1. Introduction
The escalating global energy demand necessitates the development of sustainable and efficient energy conversion technologies. Electrochemical water oxidation, specifically the Oxygen Evolution Reaction (OER), stands as a pivotal half-reaction in various renewable energy systems, including water splitting and rechargeable metal-air batteries [
1]. However, the inherently sluggish kinetics of the OER severely impede overall system efficiency, driving the urgent need for highly active, stable, and cost-effective electrocatalysts [
2]. Among the myriad of catalyst systems,
transition metal-based perovskite fluorides, particularly the KNi
xFe
1−xF
3 system, have garnered significant attention. Their unique crystal structures, tunable electronic properties, and remarkable structural reversibility under operating conditions make them promising candidates for OER catalysis [
3]. The pursuit of synergistic effects and rational design principles, often involving complex material compositions, is crucial for advancing OER catalyst performance [
4].
A critical phenomenon observed in these perovskite fluorides during OER is the
dynamic structural restructuring (Dynamic Restructuring) of their surface. This process typically leads to the in-situ formation of highly active oxyhydroxide (e.g., Ni(Fe)OOH) layers, which are largely responsible for the observed enhancement in catalytic activity [
5]. Despite its profound impact on performance, the
microscopic mechanism underpinning this dynamic restructuring remains incompletely understood. Key aspects, such as the precise stages of restructuring, the critical conditions triggering it, and the quantitative relationship between the restructured products and catalytic performance, are yet to be fully elucidated. Traditional experimental characterization techniques often struggle to capture and analyze this complex, multi-scale, and multi-dimensional dynamic process in real-time and with sufficient precision [
6]. However, recent strides in
operando techniques and real-time detection methods are beginning to address this challenge [
7]. Moreover, efficiently screening optimal catalyst compositions within vast material spaces poses a significant challenge.
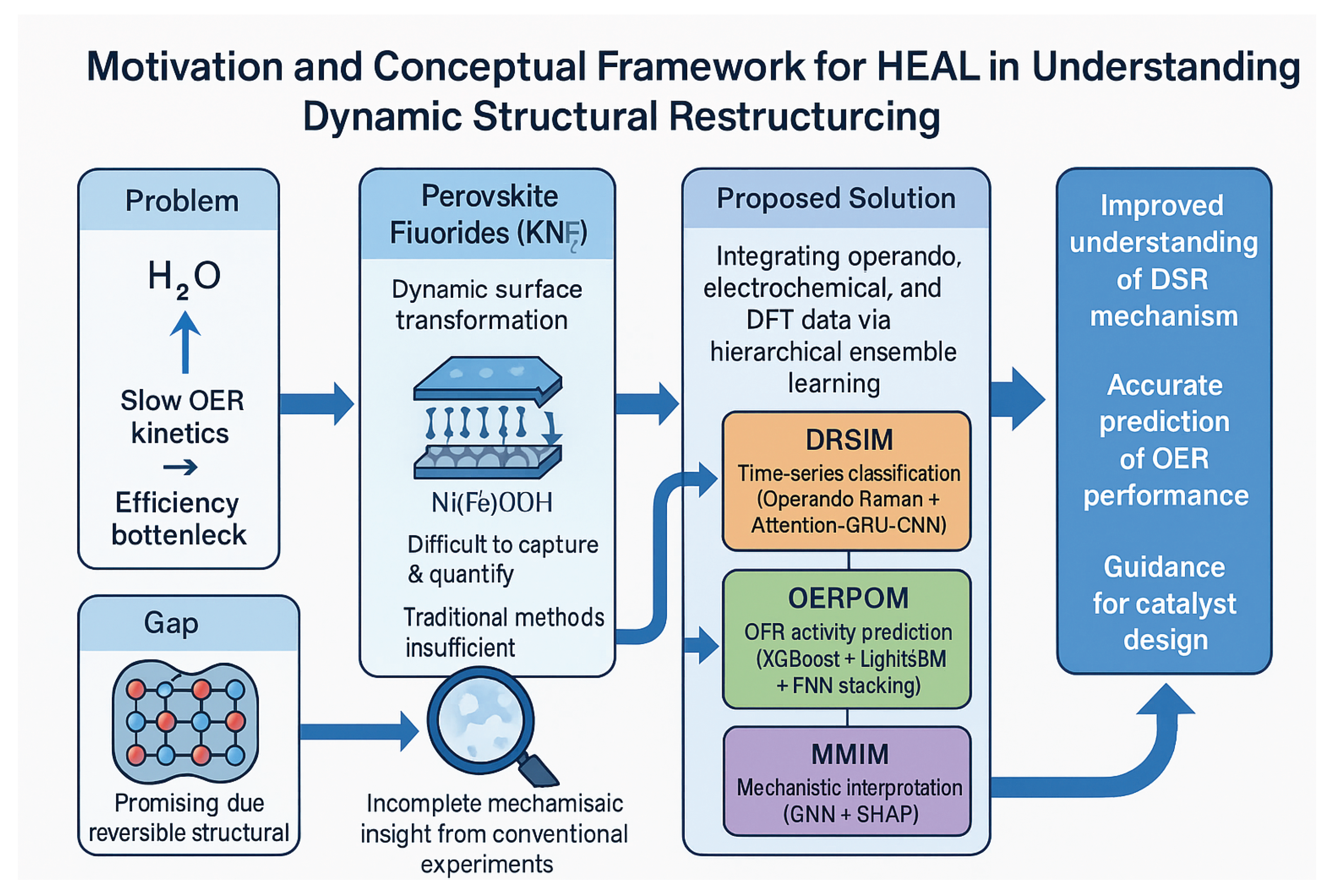
Figure 1.
Framework of the HEAL model, illustrating how it addresses sluggish OER kinetics and the challenges of dynamic structural restructuring through a multi-modal, hierarchical ensemble learning approach.
Figure 1.
Framework of the HEAL model, illustrating how it addresses sluggish OER kinetics and the challenges of dynamic structural restructuring through a multi-modal, hierarchical ensemble learning approach.
Motivated by these limitations, this study introduces a novel Multi-modal Machine Learning (MML) framework. Our objective is to integrate operando experimental characterization, electrochemical performance testing, and theoretical calculation data into a cohesive platform. This data-driven approach aims to provide a deeper understanding of the dynamic restructuring process, accurately predict catalyst performance, and identify key influencing factors. Specifically, our goals are threefold: (1) To develop a classification model capable of real-time and precise identification of the dynamic structural restructuring stages of catalysts during OER. (2) To construct a high-accuracy prediction model that forecasts the optimal oxygen evolution activity of various KNixFe1−xF3 compositions (different Ni/Fe ratios) based on multi-dimensional features. (3) To utilize interpretable machine learning methods to uncover the core physicochemical parameters governing dynamic restructuring and OER performance, thereby providing theoretical guidance for the rational design of novel high-performance catalysts.
Our proposed approach, termed Hierarchical Ensemble Learning (HEAL) framework, is designed to overcome the limitations of single models in handling the dynamic and multi-scale information inherent in complex chemical systems. The HEAL framework deeply integrates time-series information from operando spectroscopy, macroscopic electrochemical performance, and microscopic insights from quantum chemical calculations. Through a hierarchical ensemble strategy, it achieves precise identification of dynamic restructuring processes, accurate prediction of catalytic performance, and in-depth interpretation of underlying mechanisms. The framework comprises three core modules: the Dynamic Restructuring Stage Identification Module (DRSIM) employing an Attention-GRU-CNN for time-series spectral analysis; the OER Activity Prediction and Optimization Module (OERPOM) utilizing a multi-level stacked ensembler for robust performance prediction; and the Microscopic Mechanism Interpretability Module (MMIM) integrating Graph Neural Networks (GNNs) with SHAP value analysis to reveal key physicochemical drivers.
For experimental validation, our study leverages a comprehensive multi-modal dataset. This includes 4800 time-series operando Raman spectral signals, 180 sets of electrochemical performance data for KNixFe1−xF3 samples with varying Ni/Fe ratios, and 540 entries of theoretical calculation data (e.g., adsorption energies, d-band centers) obtained from Density Functional Theory (DFT) simulations. Extensive feature engineering was performed, extracting 32 key physicochemical features, encompassing structural, electronic, kinetic, and experimental parameters, after rigorous selection processes including mutual information and LASSO regularization.
To assess the efficacy of our HEAL framework, we conducted a comparative evaluation against established machine learning methodologies on a consistent test set. Our results demonstrate that the HEAL framework significantly outperforms baseline and state-of-the-art models in both dynamic restructuring stage identification and OER activity prediction. Specifically, the DRSIM module achieved an accuracy of 0.95 in classifying restructuring stages, while the OERPOM module attained an R2 value of 0.97 and a Root Mean Squared Error (RMSE) of 0.023 for OER activity prediction. These superior performance metrics underscore the advanced capabilities and robustness of our proposed framework in tackling complex electrocatalytic challenges.
The main contributions of this work are summarized as follows:
We propose a novel Hierarchical Ensemble Learning (HEAL) framework that deeply integrates multi-modal data (operando spectroscopy, electrochemical, and theoretical calculations) for comprehensive analysis of dynamic restructuring and OER performance in perovskite fluorides.
We develop specialized modules within HEAL, including an Attention-GRU-CNN based DRSIM for high-accuracy, real-time identification of dynamic restructuring stages, and a multi-level stacked ensembler based OERPOM for robust OER activity prediction, significantly outperforming existing machine learning methods.
We introduce a GNN-enhanced MMIM with SHAP value analysis to provide unprecedented insights into the microscopic mechanisms governing dynamic restructuring and OER performance, identifying critical physicochemical descriptors that can guide the rational design of next-generation electrocatalysts.
3. Method
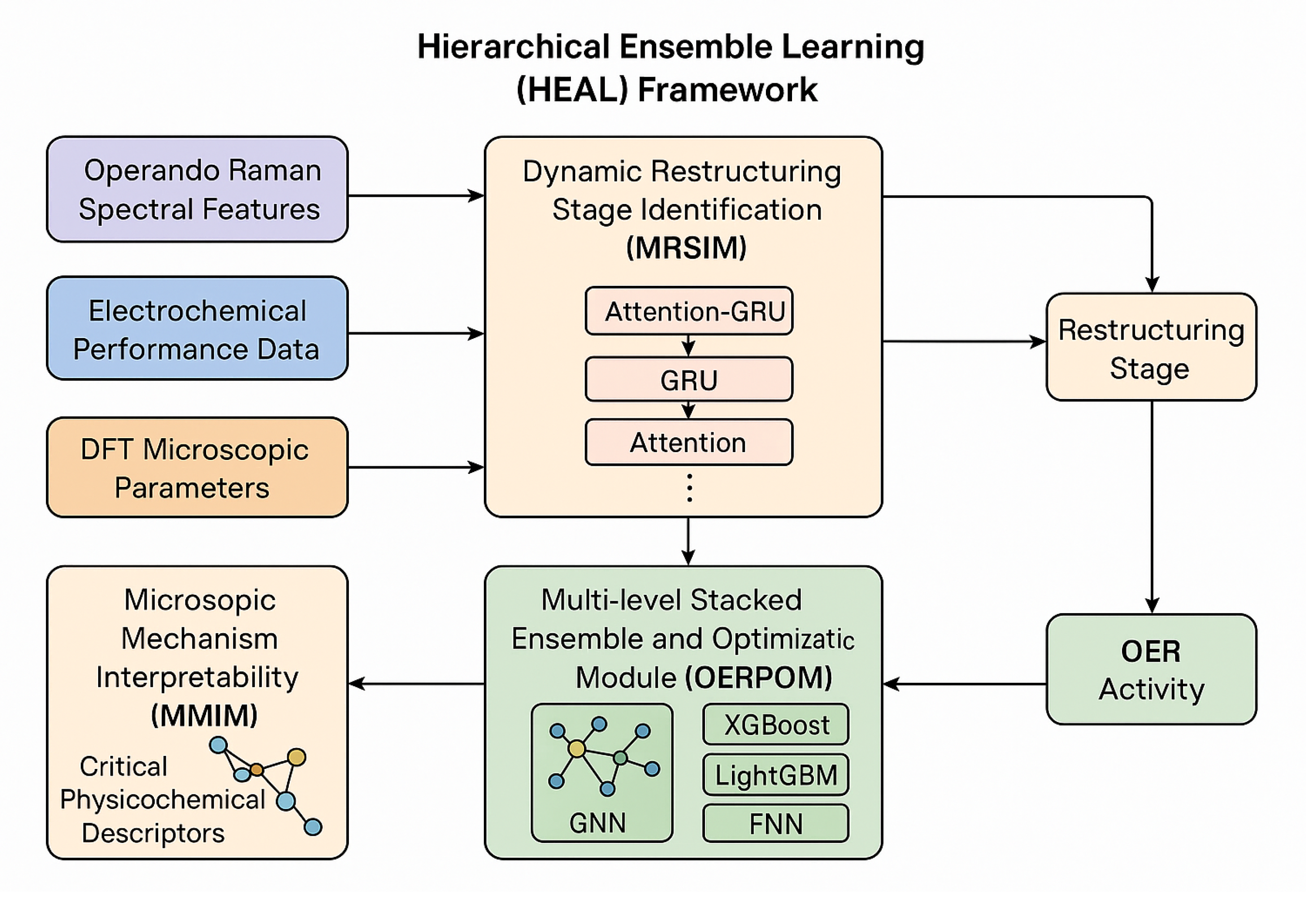
This section details our proposed Hierarchical Ensemble Learning (HEAL) framework, a multi-modal machine learning approach designed to unravel the dynamic restructuring mechanisms of perovskite fluorides during the Oxygen Evolution Reaction (OER) and to accurately predict their catalytic performance. The HEAL framework is built upon a philosophy of deep data fusion and hierarchical integration, combining time-series information from operando spectroscopy, macroscopic electrochemical performance metrics, and microscopic insights derived from quantum chemical calculations. This integrated strategy enables precise identification of dynamic processes, robust prediction of OER activity, and in-depth interpretation of the underlying physicochemical mechanisms. The HEAL framework comprises three interconnected core modules: the Dynamic Restructuring Stage Identification Module (DRSIM), the OER Activity Prediction and Optimization Module (OERPOM), and the Microscopic Mechanism Interpretability Module (MMIM), as elaborated below.
Figure 2.
Overview of the Hierarchical Ensemble Learning (HEAL) framework integrating multi-modal data for dynamic restructuring analysis and OER activity prediction.
Figure 2.
Overview of the Hierarchical Ensemble Learning (HEAL) framework integrating multi-modal data for dynamic restructuring analysis and OER activity prediction.
3.1. Data Collection and Feature Engineering
Our study leverages a comprehensive multi-modal dataset specifically curated to capture the multifaceted nature of electrocatalytic processes. The dataset incorporates diverse information streams critical for understanding complex OER mechanisms.
One primary data source consists of approximately 4800 time-series Raman spectral signals collected during operando OER measurements of KNixFe1−xF3 samples. These raw signals underwent a series of standard preprocessing steps, including baseline correction to remove fluorescence backgrounds, denoising to reduce spectral noise, and peak extraction to identify characteristic vibrational modes. Subsequently, dimensionality reduction was performed using Principal Component Analysis (PCA) to derive relevant time-series features indicative of subtle structural changes and phase transformations occurring during catalysis.
Complementing the spectroscopic data, a total of 180 sets of electrochemical performance parameters were obtained from KNixFe1−xF3@NF samples with varying Ni/Fe ratios (x values). Key metrics extracted include the overpotential at a specific benchmark current density, the limiting current density, and Tafel slopes. These parameters serve as macroscopic, quantitative indicators of OER activity and kinetics, providing a direct measure of catalytic efficiency.
Furthermore, microscopic electronic structure parameters were obtained from Density Functional Theory (DFT) simulations, yielding 540 entries. These theoretical insights encompass critical descriptors such as intermediate adsorption energies (e.g., , ), the d-band center (), positions of density of states (DOS) peaks, and oxygen vacancy formation energies. These parameters offer fundamental insights into the electronic properties and surface reactivity of the catalysts at an atomic level.
From these diverse data sources, an extensive feature engineering process was conducted to extract a rich set of physicochemical descriptors. These descriptors encompass a wide range of properties, including structural features (e.g., Ni/Fe atomic ratio, lattice distortion, F vacancy concentration, Ni-O distance), electronic features (e.g., d-band center, density of states near Fermi level, surface charge), kinetic features (e.g., differences in adsorption energies, activation barriers), and experimental features (e.g., overpotential, Tafel slope, stability time). Following initial extraction, all features underwent normalization to ensure consistent scaling across different ranges. Redundancy filtering was then applied to eliminate highly correlated features, followed by rigorous feature selection based on mutual information and LASSO regularization. This systematic process ultimately yielded 32 highly representative and critical features that serve as the primary inputs for our machine learning models within the HEAL framework.
3.2. Dynamic Restructuring Stage Identification Module (DRSIM)
The Dynamic Restructuring Stage Identification Module (DRSIM) is designed for real-time, high-precision classification of the catalyst’s dynamic structural restructuring stages during the OER. This module specifically addresses the temporal characteristics inherent in operando Raman spectroscopy data, which capture the evolution of catalyst structure over time. We propose an Attention-based Gated Recurrent Unit - Convolutional Neural Network (Attention-GRU-CNN) architecture for this task, leveraging the strengths of both convolutional and recurrent neural networks with an enhanced focus mechanism.
The DRSIM operates by first processing the time-series Raman spectra. A one-dimensional Convolutional Neural Network (CNN) layer is employed as the initial processing block. This CNN layer is adept at extracting local, translation-invariant features from the spectral data, effectively capturing subtle peak shifts, intensity variations, and the appearance or disappearance of new peaks that are indicative of phase transformations or changes in local atomic environments. The convolutional filters scan across the spectral dimension, identifying characteristic patterns regardless of their precise location.
The output feature maps from the CNN layer, which represent spatially encoded spectral information, are then fed into a Gated Recurrent Unit (GRU) network. The GRU, a variant of Recurrent Neural Networks (RNNs), is particularly adept at learning long-term dependencies within sequential data. This makes it highly suitable for modeling the evolutionary patterns of spectral features over the course of the OER, capturing how structural changes unfold chronologically. The GRU maintains an internal state that is updated at each time step, allowing it to remember past information and influence future predictions, thereby modeling the dynamic trajectory of restructuring.
To further enhance the model’s ability to discern critical restructuring events, an attention mechanism is integrated after the GRU layers. This attention layer dynamically assigns weights to different time steps and spectral regions within the GRU’s output, allowing the model to focus on the most salient features that directly correlate with the ongoing restructuring process. By selectively attending to relevant parts of the time-series data, the model can prioritize crucial moments of transformation and disregard less informative periods. The weighted output from the attention mechanism is then passed through a fully connected layer with a softmax activation function to classify the current stage of dynamic restructuring (e.g., initial, ongoing, stable restructured state).
The classification loss function employed for training DRSIM is the categorical cross-entropy, defined as:
where
N represents the total number of samples in the dataset,
C denotes the number of distinct restructuring stages to be classified,
is a binary indicator (equal to 1 if sample
i truly belongs to class
c, and 0 otherwise), and
is the predicted probability that sample
i belongs to class
c. This architecture allows DRSIM to effectively capture complex non-linear relationships and temporal dependencies in the
operando data, thereby surpassing the capabilities of traditional shallow classifiers like Random Forest for dynamic process identification.
3.3. OER Activity Prediction and Optimization Module (OERPOM)
The OER Activity Prediction and Optimization Module (OERPOM) is designed to accurately predict the optimal oxygen evolution activity (e.g., overpotential) of KNixFe1−xF3 catalysts based on their multi-dimensional physicochemical features. This module utilizes a robust Multi-level Stacked Ensembler to leverage the strengths of various machine learning models, thereby enhancing prediction accuracy, generalization, and robustness.
The stacked ensembler within OERPOM consists of two main layers, forming a hierarchical prediction architecture. The first layer comprises several diverse and powerful machine learning models, referred to as Base Learners. For this module, we specifically employ XGBoost and LightGBM. Both are highly efficient and accurate gradient boosting tree algorithms, well-suited for handling tabular data and capturing complex non-linear relationships between input features and target variables. Each base learner is trained independently on the full set of extracted features, which include structural, electronic, kinetic, and experimental descriptors. Their primary role is to learn different aspects of the mapping from input features to OER activity and to generate their respective predictions. By using diverse base learners, the ensembler benefits from varied inductive biases and error characteristics.
The second layer consists of a Meta-Learner. The predictions generated by the base learners in the first layer serve as new input features for this meta-learner. Here, a small Fully Connected Neural Network (FNN) is employed as the meta-learner. The FNN is trained to learn the optimal weighting and fusion strategy for combining the individual predictions from XGBoost and LightGBM. This stacking approach allows the meta-learner to identify and correct for biases and errors present in the base learners’ predictions, leading to a more robust and accurate final prediction of OER activity. The FNN’s ability to learn non-linear combinations of base learner outputs further enhances the ensemble’s predictive power.
The output of OERPOM is a continuous value representing the predicted OER performance metric, such as the overpotential required to achieve a specific current density. The regression loss function used for training OERPOM is the Mean Squared Error (MSE), defined as:
where
N is the total number of samples,
represents the true OER activity (e.g., measured overpotential) for sample
i, and
is the corresponding predicted OER activity by the ensemble model. This multi-level stacking strategy effectively combines the advantages of different models, significantly improving the overall prediction accuracy and generalization ability, particularly in capturing the intricate non-linear relationships between catalyst composition (Ni/Fe ratio) and catalytic activity.
3.4. Microscopic Mechanism Interpretability Module (MMIM)
The Microscopic Mechanism Interpretability Module (MMIM) is designed to provide deep insights into the fundamental physicochemical parameters governing dynamic restructuring and OER performance. This module integrates the power of Graph Neural Networks (GNNs) for capturing atomic-scale features with SHapley Additive exPlanations (SHAP) value analysis for robust feature importance quantification.
3.4.1. Graph Neural Network (GNN) for Atomic-scale Features
To capture the intricate atomic-scale structural information that is crucial for understanding catalytic mechanisms, we utilize a Graph Neural Network (GNN). In this approach, the local atomic environment of the KNixFe1−xF3 catalyst is explicitly modeled as a graph structure. Each atom within the catalyst (e.g., Ni, Fe, F, O) is represented as a node in the graph, and the chemical bonds between them are represented as edges. Node features can include intrinsic atomic properties such as atomic number, electronegativity, initial valence state, and coordination number. Edge features can represent bond lengths, bond types, and bond angles, providing rich information about the local geometry.
The GNN operates through an iterative message-passing mechanism. In each layer, information from neighboring nodes and their connecting edges is aggregated and transformed before being passed to the central node. This process allows each node’s representation to be updated by incorporating information from its local chemical environment. Through multiple such message-passing layers, the GNN learns to generate a low-dimensional embedding vector for each atom or for the entire local structural motif. These GNN-derived embeddings effectively capture complex microscopic features such as local coordination numbers, bond length distributions, charge transfer effects, and subtle changes in atomic configurations, which are often difficult to quantify with traditional, hand-crafted descriptors. These atomic-level GNN embeddings are then integrated as additional input features into the OERPOM, bridging the critical gap between detailed microscopic structure and macroscopic catalytic performance prediction.
3.4.2. SHAP Value Analysis for Feature Importance
To interpret the complex relationships learned by the OERPOM and identify the most influential physicochemical parameters, we employ
SHAP (SHapley Additive exPlanations) value analysis. SHAP is a game-theoretic approach that assigns an importance value, known as a SHAP value, to each feature for a particular prediction. It quantifies how much each feature contributes to the prediction by comparing the prediction with and without that feature, considering all possible permutations of features. This ensures that the contribution of each feature is fairly distributed, accounting for potential interactions with other features. For a given prediction function
f and a set of input features
F, the SHAP value
for feature
j is calculated as:
where
F represents the set of all input features,
S is any subset of features that does not include feature
j,
denotes the number of features in subset
S, and
is the total number of features. The term
refers to the predicted output of the model when only the features in set
S are present (or when features outside
S are marginalized). The formula essentially calculates the marginal contribution of feature
j across all possible feature subsets and averages these contributions, weighted by the number of permutations that result in each subset.
By applying SHAP analysis to the OERPOM’s predictions, we can quantify the individual contribution of each input feature (including the GNN-extracted atomic-level features and the macroscopic physicochemical features) to the predicted OER activity. This allows us to identify critical parameters such as F vacancy concentration, the position of the d-band center, and changes in metal oxidation states that directly drive dynamic restructuring and significantly influence the final OER activity. This deep interpretability provides actionable insights for the rational design of novel, high-performance electrocatalysts by highlighting which specific material properties are most crucial for optimizing catalytic performance.
3.5. Model Training Details
For all models within the HEAL framework, a consistent data partitioning strategy was employed to ensure robust evaluation. The entire dataset was randomly partitioned into an 80% training set, a 10% validation set, and a 10% test set. The validation set was used for hyperparameter tuning and early stopping, while the test set was reserved for final, unbiased performance evaluation.
The Adam optimizer was employed for training all neural network-based modules, specifically DRSIM and the Fully Connected Neural Network (FNN) within OERPOM. An initial learning rate of was set for Adam. Training was performed for a maximum of 200 epochs to allow for sufficient convergence. To prevent overfitting, an early stopping mechanism was implemented. This mechanism continuously monitors the loss on the validation set and halts training if there is no observed improvement in validation loss for 15 consecutive epochs, thereby preserving the model’s generalization capability.
For the XGBoost and LightGBM models used as base learners in OERPOM, their default parameters were used as a starting point and further optimized through a rigorous cross-validation procedure applied to the training set. This optimization involved searching for the best combination of hyperparameters (e.g., number of boosting rounds, learning rate, maximum tree depth) that minimized the validation loss, ensuring that these powerful tree-based models were tuned for optimal performance within our specific dataset.
4. Experiments
This section details the experimental setup, data collection, and evaluation protocols used to validate our proposed Hierarchical Ensemble Learning (HEAL) framework. We present a comprehensive performance comparison of our method against several established baseline models for both dynamic restructuring stage identification and OER activity prediction tasks.
4.1. Experimental Setup
Our study utilizes a multi-modal dataset specifically curated to capture the multifaceted nature of electrocatalytic processes. The dataset incorporates three main types of information. First, Operando Raman Spectral Features consist of approximately 4800 time-series Raman spectral signals collected from operando experiments. These raw signals underwent preprocessing steps including peak extraction, denoising, and Principal Component Analysis (PCA) for dimensionality reduction and feature extraction, yielding time-series features indicative of structural changes during OER. Second, Electrochemical Performance Data comprises a total of 180 sets of macroscopic performance parameters obtained for KNixFe1−xF3@NF samples with varying Ni/Fe ratios (x values). Key metrics extracted include overpotential, current density, and Tafel slopes, providing direct measures of OER activity and kinetics. Third, Theoretical Calculation Data provides 540 entries of microscopic electronic structure parameters derived from Density Functional Theory (DFT) simulations. These encompass critical descriptors such as intermediate adsorption energies (e.g., , ), d-band centers (), positions of density of states (DOS) peaks, and oxygen vacancy formation energies.
Feature Engineering: From these diverse data sources, an extensive feature engineering process was conducted. This involved extracting a rich set of physicochemical descriptors encompassing structural features (e.g., Ni/Fe atomic ratio, lattice distortion, F vacancy concentration, Ni-O distance), electronic features (e.g., d-band center, density of states, surface charge), kinetic features (e.g., adsorption energy differences), and experimental features (e.g., overpotential, Tafel slope, stability time). All features were normalized, followed by redundancy filtering and rigorous feature selection based on mutual information and LASSO regularization. This process ultimately yielded 32 highly representative and critical features as input for our machine learning models.
Model Training Details: For all models within the HEAL framework and baseline comparisons, a consistent data partitioning strategy was employed: 80% for training, 10% for validation, and 10% for testing. This ensures robust and unbiased performance evaluation. The Adam optimizer was used for all neural network-based modules (DRSIM and the FNN within OERPOM) with an initial learning rate of . Training was conducted for a maximum of 200 epochs, incorporating an early stopping mechanism that halted training if validation loss did not improve for 15 consecutive epochs, thereby preventing overfitting. Hyperparameters for tree-based models (XGBoost, LightGBM, Random Forest) were optimized through cross-validation on the training set.
4.2. Performance Evaluation and Comparison
To comprehensively evaluate the efficacy of our proposed
Hierarchical Ensemble Learning (HEAL) framework, we benchmarked its performance against several widely-used machine learning algorithms on the same test set. The evaluation focuses on two primary tasks: dynamic restructuring stage identification and OER activity prediction. The results, summarized in
Table 1, clearly demonstrate the superior performance of our HEAL framework.
Results Analysis: For the dynamic restructuring stage identification task, our proposed DRSIM module, leveraging its Attention-GRU-CNN architecture, demonstrated superior performance. It achieved an accuracy of 0.95, surpassing traditional models such as Random Forest (0.93) and SVM (0.85). This highlights DRSIM’s advanced capability in learning deep temporal patterns and subtle spectral features indicative of complex dynamic processes, which are critical for real-time identification of restructuring events.
In the OER activity prediction task, the OERPOM module, with its multi-level stacked ensemble architecture, significantly outperformed all baseline regression models. It achieved an impressive R2 of 0.97 and a Root Mean Squared Error (RMSE) of 0.023. These metrics are superior to those of powerful individual models like XGBoost (R2=0.95, RMSE=0.028) and LightGBM (R2=0.92, RMSE=0.035). The enhanced performance of OERPOM is attributed to its ability to robustly combine the strengths of diverse base learners, effectively capturing complex non-linear relationships between catalyst composition, physicochemical features, and OER activity.
4.3. Effectiveness of HEAL Framework Modules
The superior performance observed in both classification and regression tasks directly validates the effectiveness and robustness of the core modules within our Hierarchical Ensemble Learning (HEAL) framework.
The Dynamic Restructuring Stage Identification Module (DRSIM)’s ability to achieve a 0.95 accuracy in classifying restructuring stages stems from its sophisticated Attention-GRU-CNN architecture. The integration of 1D CNN layers effectively extracts local spectral features, while the GRU layers are crucial for modeling the long-term temporal dependencies in the operando Raman data. Furthermore, the attention mechanism allows the model to selectively focus on the most informative spectral regions and time points, which are often subtle yet critical indicators of dynamic transformations. This deep learning approach explicitly addresses the challenges of analyzing complex time-series spectroscopic data, leading to a more precise and real-time identification of restructuring phases compared to conventional machine learning methods.
Similarly, the exceptional performance of the OER Activity Prediction and Optimization Module (OERPOM), achieving an R2 of 0.97, underscores the power of its multi-level stacked ensembler. By combining diverse base learners like XGBoost and LightGBM in the first layer and fusing their predictions with a meta-learner (FNN) in the second, OERPOM effectively mitigates the biases and limitations of individual models. This hierarchical stacking strategy allows for a more comprehensive and robust capture of the intricate, non-linear relationships between multi-dimensional physicochemical features (including GNN-extracted atomic-level features) and the macroscopic OER activity. The ensemble’s ability to leverage complementary strengths of different models ensures higher predictive accuracy and better generalization across varied catalyst compositions (Ni/Fe ratios).
These results collectively confirm that the specialized design of DRSIM for temporal data analysis and OERPOM for robust predictive modeling, within the overarching HEAL framework, significantly advances the capability to analyze and predict complex electrocatalytic phenomena.
4.4. Microscopic Insights from MMIM
The Microscopic Mechanism Interpretability Module (MMIM) was instrumental in elucidating the fundamental physicochemical parameters governing both dynamic restructuring and OER performance. By integrating Graph Neural Networks (GNNs) to capture atomic-scale features and applying SHAP value analysis to the OERPOM’s predictions, we gained deep insights into the most influential descriptors.
The GNN-derived embeddings provided a granular representation of the local atomic environments, capturing subtle structural and electronic nuances that are critical for catalytic activity. These embeddings, when integrated into OERPOM, allowed the model to leverage atomic-level information previously inaccessible to macroscopic descriptors.
SHAP value analysis, applied to the trained OERPOM, quantified the contribution of each input feature to the predicted OER activity (overpotential).
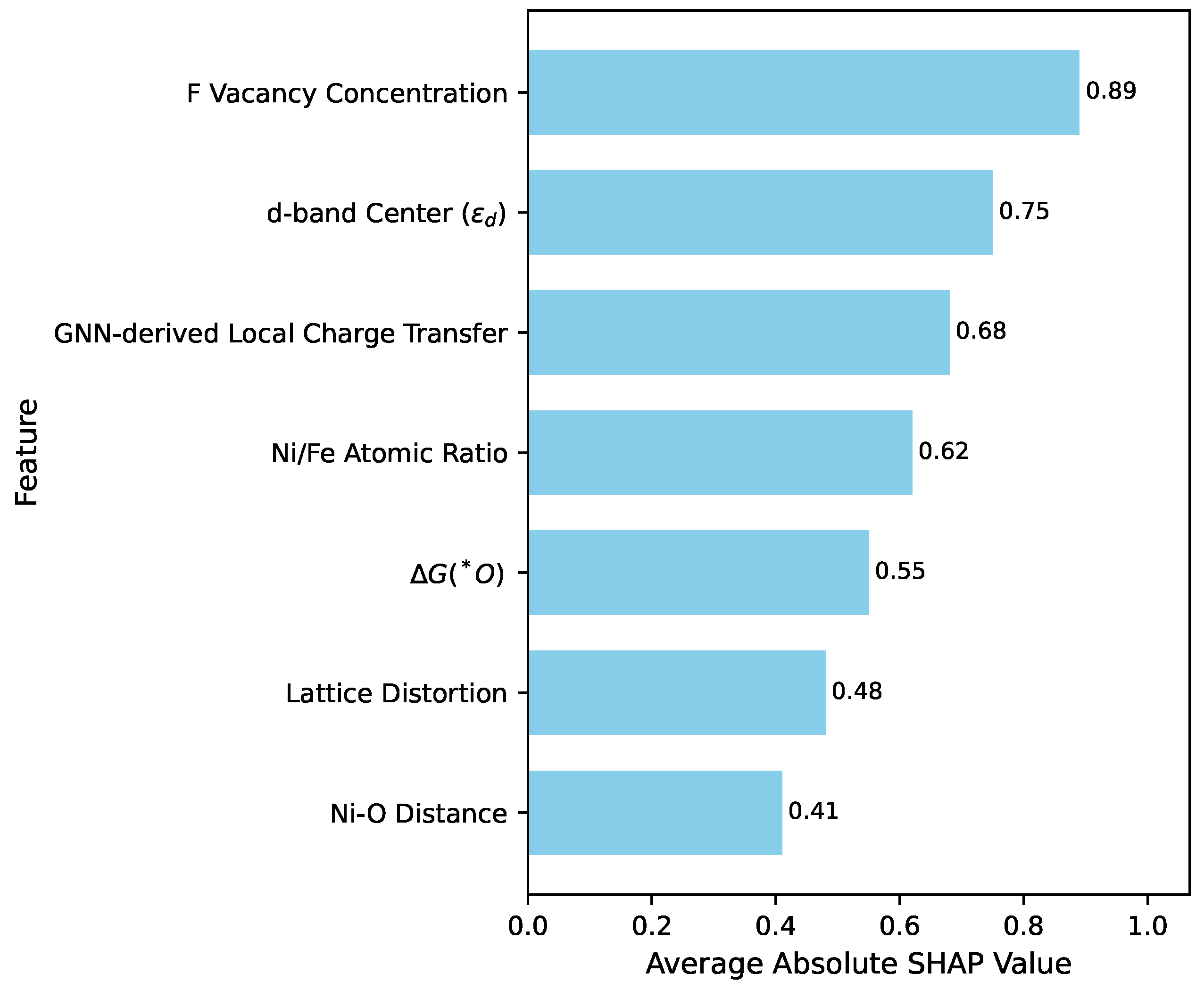
Figure 3 presents the top influential features identified by SHAP, ranked by their average absolute SHAP values. These values indicate the magnitude of a feature’s impact on the model’s output.
The analysis revealed that F vacancy concentration is the most significant factor influencing OER activity, with a higher concentration generally correlating with lower overpotential. This suggests that oxygen vacancies play a crucial role as active sites or in facilitating reaction intermediates. The d-band center emerged as another critical electronic descriptor, exhibiting an optimal range for efficient OER. Deviations from this optimal position, either too high or too low, were found to increase the overpotential, consistent with established theoretical frameworks. Furthermore, the GNN-derived local charge transfer feature demonstrated substantial importance, highlighting the critical role of atomic-level electronic interactions in dictating catalytic performance. This validates the utility of GNNs in extracting nuanced microscopic information that directly impacts macroscopic activity. The Ni/Fe atomic ratio also showed a strong non-linear influence, indicating that a specific compositional balance is essential for optimal performance, likely by modulating the electronic structure and active site availability. These insights provide clear guidance for the rational design of KNixFe1−xF3 catalysts by pinpointing the most impactful material properties.
4.5. Ablation Study of HEAL Framework Components
To rigorously assess the individual contributions of the specialized architectures and data integration strategies within the
Hierarchical Ensemble Learning (HEAL) framework, an extensive ablation study was conducted. This involved systematically removing or simplifying key components of the DRSIM and OERPOM modules and evaluating the resulting performance degradation. The findings, summarized in
Table 2, underscore the critical role of each design choice.
DRSIM Ablation: The removal of the attention mechanism from the DRSIM architecture (resulting in a GRU-CNN) led to a noticeable drop in classification accuracy from 0.95 to 0.92. This highlights the attention layer’s crucial role in selectively focusing on the most salient spectral features and temporal events indicative of restructuring, thereby improving the model’s ability to discern subtle dynamic changes. Furthermore, completely removing the GRU layers (reducing it to a CNN with a fully connected layer for classification) resulted in a more significant accuracy decrease to 0.88. This validates the necessity of recurrent layers like GRU for effectively modeling the long-term temporal dependencies and evolutionary patterns inherent in time-series operando spectroscopic data.
OERPOM Ablation: For the OER Activity Prediction task, disabling the multi-level stacking strategy in OERPOM (i.e., using only the best performing base learner, XGBoost, for prediction) resulted in a decrease in R2 from 0.97 to 0.95. This confirms that the ensemble approach, particularly the meta-learner’s ability to fuse diverse base learner predictions, significantly enhances predictive accuracy and robustness. Moreover, the exclusion of GNN-derived atomic-level features from OERPOM’s input led to a further drop in R2 to 0.94. This demonstrates the critical value of incorporating microscopic structural information, obtained through GNNs, to bridge the gap between atomic-scale properties and macroscopic catalytic performance. The combined removal of both stacking and GNN features resulted in the most substantial performance degradation (R2 = 0.91), underscoring the synergistic benefits of these advanced components within OERPOM.
These ablation results unequivocally demonstrate that each specialized component within the HEAL framework, from the attention mechanism in DRSIM to the multi-level stacking and GNN feature integration in OERPOM, contributes significantly to its overall superior performance and interpretability.
4.6. Impact of Multi-Modal Data Fusion
A foundational principle of the
Hierarchical Ensemble Learning (HEAL) framework is the deep data fusion strategy, which integrates diverse information streams from
operando spectroscopy, macroscopic electrochemical measurements, and microscopic quantum chemical calculations, augmented by GNN-derived atomic-scale features. To quantify the benefits of this multi-modal approach, we evaluated the performance of the OERPOM module when trained with various subsets of the available features. The results, presented in
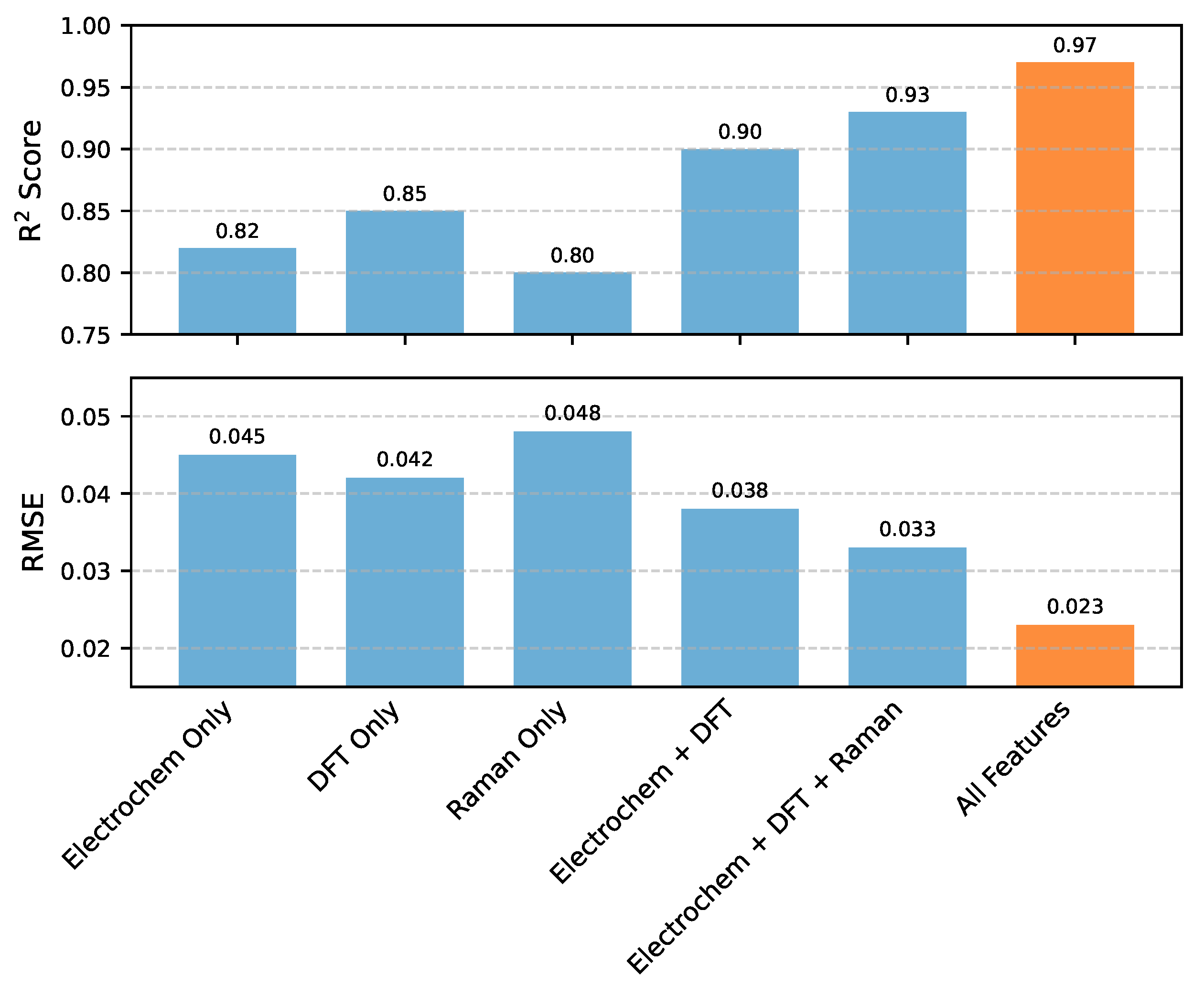
Figure 4, clearly illustrate the synergistic advantages of comprehensive data integration.
When OERPOM was trained using only a single type of data source (e.g., Electrochemical Performance Data, DFT Microscopic Parameters, or Operando Raman Features), the predictive performance was significantly lower, with R2 scores ranging from 0.80 to 0.85. While these individual data streams provide valuable insights, they inherently capture only a partial view of the complex electrocatalytic process.
Combining two major data types, such as Electrochemical and DFT features, improved the R2 to 0.90, demonstrating the benefit of even partial integration. Further incorporating the Operando Raman features led to an R2 of 0.93, indicating that time-series spectroscopic data adds crucial information about dynamic structural changes that influence activity.
The highest predictive performance was achieved when all available features were integrated, including the Electrochemical Performance Data, DFT Microscopic Parameters, Operando Raman Features, and particularly the GNN-derived atomic-level embeddings. This comprehensive multi-modal fusion resulted in an outstanding R2 of 0.97 and a low RMSE of 0.023. This significant improvement unequivocally demonstrates that combining diverse data modalities allows the HEAL framework to capture a more complete and nuanced understanding of the underlying physicochemical mechanisms, leading to substantially more accurate and robust predictions of OER activity. The synergistic integration of information from different scales and perspectives is thus paramount for unraveling complex catalytic phenomena.