Submitted:
17 October 2025
Posted:
20 October 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Gene Expression Classification Results
2.2. Pattern Analysis and Gene Functions
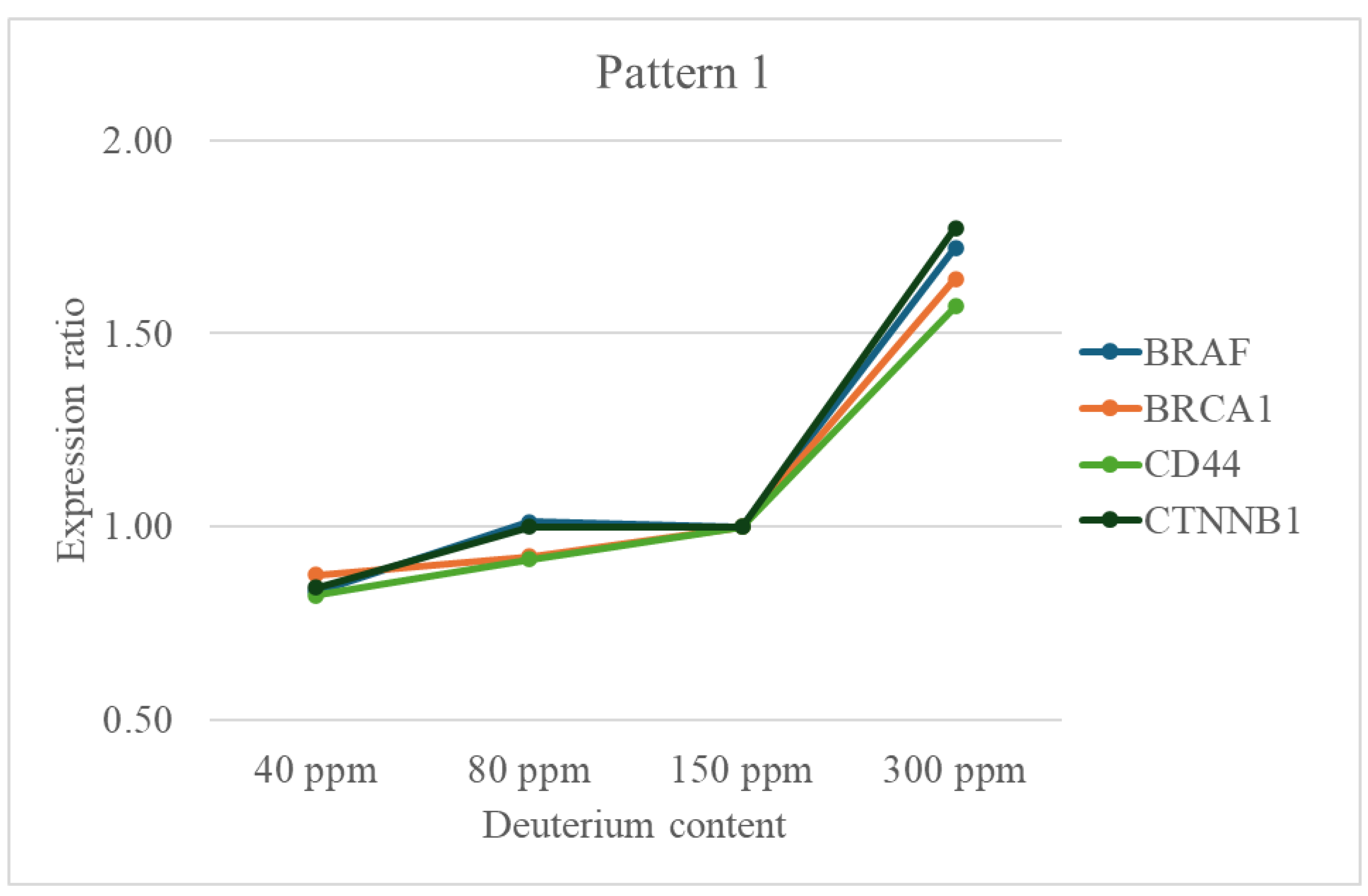
- Cell Signaling Pathways: Many are components of key growth-promoting pathways such as MAPK/ERK (BRAF, EGFR, JUN, YES1), PI3K/AKT (EGFR, ERBB3, LIF, MYC), Wnt (CTNNB1), and JAK/STAT (LIF, STAT3). Their upregulation at 300 ppm suggests an amplification of these pro-survival and pro-proliferative signals under high deuterium conditions.
- Proliferation and Survival: Oncogenes like MYC, EGFR, and ERBB3 directly promote cell cycle progression and inhibit apoptosis. SPP1 and TGFA further enhance proliferation and tumor growth.
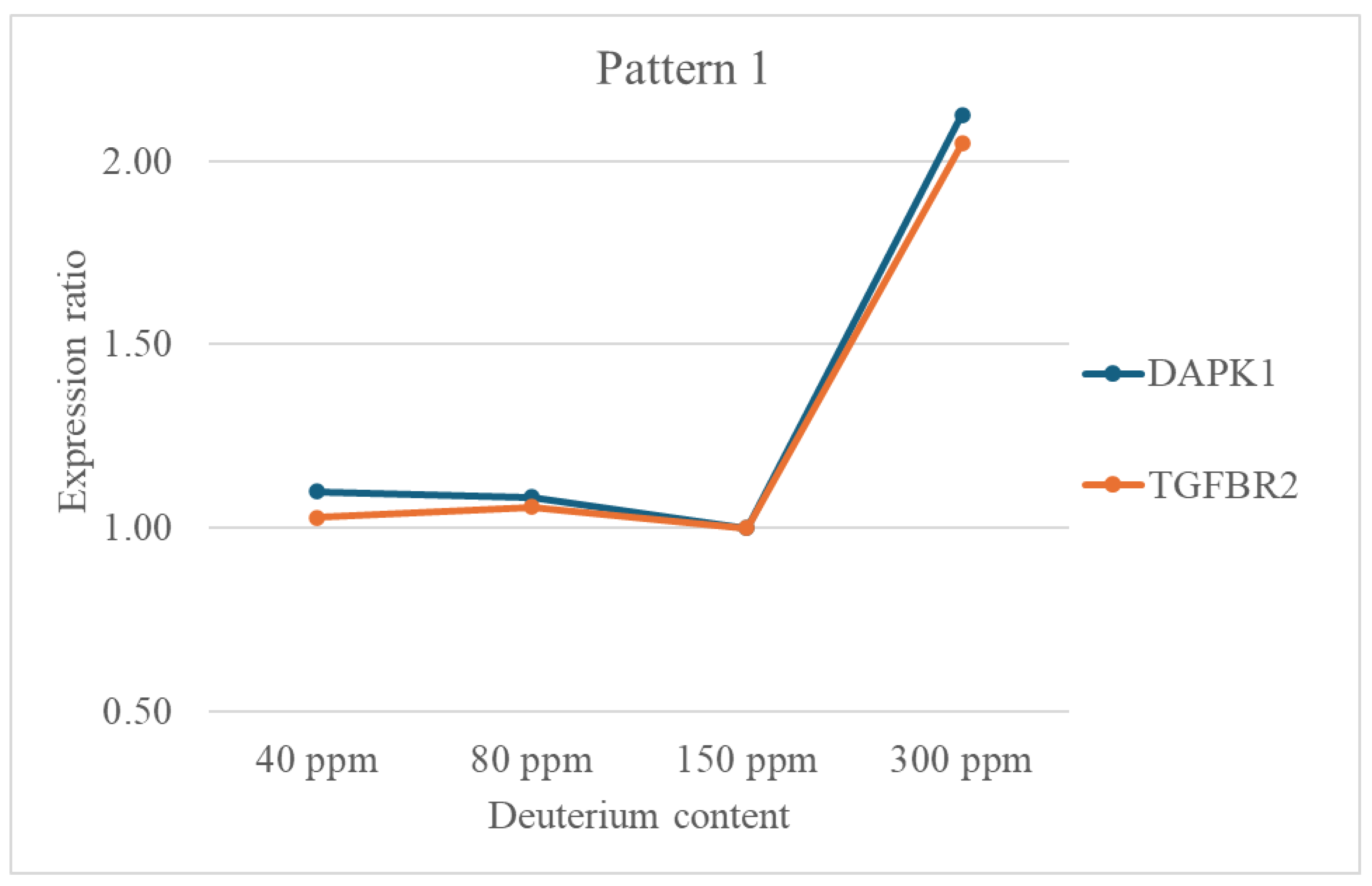
- Stress Response and DNA Repair: Genes like BRCA1, ERCC4, MSH6, and XRCC5 are crucial for maintaining genomic stability through DNA repair mechanisms. Their upregulation might indicate a cellular response to deuterium-induced stress, where increased repair capacity is needed to cope with potential DNA damage. DAPK1, a pro-apoptotic gene, may also be activated in response to stress.
- Cell Adhesion and Migration: CD44, ITGB1, LAMB1, FAT1, PLAUR, and TGFBI are integral to cell-matrix interactions and extracellular matrix (ECM) remodeling, processes critical for tumor invasion and metastasis. Their strong upregulation at 300 ppm suggests an enhanced metastatic potential.
- Immune and Inflammatory Modulation: IFNGR1 and PTGS2 (COX-2) play roles in immune responses and inflammation, which can contribute to a tumor-supportive microenvironment.
- Cell Survival and Apoptosis Regulation: Genes like BCL2, BCL2L1, AKT2, and PIM1 are anti-apoptotic, promoting cancer cell survival. FAS, a pro-apoptotic receptor, also falls into this category, suggesting a fine-tuned balance in cell death pathways.
- Cell Cycle Regulation: CCND1, CCND3, CDK6, E2F3, PCNA, and TOP2A are directly involved in cell cycle progression, particularly the G1/S transition and DNA replication. Their moderate upregulation at 300 ppm supports increased proliferative activity.
- Cell Proliferation and Growth: ERBB2 (HER2), HRAS, MET, MST1R, and PDGFA are components of various growth factors signaling pathways, driving cell proliferation.
- Oncogenic Potential: Many genes in this pattern (e.g., AKT2, BCL2, CCND1, ERBB2, HRAS) are well-known oncogenes, frequently dysregulated in cancer. Their moderate upregulation at 300 ppm further supports the pro-cancer effects of high deuterium.
- DNA Repair and Maintenance: OGG1, PCNA, TOP2A, and TYMS are crucial for DNA repair and synthesis, ensuring genomic stability necessary for rapid cancer cell proliferation.
- Cancer Progression and Survival: Most genes in this pattern are associated with hallmarks of cancer, including cell invasion (MMP9), proliferation (RAF1, RET), angiogenesis (HIF1A, SERPINE1), and cell survival (HSP90AB1, LYN).
- Deuterium Sensitivity: The moderate downregulation observed at 40 ppm and the upregulation at 300 ppm suggest that the expression of these genes is bidirectionally responsive to shifts in deuterium concentration. While these genes may not be directly involved in classical signaling pathways, their functions are nonetheless fundamental, indicating that they play a critical role in maintaining cellular homeostasis under varying D/H ratios.
- Diverse Cellular Processes: These genes span a wide range of cellular functions, including cell adhesion, transcription, signaling, matrix remodeling, DNA repair, and protein stability. This diversity indicates a broad network of cancer-related functions that are collectively modulated by deuterium's effects on cellular redox state.
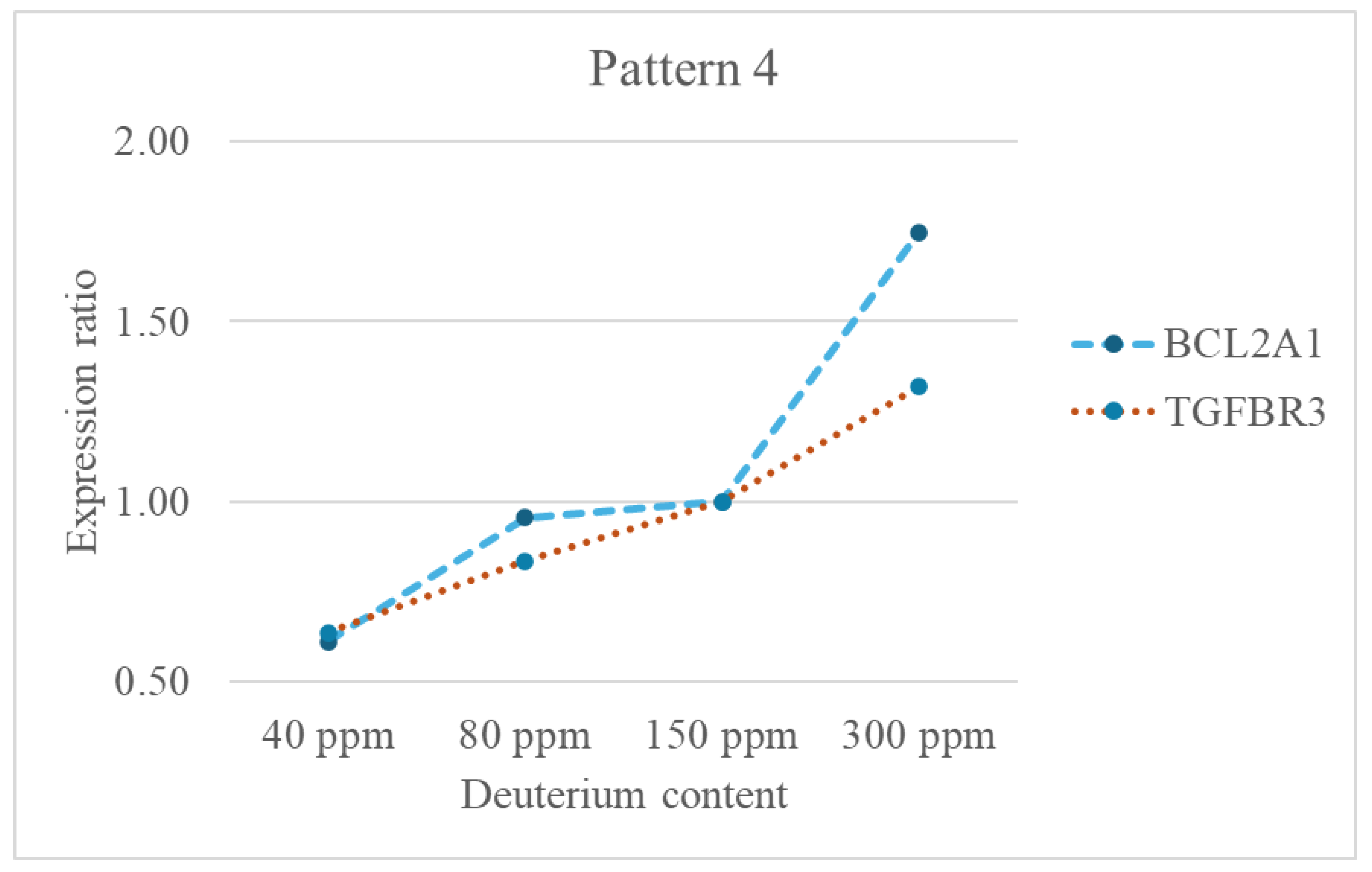
- Cell Survival and Stress Response: BCL2A1 is a potent anti-apoptotic protein, promoting cancer cell survival, especially under stress conditions like oxidative stress. TGFBR3 modulates TGF-β signaling, influencing cell survival and stress responses.
- Cancer Relevance with Context-Dependent Roles: BCL2A1 is oncogenic, supporting cancer cell survival. TGFBR3 can act as a tumor suppressor in early stages but may promote metastasis in advanced cancers.
- Deuterium Sensitivity: BCL2A1 is primarily localized to the mitochondrial outer membrane. Its location on the mitochondrial membrane is essential for its role in cell survival, particularly in conditions of stress or disease, such as in certain cancers where its overexpression contributes to chemotherapy resistance. The strong downregulation at 40 ppm suggests that very low deuterium levels suppress their expression which may contribute to the advantageous effect of DDW. The upregulation at 300 ppm aligns with the general trend of cancer gene activation under high deuterium.
- Cellular Protection and Maintenance: FANCG is involved in DNA repair, ensuring genomic integrity. MUC1 provides mucosal protection and can play roles in cell survival signaling. TFE3 regulates lysosomal biogenesis and metabolic homeostasis. PCTK1 supports cell cycle progression and neuronal differentiation.
- Limited Oncogenic Roles: While MUC1 and TFE3 have known associations with cancer, FANCG and PCTK1 are not primarily considered oncogenic drivers in the same vein as genes in Patterns 1 and 2.
- Stability Across DDW Levels: The consistent stability of these genes across all deuterium concentrations suggests that their functions are essential and tightly regulated, making them robust against variations in deuterium levels. This may indicate that these processes are fundamental for cell survival and are not significantly modulated by deuterium, or that their regulatory mechanisms are insensitive to these specific deuterium shifts.
- Oncogenic Receptor Tyrosine Kinase: FGFR4 is a receptor tyrosine kinase involved in cell proliferation, differentiation, and survival, often overexpressed in various cancers (e.g., hepatocellular carcinoma).
- Deuterium Sensitivity: The moderate downregulation at 40 ppm and 80 ppm suggests that lower deuterium levels may suppress FGFR4 expression. Its stability at 300 ppm, unlike the upregulation seen in many other oncogenes at high deuterium, might indicate tight regulatory control or a saturation of its signaling at normal/high deuterium levels.
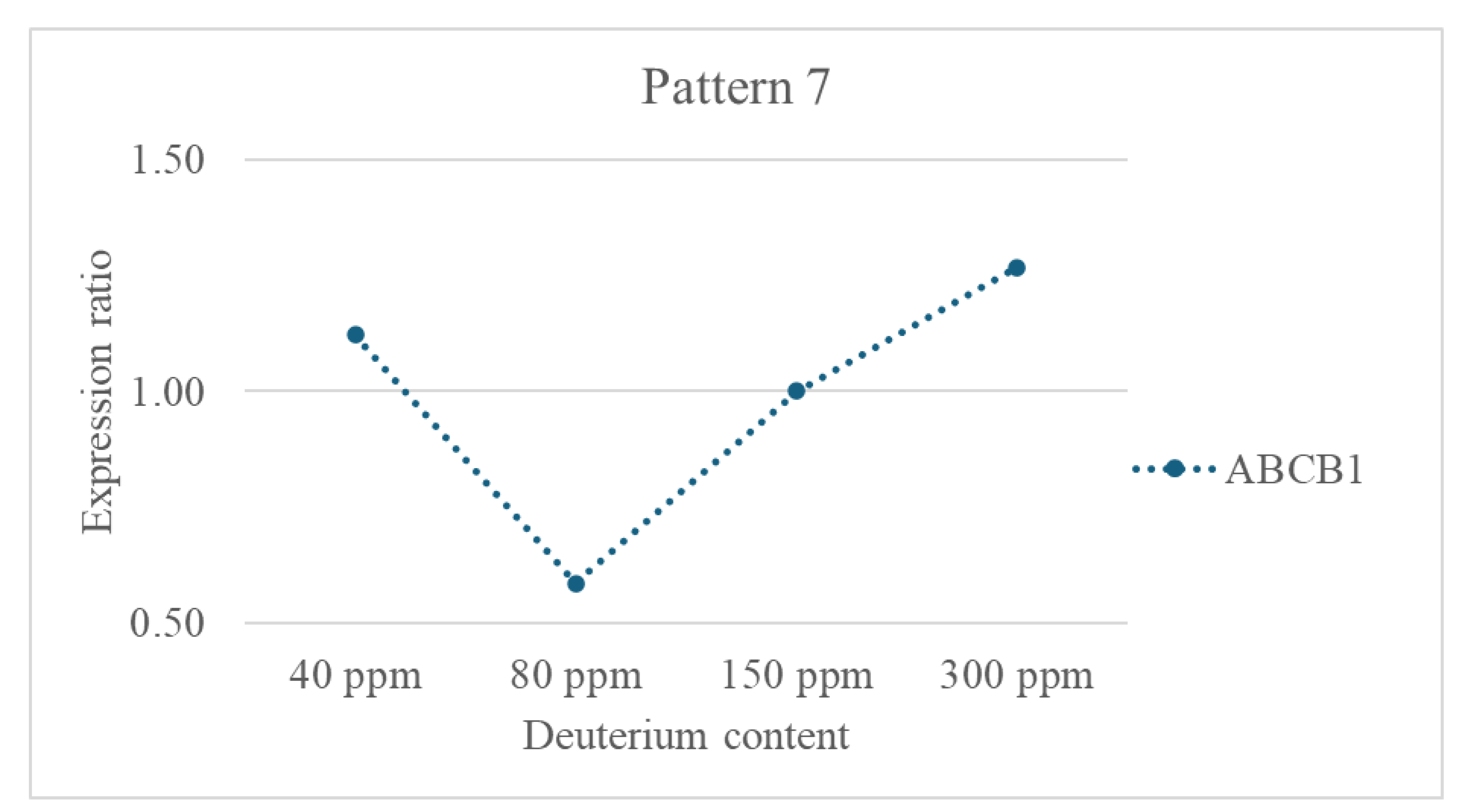
- ABCB1is a member of the (ATP-Binding Cassette) transporter superfamily.
- Pathway: Transporter Superfamily/Drug Metabolism and Transport.
- Function: It acts as an ATP-powered efflux pump on the cell membrane, actively transporting a wide variety of structurally unrelated hydrophobic molecules (including many chemotherapy drugs like Vinca alkaloids, Taxanes, and Anthracyclines) out of the cell.
- Multidrug Resistance (MDR) Efflux Pump: ABCB1 (P-glycoprotein) is a key transporter conferring multidrug resistance in cancer by actively expelling chemotherapeutic drugs from cells. Overexpression of ABCB1 is a major mechanism by which cancer cells develop resistance to a wide range of chemotherapeutic agents, making treatments ineffective (23).
- Deuterium Sensitivity: The 12% increase of its expression at 40 ppm suggests that very low deuterium concentration may not significantly perturb its expression, but it might have adverse effects. However, the downregulation at 80 ppm indicates that moderate deuterium depletion can suppress ABCB1, potentially enhancing chemotherapy sensitivity by reducing drug efflux. The moderate upregulation at 300 ppm aligns with increased ROS at high deuterium, which can induce ABCB1 expression and promote drug resistance.
- Oncogenic and Pro-Metastatic Roles: BCL3 is an oncogenic transcription co-regulator promoting cell proliferation and survival. PTK7 is a pseudokinase involved in Wnt signaling, cell polarity, and migration, often promoting invasion and metastasis.
- Unique Deuterium Sensitivity: The moderate upregulation at 40 ppm is distinct from most other patterns, suggesting a unique sensitivity to very low deuterium. Their upregulation at 300 ppm aligns with general cancer gene activation at high deuterium.
- Potent Proto-Oncogene: MYCN is a transcription factor critical for neural development but is frequently amplified or overexpressed in aggressive cancers like neuroblastoma, driving rapid tumor growth.
- Complex Deuterium Response: Its stability at 40 ppm and 300 ppm suggests tight regulatory control or context-specific roles that prevent its activation by extreme deuterium levels. However, the moderate downregulation at 80 ppm indicates that moderate deuterium depletion can suppress MYCN, potentially inhibiting tumor growth, which aligns with DDW's anti-cancer effects in neuroblastoma models.
3. Discussion
4. Materials and Methods
4.1. Experimental Setup and Cell Culture
4.2. Gene Expression Quantification
4.3. Dataset Description
4.4. Data Cleaning Criteria
- Low Average Copy Number Exclusion: Genes were excluded if their average copy number was less than 20 at the 150 ppm deuterium level. This filter targets low-expression genes, which are inherently more susceptible to technical noise and may lack significant biological relevance in this context.
- High Coefficient of Variation Exclusion: Genes were excluded if their CV exceeded 20% and |Z| > 3 at the 150 ppm deuterium level. A high CV and Z indicate excessive variability between replicate measurements, potentially stemming from experimental errors or significant biological heterogeneity that could confound interpretation.
4.5. Gene Expression Classification
- Strongly Upregulated: Ratio > 1.5 (expression is more than 50% higher than in the control medium).
- Moderately Upregulated: 1.2 < Ratio ≤ 1.5 (expression is 20% to 50% higher).
- Stable: 0.83 ≤ Ratio ≤ 1.2 (expression is within the range of -17% to +20% of the control).
- Moderately Downregulated: 0.67 ≤ Ratio < 0.83 (expression is 17% to 33% lower).
- Strongly Downregulated: Ratio < 0.67 (expression is less than two-thirds of the control).
4.6. Data Cleaning Outcomes
- Retained Genes: Genes such as BRCA1, TP53, and EGFR, known for their critical roles as tumor suppressors or oncogenes, were consistently retained. These genes exhibited stable expression (CV <10% across deuterium levels), indicating reliable measurements suitable for studying deuterium effects. The results are summarized in Table S1.
- Excluded Genes: A significant number of genes were excluded due to either low expression (e.g., AKT1, with an average copy number of 4 at 40 ppm) or high variability (e.g., AREG, with a CV of 33.33% at 40 ppm and a robust Z-score of 7.83). While the exclusion of low-expression genes helps reduce technical noise, the removal of high-variability genes, even those with high Z-scores, warrants careful consideration as they might represent genuine biological responses to deuterium-induced stress rather than mere experimental artifacts. Notably, the 40 ppm deuterium condition exhibited the largest discrepancies and highest average CV (~18% compared to ~10-14% for other levels), would have contributed disproportionately to gene exclusions. This suggests that extreme deuterium depletion may induce greater biological variability or be more susceptible to technical inconsistencies. These problems will be investigated in future studies.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
References
- Heron M, "Deaths: Leading Causes for 2019," Natl Vital Stat Rep, vol. 70(9), pp. 1-114, 2021.
- Španěl P, Shestivska V, Chippendale TWE, Smith D. Determination of the deuterium abundances in water from 156 to 10,000 ppm by SIFT-MS. J Am Soc Mass Spectrom. 2011;22(2):179–186.
- Maréchal, Y. IR spectra and dynamics of H₂O (D₂O, HDO) molecules in a still poorly known liquid: water. NATO ASI Ser C Math Phys Sci. 1993;435:149–168.
- Jancsó, G. Isotope Effects in Handbook of Nuclear Chemistry (eds. Vértes, A., Nagy, S. & Klencsár, Z.) 85-116 (Kluwer Academic Publishers: Dordrecht, Netherland, 2013).
- Sobczyk L, Obrzud M, Filarowski A. H/D isotope effects in hydrogen bonded systems. Molecules. 2013 Apr 16;18(4):4467-76. [CrossRef] [PubMed]
- Somlyai, G.; et al. Naturally occurring deuterium is essential for the normal growth rate of cells. FEBS Lett 1993, 317, 1–4. [Google Scholar] [CrossRef] [PubMed]
- G. Somlyai, B. Z. Kovács, I. Somlyai, A. Papp, L. I. Nagy and L. G. Puskás, "Deuterium depletion inhibits lung cancer cell growth and migration in vitro and results in severalfold increase of median survival time of non-small cell lung cancer patients receiving conventional therapy," Cancer Res The, vol. 9(2), pp. 12-19, 2021.
- Lu, Y. and Chen, H. Deuterium-Depleted Water in Cancer Therapy: A Systematic Review of Clinical and Experimental Trials, Nutrients, 2024 16(9), p. 1397.
- Kovács BZ, Puskás LG, Nagy LI, Papp A, Gyöngyi Z, Fórizs I, et al. Blocking the Increase of Intracellular Deuterium Concentration Prevents the Expression of Cancer-Related Genes, Tumor Development, and Tumor Recurrence in Cancer Patients. Cancer Control. 2022;10732748211068963.
- Lei, Z., Su, N., Li, M., Pan Z., Liu K., Zhang Y. Unraveling the role of deuterium in cancer: mechanisms, detection techniques, and therapeutic potential. Mol Divers (2025). [CrossRef]
- Cong F.S., Zhang Y.R., Sheng H.C., Ao Z.H., Zhang S.Y., Wang J.Y. Deuterium-Depleted Water Inhibits Human Lung Carcinoma Cell Growth by Apoptosis. Exp. Ther. Med. 1(2), 277-283.
- Boros LG, Somlyai I, Kovács BZ, Puskás LG, Nagy LI, Dux L, Farkas G, Somlyai G. Deuterium Depletion Inhibits Cell Proliferation, RNA and Nuclear Membrane Turnover to Enhance Survival in Pancreatic Cancer. Cancer Control. 2021 Jan-Dec;28:1073274821999655. [CrossRef]
- Zhang X, Gaetani M, Chernobrovkin A, Zubarev RA. Anticancer Effect of Deuterium Depleted Water – Redox Disbalance Leads to Oxidative Stress. Cancer Control. 2019;18:2373–87.
- Z. Gyöngyi, F. Budán, I. Szabó, I. Ember, K. Kiss, K. Krempels, I. Somlyai and G. Somlyai, "Deuterium depleted water effects on survival of lung cancer patients and expression of Kras, Bcl2, and Myc genes in mouse lung," Nutr Cancer, vol. 65(2), pp. 240-246, 2012.
- Gyöngyi, Z. & Somlyai, G. Deuterium depletion can decrease the expression of c-myc, Ha-ras and p53 gene in carcinogen-treated mice. In Vivo 14, 437–439; PMID.10904878 (2000).
- Fortina P, Surrey S. Digital mRNA profiling. Nat Biotechnol. 2008 Mar;26(3):293-4. [CrossRef]
- Kontomanolis EN, Koutras A, Syllaios A, Schizas D, Mastoraki A, Garmpis N, et al. Role of Oncogenes and Tumor-suppressor Genes in Carcinogenesis: A Review. Anticancer Res. 2020 Nov;40(11):6009-6015. [CrossRef]
- Carlo M. Croce, MD, Oncogenes and Cancer, 2008, N Engl J Med 2008;358:502-511. [CrossRef]
- Jang, C. W., Chen, C. H., Chen, C. C., Chen, J. Y., Su, Y. H., & Chen, R. H. (2002). TGF-β induces apoptosis through Smad-mediated expression of DAP-kinase. Nature Cell Biology, 4(1), 51–58. [CrossRef]
- Kim, N., Chen, D., Zhou, X. Z., & Lee, T. H. (2019). Death-associated protein kinase 1 phosphorylation in neuronal cell death and neurodegenerative disease. In International Journal of Molecular Sciences (Vol. 20, Issue 13). MDPI AG. [CrossRef]
- Liu, R. M., & Desai, L. P. (2015). Reciprocal regulation of TGF-β and reactive oxygen species: A perverse cycle for fibrosis. In Redox Biology (Vol. 6, pp. 565–577). Elsevier B.V. [CrossRef]
- Brown, M. F., Thurmond, R. L., Dodd, S. W., Otten, D., & Beyer, K. (2002). Elastic deformation of membrane bilayers probed by deuterium NMR relaxation. Journal of the American Chemical Society, 124(28), 8471–8484. [CrossRef]
- Xiao H, Zheng Y, Ma L, Tian L, Sun Q. Clinically-Relevant ABC Transporter for Anti-Cancer Drug Resistance. Front Pharmacol. 2021;12:648407. [CrossRef]
- Somlyai, G. Deuterium Depletion – A New Way in Curing Cancer and Preserving Health. Redwood City, CA: PublishDrive; 2021.
- Molnár M, Horváth K, Dankó T, Somlyai I, Kovács BZ, Somlyai G. Deuterium-depleted water stimulates GLUT4 translocation in the presence of insulin, which leads to decreased blood glucose concentration. Mol Cell Biochem. 2021;476(12):4507–16. [CrossRef] [PubMed]
- Somlyai G, Papp A, Somlyai I, Kovács BZ, Debrődi M. Real-World Data Confirm That the Integration of Deuterium Depletion into Conventional Cancer Therapy Multiplies the Survival Probability of Patients. Biomedicines. 2025;13:876. [CrossRef]
- Maronna RA, Martin RD, Yohai VJ. Robust Statistics: Theory and Methods (with R). 2nd ed. Chichester, UK: Wiley; 2019.
| Deuterium Content | Strong Up (>1.5) | Moderate Up (1.2–1.5) |
Stable (0.83–1.2) | Moderate Down (0.67–0.83) | Strong Down (<0.67) |
|---|---|---|---|---|---|
| 40 ppm | 0 | 2 | 66 | 17 | 2 |
| 80 ppm | 0 | 0 | 80 | 6 | 1 |
| 300 ppm | 44 | 28 | 15 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





