Submitted:
01 October 2025
Posted:
02 October 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Preface
1.2. Viral-Immune Co-Evolution
1.3. Systemic and Cellular Metabolism in Viral Control
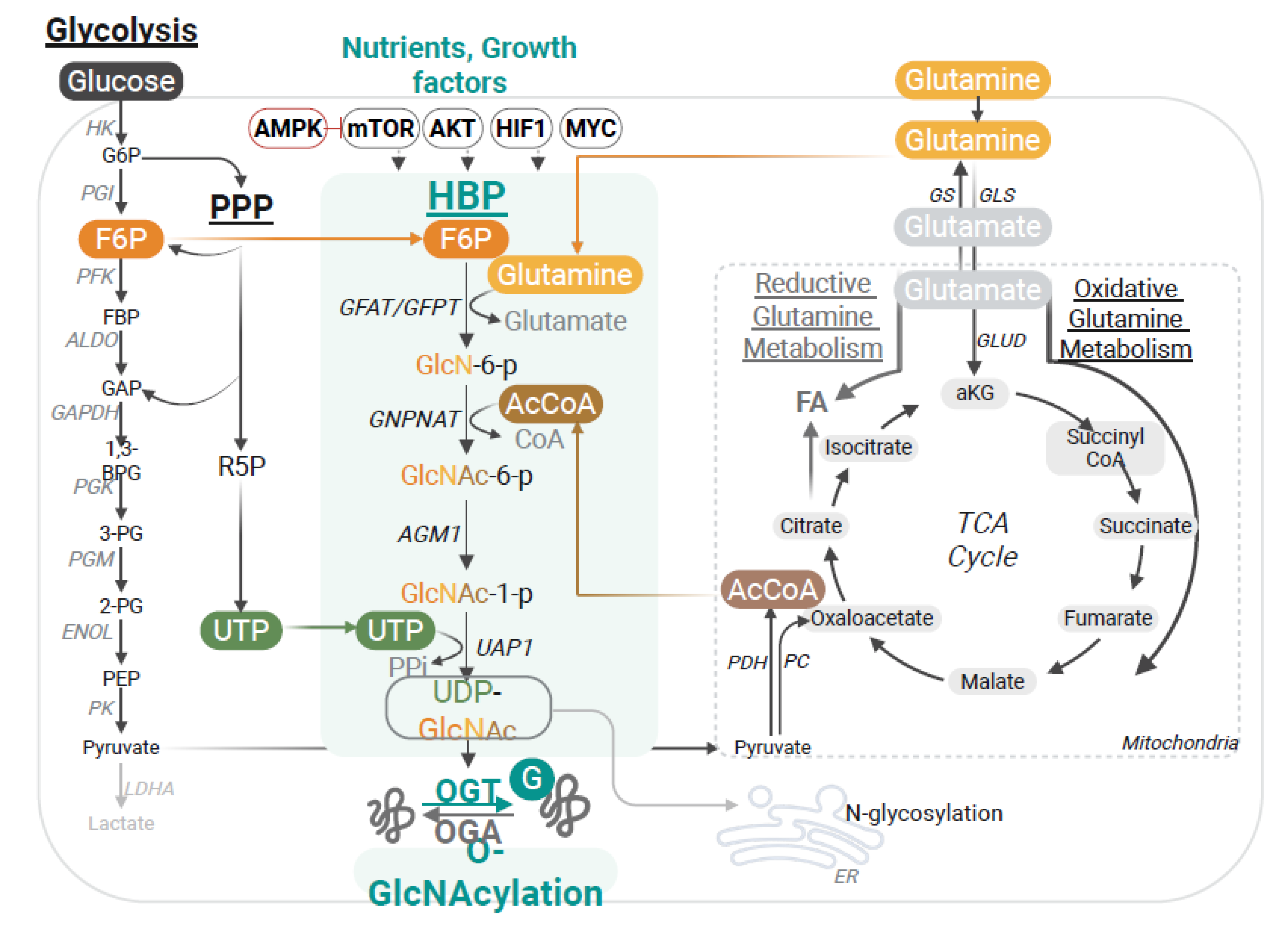
2. The Hexosamine Biosynthesis Pathway and O-GlcNAcylation: From Nutrient Sensing to the Control of Immunity
| Substrate | Identified Site | Effect on substrate | Cell type _ Functional Impact | Reference |
|---|---|---|---|---|
| RIPK3 | T467 | Inhibition of phosphorylation | Macrophages _ Reduced necroptosis & inflammation | [33] |
| S6K1 | S489 | Inhibition of phosphorylation | Macrophages _ Reduced inflammation | [34] |
| IRF5 | S430 | Activation through K63-Ub | Macrophages, PBMCs, epithelial cells _ Enhanced inflammation | [35] |
| MAVS | S366; domain 324-347 | Activation through K63-Ub | Macrophages _ Enhanced anti-viral response | [36,38] |
| S249, T250, T252, S253, S255, S256, S257 |
Inhibition | Epithelial cells and fibroblasts _ Reduced anti-viral response | [40] | |
| UBXN1 | S75, T95, S132 | Inhibition of UBXN1-MAVS interaction | Macrophages _ Enhanced MAVS anti-viral response | [41] |
| STING | T229 | Activation through K63-Ub | Fibroblasts _ Enhanced anti-viral response | [42] |
| STAT1 | T699 | Activation by inhibition of kbhb | Fibroblasts _ Enhanced anti-viral response | [43] |
| SAMHD1 | S93 | Stabilization | Macrophages, Hepatocytes _ Enhanced anti-viral response | [49] |
| YTHFD2 | S263 | Stabilization | Hepatocytes _ Enhanced proliferation | [57] |
| NFAT | n.d. | Activation | T cell activation | [24] |
| c-REL | S350 | Activation | T cell activation, FOXP3 suppression | [25,26] |
| CREB | S40 | Activation | HTLV-1 T cell _ Enhanced viral transcription | [56] |
| STAT5 | T38, S57, S58, S270, S273, |
Activation by phosphorylation | Treg _ Enhanced suppressive program | [31] |
| FOXP3 | T38, S57, S58, S270, S273 |
Stabilization | Treg _ Enhanced suppressive program | [31] |
| ACC1 | S966, S967 | Activation | Th17 _Enhanced RORγt transcriptional program | [32] |
| c-MYC | T58 | Stabilization | T cell, B cell _ Enhanced proliferation | [27,29] |
| LYN | S19 | Activation | B cell _ Enhanced BCR signaling | [28] |
| SMC1 | n.d. | Activation | B cell _ VH gene recombination | [30] |
| SMC3 | n.d. | Activation | B cell _ VH gene recombination | [30] |
| YY1 | T236 | Activation | B cell _ VH gene recombination | [30] |
| CTCF | T668 | Activation | B cell _ VH gene recombination | [30] |
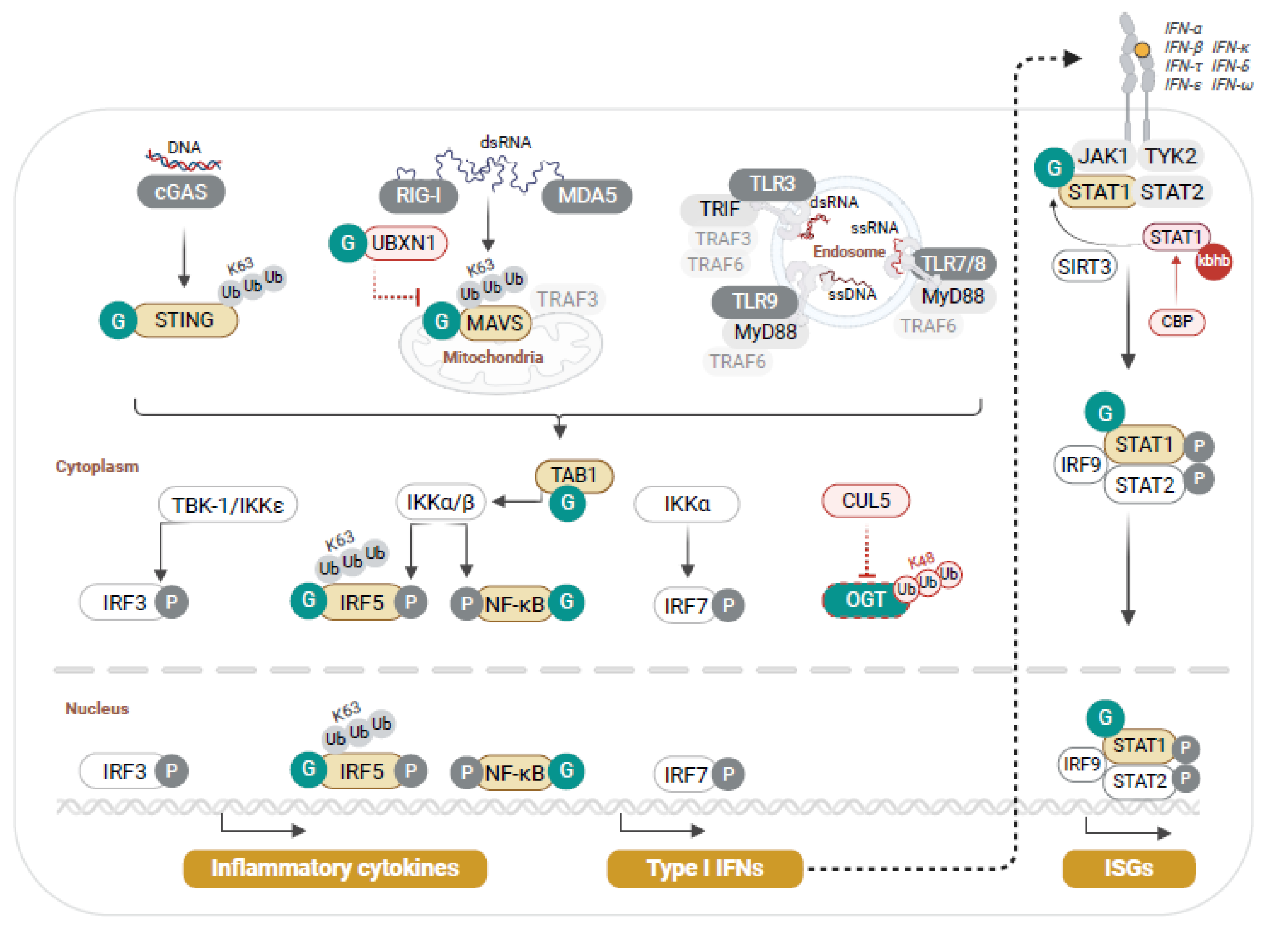
3. O-GlcNAc Modulation of Host Antiviral Innate Immunity
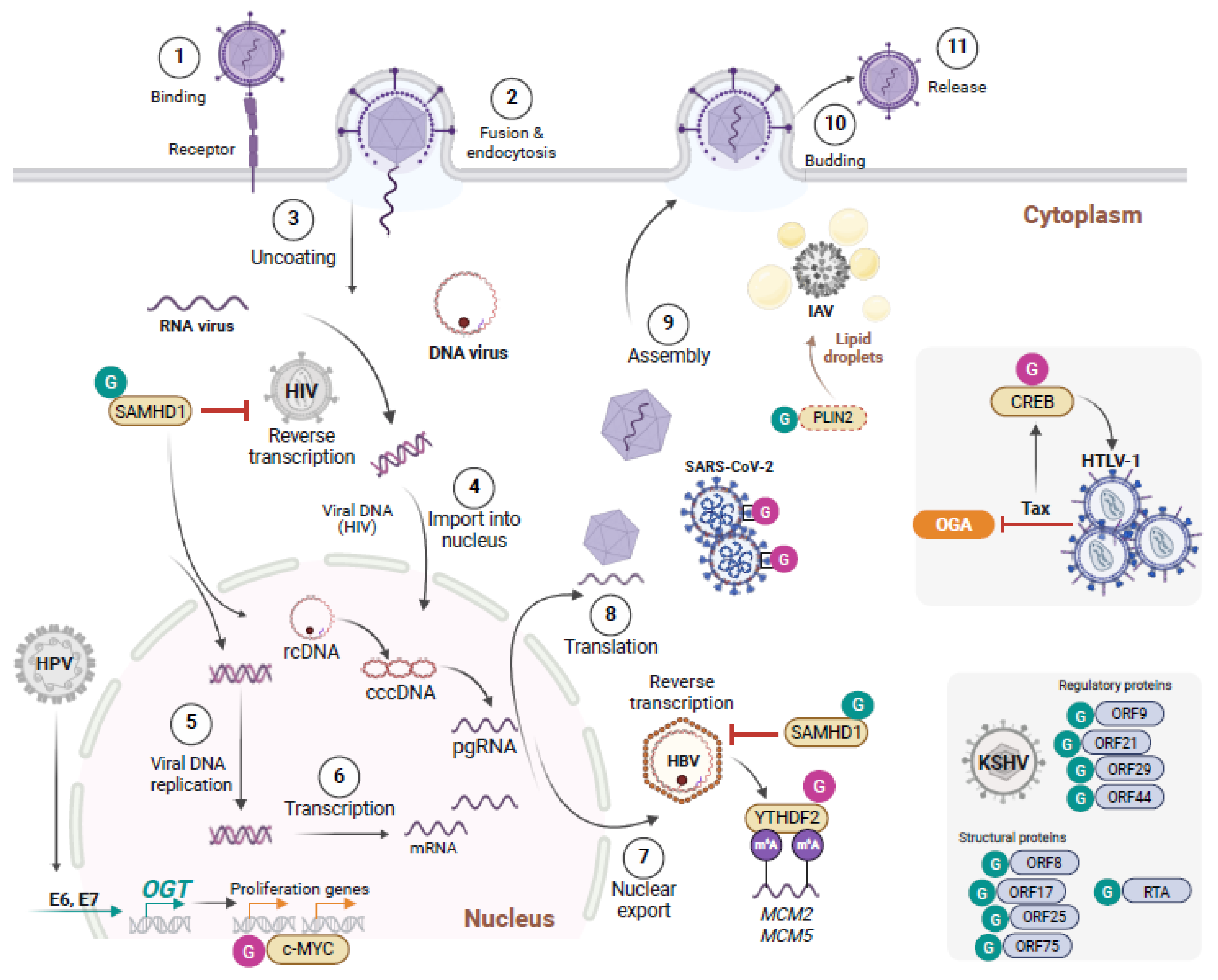
4. O-GlcNAc Interference with the Viral Machinery
5. Viral Hijacking of the O-GlcNAc Pathway to Dampen Anti-Viral Defenses, or Enhance Infectivity or Viral Oncogenic Transformation
6. Harnessing the HBP-O-GlcNAc Pathway for Clinical Applications
6.1. The HBP and O-GlcNAcylation as Potential Biomarkers of Disease Progression in Viral Infections
6.2. HBP and O-GlcNAcylation as Therapeutic Targets in Viral Infections
References
- Koonin, E.V.; Krupovic, M.; Dolja, V.V. The global virome: How much diversity and how many independent origins? Environ Microbiol 2023, 25, 40–44. [Google Scholar] [CrossRef]
- Carroll, D.; Daszak, P.; Wolfe, N.D.; Gao, G.F.; Morel, C.M.; Morzaria, S.; Pablos-Mendez, A.; Tomori, O.; Mazet, J.A.K. The Global Virome Project. Science 2018, 359, 872–874. [Google Scholar] [CrossRef]
- Roychoudhury, S.; Das, A.; Sengupta, P.; Dutta, S.; Roychoudhury, S.; Choudhury, A.P.; Ahmed, A.B.F.; Bhattacharjee, S.; Slama, P. Viral Pandemics of the Last Four Decades: Pathophysiology, Health Impacts and Perspectives. Int J Environ Res Public Health 2020, 17. [Google Scholar] [CrossRef] [PubMed]
- Marchi, J.; Lassig, M.; Walczak, A.M.; Mora, T. Antigenic waves of virus-immune coevolution. Proc Natl Acad Sci U S A 2021, 118. [Google Scholar] [CrossRef]
- Muhammad, I.; Contes, K.; Bility, M.T.; Tang, Q. Chasing Virus Replication and Infection: PAMP-PRR Interaction Drives Type I Interferon Production, Which in Turn Activates ISG Expression and ISGylation. Viruses 2025, 17. [Google Scholar] [CrossRef]
- Beachboard, D.C.; Horner, S.M. Innate immune evasion strategies of DNA and RNA viruses. Curr Opin Microbiol 2016, 32, 113–119. [Google Scholar] [CrossRef]
- Tay, D.J.W.; Lew, Z.Z.R.; Chu, J.J.H.; Tan, K.S. Uncovering Novel Viral Innate Immune Evasion Strategies: What Has SARS-CoV-2 Taught Us? Front Microbiol 2022, 13, 844447. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.K.; Gack, M.U. Viral evasion of intracellular DNA and RNA sensing. Nat Rev Microbiol 2016, 14, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 2014, 32, 513–545. [Google Scholar] [CrossRef]
- Acchioni, M.; Acchioni, C.; Hiscott, J.; Sgarbanti, M. Origin and function of anti-interferon type I viral proteins. Virology 2025, 605, 110456. [Google Scholar] [CrossRef]
- Chi, H.; Pepper, M.; Thomas, P.G. Principles and therapeutic applications of adaptive immunity. Cell 2024, 187, 2052–2078. [Google Scholar] [CrossRef]
- Mora, T.; Walczak, A.M. Quantitative Theory of Viral-Immune Coevolution May Be within Reach. PRX Life 2023, 1. [Google Scholar] [CrossRef]
- Palmer, C.S. Innate metabolic responses against viral infections. Nat Metab 2022, 4, 1245–1259. [Google Scholar] [CrossRef]
- Perakakis, N.; Harb, H.; Hale, B.G.; Varga, Z.; Steenblock, C.; Kanczkowski, W.; Alexaki, V.I.; Ludwig, B.; Mirtschink, P.; Solimena, M.; et al. Mechanisms and clinical relevance of the bidirectional relationship of viral infections with metabolic diseases. Lancet Diabetes Endocrinol 2023, 11, 675–693. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A.; Kishton, R.J.; Rathmell, J. A guide to immunometabolism for immunologists. Nat Rev Immunol 2016, 16, 553–565. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.A.; Rutter, J. Metabolites as signalling molecules. Nat Rev Mol Cell Biol 2023, 24, 355–374. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Liu, C.H.; Lei, M.; Zeng, Q.; Li, L.; Tang, H.; Zhang, N. Metabolic regulation of the immune system in health and diseases: mechanisms and interventions. Signal Transduct Target Ther 2024, 9, 268. [Google Scholar] [CrossRef]
- El Safadi, D.; Paulo-Ramos, A.; Hoareau, M.; Roche, M.; Krejbich-Trotot, P.; Viranaicken, W.; Lebeau, G. The Influence of Metabolism on Immune Response: A Journey to Understand Immunometabolism in the Context of Viral Infection. Viruses 2023, 15. [Google Scholar] [CrossRef]
- Kumar, V.; Stewart Iv, J.H. Pattern-Recognition Receptors and Immunometabolic Reprogramming: What We Know and What to Explore. J Innate Immun 2024, 16, 295–323. [Google Scholar] [CrossRef] [PubMed]
- Dsouza, L.; Pant, A.; Pope, B.; Yang, Z. Vaccinia growth factor-dependent modulation of the mTORC1-CAD axis upon nutrient restriction. J Virol 2025, 99, e0211024. [Google Scholar] [CrossRef]
- Gong, J.; Gao, X.; Ge, S.; Li, H.; Wang, R.; Zhao, L. The Role of cGAS-STING Signalling in Metabolic Diseases: from Signalling Networks to Targeted Intervention. Int J Biol Sci 2024, 20, 152–174. [Google Scholar] [CrossRef]
- Paneque, A.; Fortus, H.; Zheng, J.; Werlen, G.; Jacinto, E. The Hexosamine Biosynthesis Pathway: Regulation and Function. Genes (Basel) 2023, 14. [Google Scholar] [CrossRef]
- Issad, T.; Al-Mukh, H.; Bouaboud, A.; Pagesy, P. Protein O-GlcNAcylation and the regulation of energy homeostasis: lessons from knock-out mouse models. J Biomed Sci 2022, 29, 64. [Google Scholar] [CrossRef]
- Golks, A.; Tran, T.T.; Goetschy, J.F.; Guerini, D. Requirement for O-linked N-acetylglucosaminyltransferase in lymphocytes activation. EMBO J 2007, 26, 4368–4379. [Google Scholar] [CrossRef]
- Ramakrishnan, P.; Clark, P.M.; Mason, D.E.; Peters, E.C.; Hsieh-Wilson, L.C.; Baltimore, D. Activation of the transcriptional function of the NF-kappaB protein c-Rel by O-GlcNAc glycosylation. Sci Signal 2013, 6, ra75. [Google Scholar] [CrossRef]
- de Jesus, T.J.; Tomalka, J.A.; Centore, J.T.; Staback Rodriguez, F.D.; Agarwal, R.A.; Liu, A.R.; Kern, T.S.; Ramakrishnan, P. Negative regulation of FOXP3 expression by c-Rel O-GlcNAcylation. Glycobiology 2021, 31, 812–826. [Google Scholar] [CrossRef]
- Swamy, M.; Pathak, S.; Grzes, K.M.; Damerow, S.; Sinclair, L.V.; van Aalten, D.M.; Cantrell, D.A. Glucose and glutamine fuel protein O-GlcNAcylation to control T cell self-renewal and malignancy. Nat Immunol 2016, 17, 712–720. [Google Scholar] [CrossRef]
- Wu, J.L.; Chiang, M.F.; Hsu, P.H.; Tsai, D.Y.; Hung, K.H.; Wang, Y.H.; Angata, T.; Lin, K.I. O-GlcNAcylation is required for B cell homeostasis and antibody responses. Nat Commun 2017, 8, 1854. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Kwon, N.E.; Lee, W.J.; Lee, M.S.; Kim, D.J.; Kim, J.H.; Park, S.K. Increased O-GlcNAcylation of c-Myc Promotes Pre-B Cell Proliferation. Cells 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Jeon, B.C.; Kim, Y.J.; Park, A.K.; Song, M.R.; Na, K.M.; Lee, J.; An, D.; Park, Y.; Hwang, H.; Kim, T.D.; et al. Dynamic O-GlcNAcylation governs long-range chromatin interactions in V(D)J recombination during early B-cell development. Cell Mol Immunol 2025, 22, 68–82. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Salgado, O.C.; Singh, S.; Hippen, K.L.; Maynard, J.C.; Burlingame, A.L.; Ball, L.E.; Blazar, B.R.; Farrar, M.A.; Hogquist, K.A.; et al. The lineage stability and suppressive program of regulatory T cells require protein O-GlcNAcylation. Nat Commun 2019, 10, 354. [Google Scholar] [CrossRef]
- Machacek, M.; Saunders, H.; Zhang, Z.; Tan, E.P.; Li, J.; Li, T.; Villar, M.T.; Artigues, A.; Lydic, T.; Cork, G.; et al. Elevated O-GlcNAcylation enhances pro-inflammatory Th17 function by altering the intracellular lipid microenvironment. J Biol Chem 2019, 294, 8973–8990. [Google Scholar] [CrossRef]
- Li, X.; Gong, W.; Wang, H.; Li, T.; Attri, K.S.; Lewis, R.E.; Kalil, A.C.; Bhinderwala, F.; Powers, R.; Yin, G.; et al. O-GlcNAc Transferase Suppresses Inflammation and Necroptosis by Targeting Receptor-Interacting Serine/Threonine-Protein Kinase 3. Immunity 2019, 50, 576–590 e576. [Google Scholar] [CrossRef]
- Yang, Y.; Li, X.; Luan, H.H.; Zhang, B.; Zhang, K.; Nam, J.H.; Li, Z.; Fu, M.; Munk, A.; Zhang, D.; et al. OGT suppresses S6K1-mediated macrophage inflammation and metabolic disturbance. Proc Natl Acad Sci U S A 2020, 117, 16616–16625. [Google Scholar] [CrossRef]
- Wang, Q.; Fang, P.; He, R.; Li, M.; Yu, H.; Zhou, L.; Yi, Y.; Wang, F.; Rong, Y.; Zhang, Y.; et al. O-GlcNAc transferase promotes influenza A virus-induced cytokine storm by targeting interferon regulatory factor-5. Sci Adv 2020, 6, eaaz7086. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Li, X.; Attri, K.S.; Liu, C.; Li, L.; Herring, L.E.; Asara, J.M.; Lei, Y.L.; Singh, P.K.; Gao, C.; et al. O-GlcNAc Transferase Links Glucose Metabolism to MAVS-Mediated Antiviral Innate Immunity. Cell Host Microbe 2018, 24, 791–803 e796. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, M.; Chu, H.; Zhang, H.; Wu, H.; Song, G.; Wang, P.; Zhao, K.; Hou, J.; Wang, X.; et al. The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat Immunol 2017, 18, 214–224. [Google Scholar] [CrossRef]
- Song, N.; Qi, Q.; Cao, R.; Qin, B.; Wang, B.; Wang, Y.; Zhao, L.; Li, W.; Du, X.; Liu, F.; et al. MAVS O-GlcNAcylation Is Essential for Host Antiviral Immunity against Lethal RNA Viruses. Cell Rep 2019, 28, 2386–2396 e2385. [Google Scholar] [CrossRef]
- Zhang, H.; Xue, K.; Li, W.; Yang, X.; Gou, Y.; Su, X.; Qian, F.; Sun, L. Cullin5 drives experimental asthma exacerbations by modulating alveolar macrophage antiviral immunity. Nat Commun 2024, 15, 252. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Park, Y.S.; Kweon, T.H.; Kang, J.; Son, S.; Kim, H.B.; Seo, Y.R.; Kang, M.J.; Yi, E.C.; Lee, Y.H.; et al. O-Linked N-Acetylglucosamine Modification of Mitochondrial Antiviral Signaling Protein Regulates Antiviral Signaling by Modulating Its Activity. Front Immunol 2020, 11, 589259. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Jiang, P. p53 promotes antiviral innate immunity by driving hexosamine metabolism. Cell Rep 2024, 43, 113724. [Google Scholar] [CrossRef]
- Li, Y.; An, W.; Lu, L.; Yuan, J.; Wu, D.; Yang, Q.; Guo, J.; Yang, J.; Liu, M.; He, K.; et al. O-GlcNAc of STING mediates antiviral innate immunity. Cell Commun Signal 2024, 22, 157. [Google Scholar] [CrossRef]
- Zuo, Y.; Wang, Q.; Tian, W.; Zheng, Z.; He, W.; Zhang, R.; Zhao, Q.; Miao, Y.; Yuan, Y.; Wang, J.; et al. beta-hydroxybutyrylation and O-GlcNAc modifications of STAT1 modulate antiviral defense in aging. Cell Mol Immunol 2025, 22, 403–417. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.C.; Loo, S.L.; Nichol, K.S.; Reid, A.T.; Veerati, P.C.; Esneau, C.; Wark, P.A.B.; Grainge, C.L.; Knight, D.A.; Vincent, T.; et al. IL-25 blockade augments antiviral immunity during respiratory virus infection. Commun Biol 2022, 5, 415. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Ge, Y.; Sun, J. IL-33 in COVID-19: friend or foe? Cell Mol Immunol 2021, 18, 1602–1604. [Google Scholar] [CrossRef]
- Jochmann, R.; Thurau, M.; Jung, S.; Hofmann, C.; Naschberger, E.; Kremmer, E.; Harrer, T.; Miller, M.; Schaft, N.; Sturzl, M. O-linked N-acetylglucosaminylation of Sp1 inhibits the human immunodeficiency virus type 1 promoter. J Virol 2009, 83, 3704–3718. [Google Scholar] [CrossRef]
- Jochmann, R.; Pfannstiel, J.; Chudasama, P.; Kuhn, E.; Konrad, A.; Sturzl, M. O-GlcNAc transferase inhibits KSHV propagation and modifies replication relevant viral proteins as detected by systematic O-GlcNAcylation analysis. Glycobiology 2013, 23, 1114–1130. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.C.; Tsai, W.H.; Wang, P.W.; Wu, I.L.; Lin, S.Y.; Chen, Y.L.; Chen, J.Y.; Lin, S.F. Suppressive regulation of KSHV RTA with O-GlcNAcylation. J Biomed Sci 2012, 19, 12. [Google Scholar] [CrossRef]
- Hu, J.; Gao, Q.; Yang, Y.; Xia, J.; Zhang, W.; Chen, Y.; Zhou, Z.; Chang, L.; Hu, Y.; Zhou, H.; et al. Hexosamine biosynthetic pathway promotes the antiviral activity of SAMHD1 by enhancing O-GlcNAc transferase-mediated protein O-GlcNAcylation. Theranostics 2021, 11, 805–823. [Google Scholar] [CrossRef]
- Dong, H.; Liang, C.; Zhang, J.; Wu, W.; Kumar, N.; Liu, Z.; Sun, Y.; Liao, Z.; Cheng, X.; Yu, Y.; et al. O-GlcNAc transferase plays dual antiviral roles by integrating innate immunity and lipid metabolism. Nat Commun 2025, 16, 7721. [Google Scholar] [CrossRef]
- Fricke, J.; Koo, L.Y.; Brown, C.R.; Collins, P.L. p38 and OGT sequestration into viral inclusion bodies in cells infected with human respiratory syncytial virus suppresses MK2 activities and stress granule assembly. J Virol 2013, 87, 1333–1347. [Google Scholar] [CrossRef]
- Xu, T.; Li, J.; Lu, X.; Song, S.; Wang, S.; Li, J.; Zhang, L. O-GlcNAcylation at S659 enhances SARS-CoV-2 spike protein stability and pseudoparticle packaging efficiency. Microbiol Spectr 2025, 13, e0052725. [Google Scholar] [CrossRef] [PubMed]
- Ciraku, L.; Esquea, E.M.; Reginato, M.J. O-GlcNAcylation regulation of cellular signaling in cancer. Cell Signal 2022, 90, 110201. [Google Scholar] [CrossRef]
- Zeng, Q.; Zhao, R.X.; Chen, J.; Li, Y.; Li, X.D.; Liu, X.L.; Zhang, W.M.; Quan, C.S.; Wang, Y.S.; Zhai, Y.X.; et al. O-linked GlcNAcylation elevated by HPV E6 mediates viral oncogenesis. Proc Natl Acad Sci U S A 2016, 113, 9333–9338. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Kim, Y.S.; Choi, M.Y.; Kim, M.; Yang, J.H.; Park, H.O.; Jang, I.S.; Moon, S.H.; Kim, H.O.; Song, D.H.; et al. O-linked-N-acetylglucosamine transferase is associated with metastatic spread of human papillomavirus E6 and E7 oncoproteins to the lungs of mice. Biochem Biophys Res Commun 2017, 483, 793–802. [Google Scholar] [CrossRef]
- Groussaud, D.; Khair, M.; Tollenaere, A.I.; Waast, L.; Kuo, M.S.; Mangeney, M.; Martella, C.; Fardini, Y.; Coste, S.; Souidi, M.; et al. Hijacking of the O-GlcNAcZYME complex by the HTLV-1 Tax oncoprotein facilitates viral transcription. PLoS Pathog 2017, 13, e1006518. [Google Scholar] [CrossRef]
- Yang, Y.; Yan, Y.; Yin, J.; Tang, N.; Wang, K.; Huang, L.; Hu, J.; Feng, Z.; Gao, Q.; Huang, A. O-GlcNAcylation of YTHDF2 promotes HBV-related hepatocellular carcinoma progression in an N(6)-methyladenosine-dependent manner. Signal Transduct Target Ther 2023, 8, 63. [Google Scholar] [CrossRef] [PubMed]
- Sukkar, S.G.; Cogorno, L.; Pisciotta, L.; Pasta, A.; Vena, A.; Gradaschi, R.; Dentone, C.; Guiddo, E.; Martino, E.; Beltramini, S.; et al. Clinical efficacy of eucaloric ketogenic nutrition in the COVID-19 cytokine storm: A retrospective analysis of mortality and intensive care unit admission. Nutrition 2021, 89, 111236. [Google Scholar] [CrossRef]
- Salvatore, S.; Heuschkel, R.; Tomlin, S.; Davies, S.E.; Edwards, S.; Walker-Smith, J.A.; French, I.; Murch, S.H. A pilot study of N-acetyl glucosamine, a nutritional substrate for glycosaminoglycan synthesis, in paediatric chronic inflammatory bowel disease. Aliment Pharmacol Ther 2000, 14, 1567–1579. [Google Scholar] [CrossRef]
- Grigorian, A.; Araujo, L.; Naidu, N.N.; Place, D.J.; Choudhury, B.; Demetriou, M. N-acetylglucosamine inhibits T-helper 1 (Th1)/T-helper 17 (Th17) cell responses and treats experimental autoimmune encephalomyelitis. J Biol Chem 2011, 286, 40133–40141. [Google Scholar] [CrossRef]
- Richter, J.; Capkova, K.; Hribalova, V.; Vannucci, L.; Danyi, I.; Maly, M.; Fiserova, A. Collagen-induced arthritis: severity and immune response attenuation using multivalent N-acetyl glucosamine. Clin Exp Immunol 2014, 177, 121–133. [Google Scholar] [CrossRef]
- Azuma, K.; Osaki, T.; Wakuda, T.; Tsuka, T.; Imagawa, T.; Okamoto, Y.; Minami, S. Suppressive effects of N-acetyl-D-glucosamine on rheumatoid arthritis mouse models. Inflammation 2012, 35, 1462–1465. [Google Scholar] [CrossRef]
- Lee, S.U.; Li, C.F.; Mortales, C.L.; Pawling, J.; Dennis, J.W.; Grigorian, A.; Demetriou, M. Increasing cell permeability of N-acetylglucosamine via 6-acetylation enhances capacity to suppress T-helper 1 (TH1)/TH17 responses and autoimmunity. PloS one 2019, 14, e0214253. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.E. An observational cohort study to assess N-acetylglucosamine for COVID-19 treatment in the inpatient setting. Ann Med Surg (Lond) 2021, 68, 102574. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, M.; De Berardis, B.; Bigioni, I.; Mariano, A.; Superti, F.; Scotto d’Abusco, A. In Vitro Antiviral and Anti-Inflammatory Activities of N-Acetylglucosamine: Development of an Alternative and Safe Approach to Fight Viral Respiratory Infections. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef] [PubMed]
- Kielbasa, W.; Goldsmith, P.; Donnelly, K.B.; Nuthall, H.N.; Shcherbinin, S.; Fleisher, A.S.; Hendle, J.; DuBois, S.L.; Lowe, S.L.; Zhang, F.F.; et al. Discovery and clinical translation of ceperognastat, an O-GlcNAcase (OGA) inhibitor, for the treatment of Alzheimer’s disease. Alzheimers Dement (N Y) 2024, 10, e70020. [Google Scholar] [CrossRef] [PubMed]
- Permanne, B.; Sand, A.; Ousson, S.; Neny, M.; Hantson, J.; Schubert, R.; Wiessner, C.; Quattropani, A.; Beher, D. O-GlcNAcase Inhibitor ASN90 is a Multimodal Drug Candidate for Tau and alpha-Synuclein Proteinopathies. ACS Chem Neurosci 2022, 13, 1296–1314. [Google Scholar] [CrossRef]
- Selnick, H.G.; Hess, J.F.; Tang, C.; Liu, K.; Schachter, J.B.; Ballard, J.E.; Marcus, J.; Klein, D.J.; Wang, X.; Pearson, M.; et al. Discovery of MK-8719, a Potent O-GlcNAcase Inhibitor as a Potential Treatment for Tauopathies. J Med Chem 2019, 62, 10062–10097. [Google Scholar] [CrossRef]
- Pradeu, T.; Thomma, B.; Girardin, S.E.; Lemaitre, B. The conceptual foundations of innate immunity: Taking stock 30 years later. Immunity 2024, 57, 613–631. [Google Scholar] [CrossRef]
- Wells, A.I.; Coyne, C.B. Type III Interferons in Antiviral Defenses at Barrier Surfaces. Trends Immunol 2018, 39, 848–858. [Google Scholar] [CrossRef]
- Tomalka, J.A.; Suthar, M.S.; Diamond, M.S.; Sekaly, R.P. Innate antiviral immunity: how prior exposures can guide future responses. Trends Immunol 2022, 43, 696–705. [Google Scholar] [CrossRef]
- Vivier, E.; Rebuffet, L.; Narni-Mancinelli, E.; Cornen, S.; Igarashi, R.Y.; Fantin, V.R. Natural killer cell therapies. Nature 2024, 626, 727–736. [Google Scholar] [CrossRef]
- Ngo, C.; Garrec, C.; Tomasello, E.; Dalod, M. The role of plasmacytoid dendritic cells (pDCs) in immunity during viral infections and beyond. Cell Mol Immunol 2024, 21, 1008–1035. [Google Scholar] [CrossRef] [PubMed]
- Embgenbroich, M.; Burgdorf, S. Current Concepts of Antigen Cross-Presentation. Front Immunol 2018, 9, 1643. [Google Scholar] [CrossRef] [PubMed]
- Carroll, S.L.; Pasare, C.; Barton, G.M. Control of adaptive immunity by pattern recognition receptors. Immunity 2024, 57, 632–648. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Wang, X.; Ho, W.Z. Roles of Macrophages in Viral Infections. Viruses 2024, 16. [Google Scholar] [CrossRef]
- Brancewicz, J.; Wojcik, N.; Sarnowska, Z.; Robak, J.; Krol, M. The Multifaceted Role of Macrophages in Biology and Diseases. Int J Mol Sci 2025, 26. [Google Scholar] [CrossRef]
- Rodrigo-Munoz, J.M.; Sastre, B.; Canas, J.A.; Gil-Martinez, M.; Redondo, N.; Del Pozo, V. Eosinophil Response Against Classical and Emerging Respiratory Viruses: COVID-19. J Investig Allergol Clin Immunol 2021, 31, 94–107. [Google Scholar] [CrossRef]
- Rupani, H.; Busse, W.W.; Howarth, P.H.; Bardin, P.G.; Adcock, I.M.; Konno, S.; Jackson, D.J. Therapeutic relevance of eosinophilic inflammation and airway viral interactions in severe asthma. Allergy 2024, 79, 2589–2604. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, Y.; Zhu, L. Role of neutrophils in acute viral infection. Immun Inflamm Dis 2021, 9, 1186–1196. [Google Scholar] [CrossRef]
- Sasaki, H.; Miyata, J.; Kawana, A.; Fukunaga, K. Antiviral roles of eosinophils in asthma and respiratory viral infection. Front Allergy 2025, 6, 1548338. [Google Scholar] [CrossRef] [PubMed]
- Gebremeskel, S.; Schanin, J.; Coyle, K.M.; Butuci, M.; Luu, T.; Brock, E.C.; Xu, A.; Wong, A.; Leung, J.; Korver, W.; et al. Mast Cell and Eosinophil Activation Are Associated With COVID-19 and TLR-Mediated Viral Inflammation: Implications for an Anti-Siglec-8 Antibody. Front Immunol 2021, 12, 650331. [Google Scholar] [CrossRef] [PubMed]
| Virus | Substrate | Identified Site | Functional impact | Reference |
|---|---|---|---|---|
| KSHV | ORF3 | S278 | O-GlcNAc transferase inhibits KSHV propagation and modifies replication relevant viral proteins as detected by systematic O-GlcNAcylation analysis. | [47] |
| ORF10 | T225, T338, S594, T632, T709 | |||
| ORF8 | S92 | |||
| ORF44 | S727 | |||
| ORF21 | S62 | |||
| ORF29 | n.d. | |||
| ORF75 | n.d. | |||
| RTA (ORF50) | T366, T367 | O-GlcNAc suppresses transactivation & lytic reactivation | [48] | |
| SARS-CoV-2 | Spike | S659 | Spike stability and pseudo-particle packaging | [52] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




