs must consider not only the target snoRNA sequence but also its structure and molecular context. Computational tools such as PLEXY, developed by Kehr and colleagues (2011), have significantly facilitated prediction of box C/D snoRNA targets, allowing more rational design of therapeutic interventions [
70].
As an alternative to ASOs, CRISPR-based approaches offer precise modulation of snoRNA expression with the advantage of providing lasting effects rather than requiring continuous administration. Given that most mammalian snoRNAs reside within introns of protein-coding or non-coding host genes [
71], developing CRISPR systems capable of targeting snoRNAs without disrupting host gene function is particularly important. Selective editing of individual snoRNAs can be achieved without affecting other snoRNAs within the same host gene [
72], highlighting the feasibility of this approach.
Beyond direct targeting methods, an alternative strategy involves modulating factors that regulate snoRNA expression or function. Epigenetic modulators affecting transcription of snoRNA host genes have shown promise in preclinical studies. DNA methyltransferase inhibitors can restore expression of tumor suppressor snoRNAs silenced by promoter hypermethylation. Cancer-specific hypermethylation of CpG islands associated with snoRNAs SNORD123, U70C, and ACA59B has been demonstrated in colorectal and lung cancer cell lines, resulting in transcriptional silencing [
73].
While targeting snoRNAs as monotherapy holds promise, the complexity of cancer biology suggests that integrating snoRNA-targeted approaches with conventional treatments may be essential for achieving optimal outcomes. This rationale is supported by mounting evidence that snoRNAs function as modulators of treatment response across multiple therapeutic modalities. SnoRNA expression levels can predict sensitivity to chemotherapy and influence therapeutic efficacy. Plasma levels of SNORD33 serve as an independent predictor of platinum-based chemotherapy response in metastatic triple-negative breast cancer, with low SNORD33 expression correlating with shorter progression-free survival (6.6 versus 10.1 months; P = 0.005) and reduced overall survival in patients receiving first-line platinum regimens [
74]. The role of SNORA47 in chemotherapy resistance through the EBF3/RPL11/c-Myc axis further illustrates how mechanistic understanding can identify combinatorial strategies to enhance treatment sensitivity in specific contexts such as luminal A breast cancer [
18]. Beyond chemosensitization, snoRNAs also influence immunotherapy efficacy by regulating expression of immune checkpoint molecules and cytokines that modulate antitumor immune responses. Their modulation could convert immunologically “cold” tumors into “hot” tumors more susceptible to checkpoint inhibitors. These findings underscore that snoRNA-based therapeutics may achieve their greatest clinical impact not as standalone interventions but as integral components of multimodal treatment regimens tailored to exploit cancer-specific vulnerabilities.
6. Conclusions and Perspectives
The field of snoRNAs in cancer stands at a critical juncture between fundamental discovery and clinical translation. Over the past two decades, these molecules have evolved from simple rRNA modification guides into recognized regulators of cancer biology, participating in virtually all aspects of oncogenesis—from translational machinery reprogramming through specialized ribosomes to modulation of signaling networks and epigenetic landscapes. This accumulated evidence demonstrates their profound involvement in tumorigenesis, yet transforming these insights into clinical applications presents both formidable challenges and unprecedented opportunities that define the current state of the field.
The path toward clinical implementation confronts intertwined technical and biological obstacles that together shape the translational landscape. Accurate detection of snoRNAs remains problematic due to their stable secondary structures interfering with conventional extraction and amplification methods, while their biological complexity—simultaneously participating in multiple cellular processes—makes predicting consequences of their modulation difficult. The context-dependent duality observed with specific snoRNAs exemplifies this challenge particularly well. SNORA24 functions as a tumor suppressor in RAS-driven hepatocellular carcinoma by maintaining ribosomal translation fidelity [
19], while SNORD50A and SNORD50B demonstrate opposing roles depending on p53 status, acting as oncogenes in p53 wild-type breast cancers through TRIM21-GMPS-mediated p53 degradation but functioning as tumor suppressors in other contexts [
50]. This functional plasticity underscores the necessity for cancer-type-specific and molecular context-specific therapeutic strategies. Tumor heterogeneity compounds these difficulties, as different cellular populations may show distinct dependencies on specific snoRNAs, potentially leading to heterogeneous therapeutic responses.
However, these very complexities are driving innovation in ways that promise to overcome current limitations. Emerging single-cell sequencing technologies are beginning to reveal dynamic snoRNA expression patterns during tumor progression and therapy response at unprecedented resolution, while artificial intelligence and machine learning approaches are accelerating discovery of clinically relevant snoRNA signatures that capture biological complexity impossible to detect through traditional analyses [
69]. Development of international consortia and comprehensive databases like snoDB, recently enhanced as snoDB 2.0 with improved snoRNA-RNA target prediction and expression profiling capabilities, will prove crucial for standardizing methodologies, sharing data across diverse cancer types, and accelerating clinical translation. [
4]. [
75] (
https://bioinfo-scottgroup.med.usherbrooke.ca/snoDB/)?
Progress toward clinical implementation has been particularly notable for biomarkers, where circulating snoRNA panels demonstrate diagnostic capacity comparable or superior to conventional markers. The therapeutic potential of snoRNAs, though in earlier developmental stages, is accelerating through advances in oligonucleotide technology, delivery systems, and genomic editing. As the field matures, snoRNAs are positioned to occupy an increasingly prominent place in precision cancer medicine, bridging fundamental cell biology with clinical applications and offering new paradigms for understanding and treating cancer.:
References
- Matera, A.G.; Terns, R.M.; Terns, M.P. Non-Coding RNAs: Lessons from the Small Nuclear and Small Nucleolar RNAs. Nat Rev Mol Cell Biol 2007, 8, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Kiss, T. Small Nucleolar RNAs: An Abundant Group of Noncoding RNAs with Diverse Cellular Functions. Cell 2002, 109, 145–148. [Google Scholar] [CrossRef] [PubMed]
- Bachellerie, J.P.; Cavaillé, J.; Hüttenhofer, A. The Expanding snoRNA World. Biochimie 2002, 84, 775–790. [Google Scholar] [CrossRef] [PubMed]
- Bouchard-Bourelle, P.; Desjardins-Henri, C.; Mathurin-St-Pierre, D.; Deschamps-Francoeur, G.; Fafard-Couture, É.; Garant, J.-M.; Elela, S.A.; Scott, M.S. snoDB: An Interactive Database of Human snoRNA Sequences, Abundance and Interactions. Nucleic Acids Res 2020, 48, D220–D225. [Google Scholar] [CrossRef]
- Kishore, S.; Khanna, A.; Zhang, Z.; Hui, J.; Balwierz, P.J.; Stefan, M.; Beach, C.; Nicholls, R.D.; Zavolan, M.; Stamm, S. The snoRNA MBII-52 (SNORD 115) Is Processed into Smaller RNAs and Regulates Alternative Splicing. Hum Mol Genet 2010, 19, 1153–1164. [Google Scholar] [CrossRef]
- McMahon, M.; Contreras, A.; Ruggero, D. Small RNAs with Big Implications: New Insights into H/ACA snoRNA Function and Their Role in Human Disease. Wiley Interdiscip Rev RNA 2015, 6, 173–189. [Google Scholar] [CrossRef]
- Sharma, E.; Sterne-Weiler, T.; O’Hanlon, D.; Blencowe, B.J. Global Mapping of Human RNA-RNA Interactions. Mol Cell 2016, 62, 618–626. [Google Scholar] [CrossRef]
- Huo, M.; Rai, S.K.; Nakatsu, K.; Deng, Y.; Jijiwa, M. Subverting the Canon: Novel Cancer-Promoting Functions and Mechanisms for snoRNAs. Int J Mol Sci 2024, 25, 2923. [Google Scholar] [CrossRef]
- Gong, J.; Li, Y.; Liu, C.-J.; Xiang, Y.; Li, C.; Ye, Y.; Zhang, Z.; Hawke, D.H.; Park, P.K.; Diao, L.; et al. A Pan-Cancer Analysis of the Expression and Clinical Relevance of Small Nucleolar RNAs in Human Cancer. Cell Rep 2017, 21, 1968–1981. [Google Scholar] [CrossRef]
- Liang, J.; Wen, J.; Huang, Z.; Chen, X.-P.; Zhang, B.-X.; Chu, L. Small Nucleolar RNAs: Insight Into Their Function in Cancer. Front Oncol 2019, 9, 587. [Google Scholar] [CrossRef]
- Wu, F.; Zhang, L.; Wu, P.; Wu, Y.; Zhang, T.; Zhang, D.; Tian, J. The Potential Role of Small Nucleolar RNAs in Cancers - An Evidence Map. Int J Gen Med 2022, 15, 3851–3864. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Toden, S.; Weng, W.; Shigeyasu, K.; Miyoshi, J.; Turner, J.; Nagasaka, T.; Ma, Y.; Takayama, T.; Fujiwara, T.; et al. SNORA21 - An Oncogenic Small Nucleolar RNA, with a Prognostic Biomarker Potential in Human Colorectal Cancer. EBioMedicine 2017, 22, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.-Z.; Wu, Z.; Chen, W.-J.; Fang, Z.-X.; Yu, X.-N.; Wu, H.-T.; Liu, J. Small Nucleolar RNA and Its Potential Role in the Oncogenesis and Development of Colorectal Cancer. World J Gastroenterol 2024, 30, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Nait Slimane, S.; Marcel, V.; Fenouil, T.; Catez, F.; Saurin, J.-C.; Bouvet, P.; Diaz, J.-J.; Mertani, H.C. Ribosome Biogenesis Alterations in Colorectal Cancer. Cells 2020, 9, 2361. [Google Scholar] [CrossRef]
- Kampen, K.R.; Sulima, S.O.; De Keersmaecker, K. Rise of the Specialized Onco-Ribosomes. Oncotarget 2018, 9, 35205–35206. [Google Scholar] [CrossRef]
- He, J.-Y.; Liu, X.; Qi, Z.-H.; Wang, Q.; Lu, W.-Q.; Zhang, Q.-T.; He, S.-Y.; Wang, Z.-D. Small Nucleolar RNA, C/D Box 16 (SNORD16) Acts as a Potential Prognostic Biomarker in Colon Cancer. Dose Response 2020, 18, 1559325820917829. [Google Scholar] [CrossRef]
- Yang, T.-F.; Li, X.-R.; Kong, M.-W. Molecular Mechanisms Underlying Roles of Long Non-Coding RNA Small Nucleolar RNA Host Gene 16 in Digestive System Cancers. World J Gastrointest Oncol 2024, 16, 4300–4308. [Google Scholar] [CrossRef]
- Han, Q.; Zhou, Y.; Dong, Z.; Wang, W.; Wang, M.; Pang, M.; Song, X.; Chen, B.; Zheng, A. SNORA47 Affects Stemness and Chemotherapy Sensitivity via EBF3/RPL11/c-Myc Axis in Luminal A Breast Cancer. Mol Med 2025, 31, 150. [Google Scholar] [CrossRef]
- McMahon, M.; Contreras, A.; Holm, M.; Uechi, T.; Forester, C.M.; Pang, X.; Jackson, C.; Calvert, M.E.; Chen, B.; Quigley, D.A.; et al. A Single H/ACA Small Nucleolar RNA Mediates Tumor Suppression Downstream of Oncogenic RAS. Elife 2019, 8, e48847. [Google Scholar] [CrossRef]
- Yuan, S.; Wu, Y.; Wang, Y.; Chen, J.; Chu, L. An Oncolytic Adenovirus Expressing SNORD44 and GAS5 Exhibits Antitumor Effect in Colorectal Cancer Cells. Hum Gene Ther 2017, 28, 690–700. [Google Scholar] [CrossRef]
- Xu, G.; Yang, F.; Ding, C.-L.; Zhao, L.-J.; Ren, H.; Zhao, P.; Wang, W.; Qi, Z.-T. Small Nucleolar RNA 113-1 Suppresses Tumorigenesis in Hepatocellular Carcinoma. Mol Cancer 2014, 13, 216. [Google Scholar] [CrossRef] [PubMed]
- Siprashvili, Z.; Webster, D.E.; Johnston, D.; Shenoy, R.M.; Ungewickell, A.J.; Bhaduri, A.; Flockhart, R.; Zarnegar, B.J.; Che, Y.; Meschi, F.; et al. The Noncoding RNAs SNORD50A and SNORD50B Bind K-Ras and Are Recurrently Deleted in Human Cancer. Nat Genet 2016, 48, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.-Y.; Rodriguez, C.; Guo, P.; Sun, X.; Talbot, J.T.; Zhou, W.; Petros, J.; Li, Q.; Vessella, R.L.; Kibel, A.S.; et al. SnoRNA U50 Is a Candidate Tumor-Suppressor Gene at 6q14.3 with a Mutation Associated with Clinically Significant Prostate Cancer. Hum Mol Genet 2008, 17, 1031–1042. [Google Scholar] [CrossRef]
- Dong, X.-Y.; Guo, P.; Boyd, J.; Sun, X.; Li, Q.; Zhou, W.; Dong, J.-T. Implication of snoRNA U50 in Human Breast Cancer. J Genet Genomics 2009, 36, 447–454. [Google Scholar] [CrossRef]
- Xu, B.; Ye, M.-H.; Lv, S.-G.; Wang, Q.-X.; Wu, M.-J.; Xiao, B.; Kang, C.-S.; Zhu, X.-G. SNORD47, a Box C/D snoRNA, Suppresses Tumorigenesis in Glioblastoma. Oncotarget 2017, 8, 43953–43966. [Google Scholar] [CrossRef]
- Wu, L.; Chang, L.; Wang, H.; Ma, W.; Peng, Q.; Yuan, Y. Clinical Significance of C/D Box Small Nucleolar RNA U76 as an Oncogene and a Prognostic Biomarker in Hepatocellular Carcinoma. Clin Res Hepatol Gastroenterol 2018, 42, 82–91. [Google Scholar] [CrossRef]
- Liu, X.; Xie, W.; Meng, S.; Kang, X.; Liu, Y.; Guo, L.; Wang, C. Small Nucleolar RNAs and Their Comprehensive Biological Functions in Hepatocellular Carcinoma. Cells 2022, 11, 2654. [Google Scholar] [CrossRef]
- Chen, L.; Han, L.; Wei, J.; Zhang, K.; Shi, Z.; Duan, R.; Li, S.; Zhou, X.; Pu, P.; Zhang, J.; et al. SNORD76, a Box C/D snoRNA, Acts as a Tumor Suppressor in Glioblastoma. Sci Rep 2015, 5, 8588. [Google Scholar] [CrossRef]
- Watkins, N.J.; Bohnsack, M.T. The Box C/D and H/ACA snoRNPs: Key Players in the Modification, Processing and the Dynamic Folding of Ribosomal RNA. Wiley Interdiscip Rev RNA 2012, 3, 397–414. [Google Scholar] [CrossRef]
- Janin, M.; Coll-SanMartin, L.; Esteller, M. Disruption of the RNA Modifications That Target the Ribosome Translation Machinery in Human Cancer. Mol Cancer 2020, 19, 70. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, S.; Zhang, H.; Lee, J.-S.; Ni, C.; Guo, J.; Chen, E.; Wang, S.; Acharya, A.; Chang, T.-C.; et al. A Non-Canonical Role for a Small Nucleolar RNA in Ribosome Biogenesis and Senescence. Cell 2024, 187, 4770–4789.e23. [Google Scholar] [CrossRef] [PubMed]
- Ramalho, S.; Dopler, A.; Faller, W.J. Ribosome Specialization in Cancer: A Spotlight on Ribosomal Proteins. NAR Cancer 2024, 6, zcae029. [Google Scholar] [CrossRef] [PubMed]
- Ebright, R.Y.; Lee, S.; Wittner, B.S.; Niederhoffer, K.L.; Nicholson, B.T.; Bardia, A.; Truesdell, S.; Wiley, D.F.; Wesley, B.; Li, S.; et al. Deregulation of Ribosomal Protein Expression and Translation Promotes Breast Cancer Metastasis. Science 2020, 367, 1468–1473. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Qin, W.; Lu, S.; Wang, X.; Zhang, J.; Sun, T.; Hu, X.; Li, Y.; Chen, Q.; Wang, Y.; et al. Long Noncoding RNA ZFAS1 Promoting Small Nucleolar RNA-Mediated 2’-O-Methylation via NOP58 Recruitment in Colorectal Cancer. Mol Cancer 2020, 19, 95. [Google Scholar] [CrossRef]
- Zacchini, F.; Barozzi, C.; Venturi, G.; Montanaro, L. How snoRNAs Can Contribute to Cancer at Multiple Levels. NAR Cancer 2024, 6, zcae005. [Google Scholar] [CrossRef]
- Anders, S.; Reyes, A.; Huber, W. Detecting Differential Usage of Exons from RNA-Seq Data. Genome Res 2012, 22, 2008–2017. [Google Scholar] [CrossRef]
- Kishore, S.; Stamm, S. The snoRNA HBII-52 Regulates Alternative Splicing of the Serotonin Receptor 2C. Science 2006, 311, 230–232. [Google Scholar] [CrossRef]
- Bazeley, P.S.; Shepelev, V.; Talebizadeh, Z.; Butler, M.G.; Fedorova, L.; Filatov, V.; Fedorov, A. snoTARGET Shows That Human Orphan snoRNA Targets Locate Close to Alternative Splice Junctions. Gene 2008, 408, 172–179. [Google Scholar] [CrossRef]
- Han, C.; Sun, L.-Y.; Luo, X.-Q.; Pan, Q.; Sun, Y.-M.; Zeng, Z.-C.; Chen, T.-Q.; Huang, W.; Fang, K.; Wang, W.-T.; et al. Chromatin-Associated Orphan snoRNA Regulates DNA Damage-Mediated Differentiation via a Non-Canonical Complex. Cell Rep 2022, 38, 110421. [Google Scholar] [CrossRef]
- Shi, Y.; Shi, Q.; Shen, Q.; Zhang, Q.; Cao, X. Dicer-Independent snRNA/snoRNA-Derived Nuclear RNA 3 Regulates Tumor-Associated Macrophage Function by Epigenetically Repressing Inducible Nitric Oxide Synthase Transcription. Cancer Commun (Lond) 2021, 41, 140–153. [Google Scholar] [CrossRef]
- Lu, B.; Chen, X.; Liu, X.; Chen, J.; Qin, H.; Chen, S.; Zhao, Y. C/D Box Small Nucleolar RNA SNORD104 Promotes Endometrial Cancer by Regulating the 2’-O-Methylation of PARP1. J Transl Med 2022, 20, 618. [Google Scholar] [CrossRef] [PubMed]
- Taft, R.J.; Glazov, E.A.; Lassmann, T.; Hayashizaki, Y.; Carninci, P.; Mattick, J.S. Small RNAs Derived from snoRNAs. RNA 2009, 15, 1233–1240. [Google Scholar] [CrossRef]
- Ender, C.; Krek, A.; Friedländer, M.R.; Beitzinger, M.; Weinmann, L.; Chen, W.; Pfeffer, S.; Rajewsky, N.; Meister, G. A Human snoRNA with microRNA-like Functions. Mol Cell 2008, 32, 519–528. [Google Scholar] [CrossRef]
- Michel, C.I.; Holley, C.L.; Scruggs, B.S.; Sidhu, R.; Brookheart, R.T.; Listenberger, L.L.; Behlke, M.A.; Ory, D.S.; Schaffer, J.E. Small Nucleolar RNAs U32a, U33, and U35a Are Critical Mediators of Metabolic Stress. Cell Metab 2011, 14, 33–44. [Google Scholar] [CrossRef]
- Abel, Y.; Rederstorff, M. SnoRNAs and the Emerging Class of sdRNAs: Multifaceted Players in Oncogenesis. Biochimie 2019, 164, 17–21. [Google Scholar] [CrossRef]
- Patterson, D.G.; Roberts, J.T.; King, V.M.; Houserova, D.; Barnhill, E.C.; Crucello, A.; Polska, C.J.; Brantley, L.W.; Kaufman, G.C.; Nguyen, M.; et al. Human snoRNA-93 Is Processed into a microRNA-like RNA That Promotes Breast Cancer Cell Invasion. NPJ Breast Cancer 2017, 3, 25. [Google Scholar] [CrossRef]
- Martens-Uzunova, E.S.; Hoogstrate, Y.; Kalsbeek, A.; Pigmans, B.; Vredenbregt-van den Berg, M.; Dits, N.; Nielsen, S.J.; Baker, A.; Visakorpi, T.; Bangma, C.; et al. C/D-Box snoRNA-Derived RNA Production Is Associated with Malignant Transformation and Metastatic Progression in Prostate Cancer. Oncotarget 2015, 6, 17430–17444. [Google Scholar] [CrossRef]
- Wu, W.; Chen, X.; Liu, X.; Bao, H.-J.; Li, Q.-H.; Xian, J.-Y.; Lu, B.-F.; Zhao, Y.; Chen, S. SNORD60 Promotes the Tumorigenesis and Progression of Endometrial Cancer through Binding PIK3CA and Regulating PI3K/AKT/mTOR Signaling Pathway. Mol Carcinog 2023, 62, 413–426. [Google Scholar] [CrossRef]
- Hu, X.; Cui, W.; Liu, M.; Zhang, F.; Zhao, Y.; Zhang, M.; Yin, Y.; Li, Y.; Che, Y.; Zhu, X.; et al. SnoRNAs: The Promising Targets for Anti-Tumor Therapy. J Pharm Anal 2024, 14, 101064. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Feng, C.; Wang, S.; Shi, L.; Gu, Q.; Zhang, H.; Lan, X.; Zhao, Y.; Qiang, W.; Ji, M.; et al. The Noncoding RNAs SNORD50A and SNORD50B-Mediated TRIM21-GMPS Interaction Promotes the Growth of P53 Wild-Type Breast Cancers by Degrading P53. Cell Death Differ 2021, 28, 2450–2464. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Feng, X.; Liu, M.; Gong, H.; Zhou, X. SnoRNA and lncSNHG: Advances of Nucleolar Small RNA Host Gene Transcripts in Anti-Tumor Immunity. Front Immunol 2023, 14, 1143980. [Google Scholar] [CrossRef] [PubMed]
- Shav-Tal, Y.; Blechman, J.; Darzacq, X.; Montagna, C.; Dye, B.T.; Patton, J.G.; Singer, R.H.; Zipori, D. Dynamic Sorting of Nuclear Components into Distinct Nucleolar Caps during Transcriptional Inhibition. Mol Biol Cell 2005, 16, 2395–2413. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Bae, B.; Schnabl, S.; Yuan, F.; De Zoysa, T.; Akinyi, M.V.; Le Roux, C.A.; Choquet, K.; Whipple, A.J.; Van Nostrand, E.L. Mapping snoRNA-Target RNA Interactions in an RNA-Binding Protein-Dependent Manner with Chimeric eCLIP. Genome Biol 2025, 26, 39. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.-J.; Chen, X.; Liu, X.; Wu, W.; Li, Q.-H.; Xian, J.-Y.; Zhao, Y.; Chen, S. Box C/D snoRNA SNORD89 Influences the Occurrence and Development of Endometrial Cancer through 2’-O-Methylation Modification of Bim. Cell Death Discov 2022, 8, 309. [Google Scholar] [CrossRef]
- Li, X.; Zhao, X.; Xie, L.; Song, X.; Song, X. Identification of Four snoRNAs (SNORD16, SNORA73B, SCARNA4, and SNORD49B) as Novel Non-Invasive Biomarkers for Diagnosis of Breast Cancer. Cancer Cell Int 2024, 24, 55. [Google Scholar] [CrossRef]
- Cao, F.; You, Q.; Zhu, F.; Zhang, Y. Serum Exosomal Small Nucleolar RNA (snoRNA) Signatures as a Predictive Biomarker for Benign and Malignant Pulmonary Nodules. Cancer Cell Int 2024, 24, 341. [Google Scholar] [CrossRef]
- Gómez-Matas, J.; Duran-Sanchon, S.; Lozano, J.-J.; Ferrero, G.; Tarallo, S.; Pardini, B.; Naccarati, A.; Castells, A.; Gironella, M. SnoRNA Profiling in Colorectal Cancer and Assessment of Non-Invasive Biomarker Capacity by ddPCR in Fecal Samples. iScience 2024, 27, 109283. [Google Scholar] [CrossRef]
- Zhang, Y.; Shang, X.; Yu, M.; Bi, Z.; Wang, K.; Zhang, Q.; Xie, L.; Song, X.; Song, X. A Three-snoRNA Signature: SNORD15A, SNORD35B and SNORD60 as Novel Biomarker for Renal Cell Carcinoma. Cancer Cell Int 2023, 23, 136. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, X.; Zhang, W.; Deng, J.; Lin, C.; Qi, Z.; Li, Y.; Gu, Y.; Wang, Q.; Shen, L.; et al. Oncogene SCARNA12 as a Potential Diagnostic Biomarker for Colorectal Cancer. Mol Biomed 2023, 4, 37. [Google Scholar] [CrossRef]
- Duan, C.; Abola, Y.; Zhao, J.; Wang, Y. Small Nucleolar RNAs in Head and Neck Squamous Cell Carcinomas. J Dent Res 2025, 104, 5–16. [Google Scholar] [CrossRef]
- Yang, H.; Lin, P.; Wu, H.-Y.; Li, H.-Y.; He, Y.; Dang, Y.-W.; Chen, G. Genomic Analysis of Small Nucleolar RNAs Identifies Distinct Molecular and Prognostic Signature in Hepatocellular Carcinoma. Oncol Rep 2018, 40, 3346–3358. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, P.; Ghosh, S.; Wang, B.; Heyns, M.; Graham, K.; Mackey, J.R.; Kovalchuk, O.; Damaraju, S. Profiling of Small Nucleolar RNAs by Next Generation Sequencing: Potential New Players for Breast Cancer Prognosis. PLoS One 2016, 11, e0162622. [Google Scholar] [CrossRef]
- Valleron, W.; Laprevotte, E.; Gautier, E.-F.; Quelen, C.; Demur, C.; Delabesse, E.; Agirre, X.; Prósper, F.; Kiss, T.; Brousset, P. Specific Small Nucleolar RNA Expression Profiles in Acute Leukemia. Leukemia 2012, 26, 2052–2060. [Google Scholar] [CrossRef]
- Ronchetti, D.; Mosca, L.; Cutrona, G.; Tuana, G.; Gentile, M.; Fabris, S.; Agnelli, L.; Ciceri, G.; Matis, S.; Massucco, C.; et al. Small Nucleolar RNAs as New Biomarkers in Chronic Lymphocytic Leukemia. BMC Med Genomics 2013, 6, 27. [Google Scholar] [CrossRef]
- Valleron, W.; Ysebaert, L.; Berquet, L.; Fataccioli, V.; Quelen, C.; Martin, A.; Parrens, M.; Lamant, L.; de Leval, L.; Gisselbrecht, C.; et al. Small Nucleolar RNA Expression Profiling Identifies Potential Prognostic Markers in Peripheral T-Cell Lymphoma. Blood 2012, 120, 3997–4005. [Google Scholar] [CrossRef]
- Gao, L.; Ma, J.; Mannoor, K.; Guarnera, M.A.; Shetty, A.; Zhan, M.; Xing, L.; Stass, S.A.; Jiang, F. Genome-Wide Small Nucleolar RNA Expression Analysis of Lung Cancer by next-Generation Deep Sequencing. Int J Cancer 2015, 136, E623–629. [Google Scholar] [CrossRef]
- Mannoor, K.; Shen, J.; Liao, J.; Liu, Z.; Jiang, F. Small Nucleolar RNA Signatures of Lung Tumor-Initiating Cells. Mol Cancer 2014, 13, 104. [Google Scholar] [CrossRef]
- Okugawa, Y.; Toiyama, Y.; Toden, S.; Mitoma, H.; Nagasaka, T.; Tanaka, K.; Inoue, Y.; Kusunoki, M.; Boland, C.R.; Goel, A. Clinical Significance of SNORA42 as an Oncogene and a Prognostic Biomarker in Colorectal Cancer. Gut 2017, 66, 107–117. [Google Scholar] [CrossRef]
- Pan, X.; Chen, L.; Feng, K.-Y.; Hu, X.-H.; Zhang, Y.-H.; Kong, X.-Y.; Huang, T.; Cai, Y.-D. Analysis of Expression Pattern of snoRNAs in Different Cancer Types with Machine Learning Algorithms. Int J Mol Sci 2019, 20, 2185. [Google Scholar] [CrossRef] [PubMed]
- Kehr, S.; Bartschat, S.; Stadler, P.F.; Tafer, H. PLEXY: Efficient Target Prediction for Box C/D snoRNAs. Bioinformatics 2011, 27, 279–280. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Steitz, J.A. Position within the Host Intron Is Critical for Efficient Processing of Box C/D snoRNAs in Mammalian Cells. Proc Natl Acad Sci U S A 2001, 98, 12914–12919. [Google Scholar] [CrossRef]
- Filippova, J.A.; Matveeva, A.M.; Zhuravlev, E.S.; Balakhonova, E.A.; Prokhorova, D.V.; Malanin, S.J.; Shah Mahmud, R.; Grigoryeva, T.V.; Anufrieva, K.S.; Semenov, D.V.; et al. Are Small Nucleolar RNAs “CRISPRable”? A Report on Box C/D Small Nucleolar RNA Editing in Human Cells. Front Pharmacol 2019, 10, 1246. [Google Scholar] [CrossRef]
- Ferreira, H.J.; Heyn, H.; Moutinho, C.; Esteller, M. CpG Island Hypermethylation-Associated Silencing of Small Nucleolar RNAs in Human Cancer. RNA Biol 2012, 9, 881–890. [Google Scholar] [CrossRef]
- Wang, B.; Zhao, Y.; Li, Y.; Xu, Y.; Chen, Y.; Jiang, Q.; Yao, D.; Zhang, L.; Hu, X.; Fu, C.; et al. A Plasma SNORD33 Signature Predicts Platinum Benefit in Metastatic Triple-Negative Breast Cancer Patients. Mol Cancer 2022, 21, 22. [Google Scholar] [CrossRef]
- Bergeron, D.; Paraqindes, H.; Fafard-Couture, É.; Deschamps-Francoeur, G.; Faucher-Giguère, L.; Bouchard-Bourelle, P.; Abou Elela, S.; Catez, F.; Marcel, V.; Scott, M.S. snoDB 2.0: An Enhanced Interactive Database, Specializing in Human snoRNAs. Nucleic Acids Res 2023, 51, D291–D296. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).