1. Introduction
Amyotrophic Lateral Sclerosis (ALS) is a fatal neurodegenerative disease specifically affecting motor neurons with no known cures or effective treatments. The disease is heterogeneous in its pathophysiology, but is uniformly defined by the continual degeneration of upper and lower motor neurons causing the progressive reduction of muscle function, inevitably culminating in the loss of respiratory muscle function and fatal respiratory failure. Approximately 90% of ALS cases are sporadic (sALS) with no known inherited genetic mutations, and the remaining 10% of familial cases (fALS) are causally linked to genetic mutations which are usually inherited in an autosomal-dominant manner [
1,
2]. Mutations to the
sod1 gene encoding Cu/Zn superoxide dismutase (SOD1) account for 20% of fALS cases [
3]. SOD1 is a ubiquitously expressed, primarily cytosolic, oxireductase metalloenzyme that catalyzes the disproportionation of harmful superoxide radicals to mitigate oxidative stress. The native structure of the wild-type (WT) enzyme is a 32 kDa homodimer consisting of two 153 amino acid monomers, each including one catalytic Cu
1+/2+ ion cofactor, one structurally stabilizing Zn
2+ ion cofactor, and one intramonomeric disulfide bond [
4]. WT holo SOD1 is one of the most stable naturally occurring proteins with a melting point of up to 95°C and with enzymatic activity conserved even in the presence of strong denaturing agents such as urea or guanidine hydrochloride [
4]. The notable stability of SOD1 is a result of the intramonomeric disulfide bonds and incorporation of the metal ion cofactors, particularly the zinc cofactor which facilitates a hydrogen bonding network, and finally dimerization through hydrophobic intermonomeric contacts. In contrast to dimeric holo-SOD1, monomeric SOD1 lacks most or all of these structure-stabilizing elements, is intrinsically unstable with a greatly reduced melting point (42.9°C), and has a tendency to adopt non-native conformations and associate with other misfolded SOD1 species [
5].
More than 150 fALS-causative mutations to SOD1 have been identified, the majority of which are amino acid substitutions [
6]. Mutations influence the folding, stability, maturation, and enzymatic function of SOD1 to varying degrees, disparately altering the SOD1 conformational landscape and leading to accumulation of SOD1 aggregates in the spinal cords of patients [
2,
6,
7]. Despite the fact that these mutations all lead to an ALS disease phenotype, albeit to different degrees of severity, fALS mutation sites are scattered throughout the SOD1 sequence, disrupting various aspects of the proteins structure [
6,
8,
9]. Mutants of SOD1 are categorized into two groups according to the type of structural distortion they exert on the protein. Mutations that produce relatively subtle changes to the protein’s native fold are called wild-type like (WTL) mutations while mutations that directly distort the metal binding region are called metal-binding region (MBR) mutants. The changes to the structure, metal binding, and enzymatic activity observed in WTL and MBR mutants differ significantly, and there is also significant diversity in the biophysical properties of mutants within each category [
4]. Further, the indirect modulation of metal binding region residues through allosteric pathways impacts the metal binding affinity of various WTL mutants as well.
The ALS-associated pathogenicity of misfolded SOD1 has been extensively proven to result from a gain of toxic function rather than the loss of enzymatic activity. Many WTL mutants, for example, retain some degree of enzymatic activity and SOD1 knockout mice do not exhibit ALS-like motor neuron degeneration [
4,
7,
10,
11,
12]. Insoluble aggregates of mutant SOD1 were found to accumulate in fALS motor neurons and early studies suggested that motor neuron degeneration could be a direct result of large aggregate formation.[
10] Further research, however, has disproven this hypothesis as growing evidence suggests that the toxic species are a variety of soluble oligomers of misfolded SOD1, while disease-correlated aggregates instead serve a protective function by sequestering these toxic species [
13,
14,
15,
16,
17]. Additionally, pathogenic misfolding of WT-SOD1 has been reported in sALS cases by post-mortem analyses.[
3,
18,
19,
20].Soluble oligomeric species of misfolded WT-SOD1 in sALS spinal tissue have been shown to react with antibodies that specifically recognize conformational epitopes present in misfolded species of fALS mutant SOD1 [
3,
18]. These discoveries suggest that pathogenic SOD1 species in familial and sporadic cases can adopt similar aberrant conformations.
Considering that a common ALS phenotype is achieved as a result of dissimilar structural perturbations to SOD1, and that similarly misfolded SOD1 species are present in fALS and sALS patients, the analysis of non-genetic factors including post-translational modifications (PTMs) that influence SOD1 folding is vital to sALS research. SOD1 is ubiquitously expressed throughout the body and it is still not understood why motor neurons are the targeted cell type. We hypothesized that there could be structure disrupting PTMs to SOD1 that are exclusive to motor neurons and may be the reason for cell type specificity in ALS. Several asparagine deamidated variants of SOD1, for example, have been investigated by computational methods and found to have a significant influence on the propensity to misfold and adopt mutant-like ALS relevant isoforms [
21]. Some of such deamidated forms have been identified with increased frequency in fALS cases [
22].
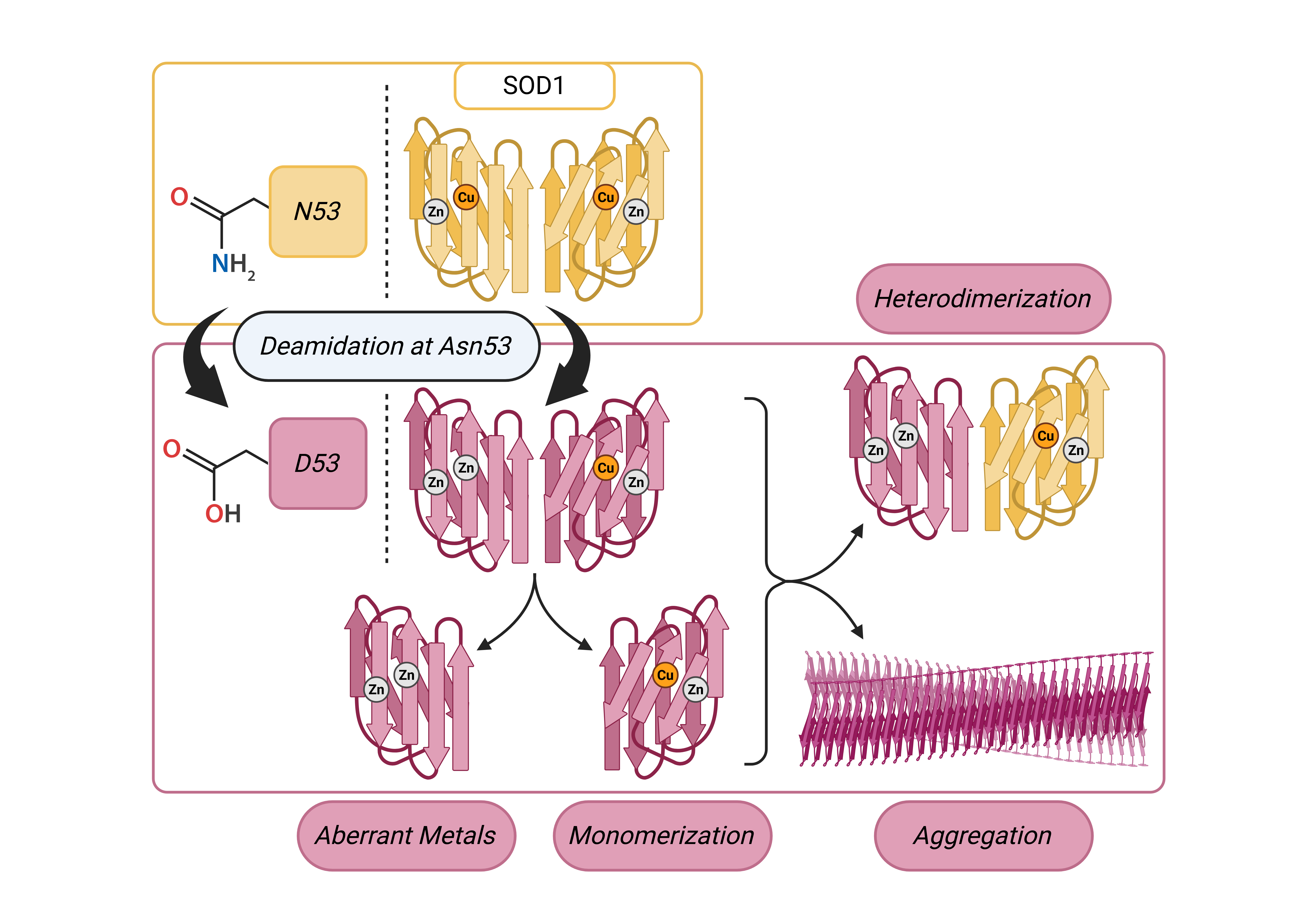
One possible target for asparagine deamidation, Asn53 (N53), is both predicted to disrupt dimer stability and is detected with a significantly higher frequency in fALS cases [
21,
22]. The N53 residue is located near the dimer interface within both the Zn binding loop and the GDNT motif of the disulfide loop through Cys57, which is involved in copper chaperone association. (
Figure 1) The deamidation of N53 introduces a non-native negative charge, thereby modifying the electrostatic environment of this region vital for the structural stabilization of SOD1.
Here we describe the identification of deamidation of SOD1 at N53 in WT human induced pluripotent stem cell (iPSC)-derived motor neurons and characterize how this PTM affects SOD1 structural stability. This deamidation was not detected in SOD1 extracted from non-neuronal cultures differentiated from the same iPSC line, suggesting that the PTM may be specific to or may occur with greater frequency in motor neurons. Using recombinantly expressed N53D SOD1, a mutant mimicking N53 deamidated WT SOD1, we measured increased formation of monomer, decreased thermal stability, and increased aggregation propensity when compared to WT SOD1. Investigation of SOD1 metal cofactors showed that N53D SOD1 binds more Zn than WT, exceeding the quantity of Zn binding sites, suggesting longer range structural perturbations affecting the Cu binding site. Molecular dynamic simulations echo these findings showing more conformational flexibility in the N53D mutant. Interestingly, modeling predicts the N53D-WT heterodimer to be more stable than the WT-WT homodimer suggesting another route to toxicity in vivo. Through the identification and characterization of SOD1 PTMs, we gain a better understanding of the in vivo landscape that can lead to SOD1 misfolding and ultimately ALS.
2. Materials and Methods
Cell Culture and SOD1 Immunoprecipitation
Our research uses de-identified, established human induced pluripotent stem cell (iPSC) lines. The creators of the lines, Harvard University (Harvard Stem Cell Institute) maintain the signed donor consent forms. All research has a “non-human subject” designation from Wesleyan’s Institutional Review Board and approval from Wesleyan’s Embryonic Stem Cell Research Oversight committee.
iPSCs were differentiated to motor neurons and astrocytes using established protocols with no modifications.[
23,
24] The WT 1016a iPSC and the SOD1 A4V patient derived iPSC line were obtained from the Harvard Stem Cell Core. iPSC were maintained on ESC qualified Matrigel (Corning) coated dishes and mTesr Plus medium (Stem Cell Technologies) before differentiation. Post differentiation cellular identity was confirmed with immunofluorescent labeling.(
Supplemental Figure S1)
To detach adherent cells, culture medium was aspirated from the plate and immediately replaced with Accutase at appropriate volume to fully cover cells. The plate was then incubated at 37°C for 5 minutes. Detached cells in Accutase were added to media, gently pipetted to create a single-cell suspension, and centrifuged at 300 g for 5 minutes to pellet. The cell pellet was washed 2x with sterile PBS to remove media before being stored at -80°C.
Frozen cell pellets were first lysed with radioimmunoprecipitation assay buffer (RIPA) supplemented with 10 µL/mL Halt protease/phosphatase inhibitor, and 2 µL/mL DNAse for 30 minutes on ice (Thermo Scientific). Lysates were next subjected to three freeze/thaw cycles before clarification by centrifugation at 16,000 x g x 10 minutes. The supernatant was collected, and total protein concentration was determined via BCA analysis. A SOD1 antibody cocktail consisting of 10 µg each of two antibodies (ProteinTech cat # 10269-1-AP and Enzo cat # ADI-SOD-100-J) was added to normalized lysate and the mixture was incubated at 4°C overnight while mixing. The next day, 30 ug of Sera-Mag™ SpeedBeads (Magnetic Protein A/G beads) were added to the mixture and incubated for 1 hour at room temperature. Beads were collected with a magnetic stand and the supernatant was discarded. Washing and binding was performed following the manufacturer’s recommendations.
Bottom-Up Untargeted Protein Identification and Label-Free Quantification by Tandem Mass Spectrometry
Immunoprecipitated samples of cellular SOD1 were analyzed alongside commercial SOD1 protein standard for coverage reference and comparison.. All samples were reduced with 5 mM fresh DTT for 1.5 hours at room temperature, alkylated with 10 mM fresh iodoacetamide for 45 minutes at room temperature protected from light, and digested with trypsin (Promega cat # V5113) at 1:20 enzyme:protein (w/w) overnight at 37°C. Peptide-containing supernatant was removed from beads for IP samples. All peptide solutions were acidified and desalted over C18 resin spin columns (Pierce cat # 89870). Samples were dried to completion, resuspended in a minimal volume of Fisher Optima grade water with 0.1% formic acid, and quantified by absorbance at 280 nm. The injection amount was normalized across all samples.
Samples were analyzed using a Thermo Scientific Ultimate 3000 RSLCnano ultra-high performance liquid chromatography (UPLC) system coupled to a high-resolution Thermo Scientific Q-Exactive HF mass spectrometer. Each sample was injected onto a nanoEase M/Z Peptide BEH C18 column (1.7 μm, 75 μm x 250 mm, Waters Corporation) and separated by reversed-phase UPLC using a gradient of 4-30% Solvent B (0.1% formic acid in Fisher Optima LC/MS grade acetonitrile) over a 50 minute gradient at 300 nL/min flow, followed by a ramp of 30-90% Solvent B over 10 minutes, for a 60 minute total gradient.
Peptides were eluted directly into the Q Exactive HF using positive mode nanoflow electrospray ionization. MS1 scans were acquired at 60,000 resolution, with an AGC target of 1e6, maximum ion time of 60 ms, and a scan range of 300-1800 m/z. Data-dependent MS2 scans were acquired at 15,000 resolution, with an AGC target of 1e5, maximum ion time of 40 ms, isolation window of 2.0 m/z, loop count of 15, normalized collision energy of 27, dynamic exclusion window of 30 s, and charge exclusion applied for unassigned, +1, and > +8 charged species.
Raw data were searched using MaxQuant v1.6.10.43 and its embedded Andromeda search engine, and quantified using label-free quantification [
25]. Databases included the Homo sapiens reference proteome downloaded from UniProt (UP000005640, downloaded Sept 11, 2020), the MaxQuant contaminants database, and a custom user-provided SOD1 sequence. Minimum peptide length was set to 7 residues and maximum mass to 4600 Da. Up to 5 variable modifications per peptide were allowed from the following list: oxidation (M), acetylation (protein N-term), acetylation (K), deamidation (N/Q), GlyGly (K), and phosphorylation (S/T/Y). Carbamidomethylation was fixed for cysteine residues. Match between runs was enabled, with a matching time window of 0.7 minutes. All results were filtered to a 1% false discovery rate at the peptide and protein levels using the target-decoy approach; all other parameters were left as default values. MaxQuant output files were imported into Scaffold (Proteome Software, Inc.) for data visualization and subsequent analyses.
Expression and Purification of Recombinant GST-Fusion SOD1
A pCW vector encoding GST-fusion H46R-SOD1, originally provided by Biogen, Inc., was used as the backbone for the construction of GST-fusion WT- and N53D-SOD1 expression plasmids. The pCW plasmid was modified by site directed mutagenesis (SDM) using 18-22 nucleotide primers to replace a single nucleotide per reaction, introducing a single amino acid substitution. SDM was first performed to generate a GST-WT-SOD1 construct by R46H (CGT to CAT) substitution. After the GST-WT-SOD1 construct was transformed into DH5α cells for cloning, extracted plasmid was purified from and sequenced before further SDM. The N53D (AAT to GAT) substitution was then similarly introduced to the WT construct, cloned, and sequence confirmed. BL21-Codon+ E. Coli were transformed with the GST-SOD1 plasmids and the recombinant protein was expressed in LB media containing 100 µg/mL ampicillin at room temperature with 200 rpm shaking. Protein expression was triggered with the addition of IPTG to a final concentration of 0.2 mM at OD
600 ≈ 0.6. Expression continued for 8-16 hours before cells were harvested by 10 minutes of centrifugation at 5,000 x g. Cell pellets were lysed by first performing a freeze-thaw cycle in liquid nitrogen and warm water, then again after resuspension in 3-5 mL lysis buffer per gram (50 mM Tris, 400 mM NaCl, 1 mM EDTA, 5 mM DTT, 1 mM PMSF, 1% Triton X-100, pH 8.0). Cells were further lysed by sonication on ice at 80% amplitude for 10 cycles of 15 seconds on, 50 seconds off. Benzonase nuclease was then added to the lysates at approximately 1 unit per milliliter of lysis buffer before mixing at room temperature for 30-60 minutes, or until sufficient digestion of DNA was observed. Lysates were then clarified by centrifugation at 10,000 x g for 30 minutes. After pre-equilibrating with 3X bed volumes of lysis buffer, the supernatants were pumped at 0.5-1 mL/min over a column packed with 15 mL of GST affinity glutathione agarose resin (GoldBio cat # G-250) using an AKTA Explorer FPLC P-960 sample pump at room temperature. The GST affinity resin was then thoroughly washed with 5-10X bed volumes of wash buffer (50 mM Tris, 400 mM NaCl, 1 mM EDTA, pH 8.0) at 1-3 mL/min before resin-bound SOD1-GST was eluted with 7-15X bed volumes of elution buffer (50 mM Tris, 400 mM NaCl, 1 mM EDTA, 10 mM L-Glutathione, pH 8.0). Eluted protein was collected in 10 mL fractions and SDS-PAGE of each fraction was performed to identify peak fractions of SOD1-GST fusion protein and assess the purity of the protein samples. (
Supplemental Figure S2) The GST affinity resin was regenerated according to manufacturer’s specifications after each use.
Processing and Maturation of Recombinant SOD1-GST
Peak fractions of SOD1-GST fusion protein were pooled and 10 mM DTT was added to the solution to reduce disulfide bonds to glutathione. The reduced protein was then subjected to dialysis against 4 L of wash buffer at 4°C in 6-8 kDA MWCO tubing (Spectra/Por®) to remove reduced glutathione from solution. The dialysis membrane was then buried in powdered PEG 20,000 at 4°C to concentrate the solution by dehydration, and concentrated samples were then subjected to another round of dialysis against GST wash buffer in a fresh dialysis membrane. During this dialysis, the GST tag was cleaved from SOD1 by the addition of HRV-2C Protease according to manufacturer’s specifications (ACRO Biosystems cat # 3CC-N3136). Once total tag cleavage was confirmed by SDS-PAGE after 16-24 hours, the solution was pumped through the regenerated GST affinity resin column to remove the cleaved GST tag and GST-tagged protease. After GST removal was confirmed by SDS-PAGE, the SOD1 solution was subjected to a series of dialysis steps to mature the apoprotein. (
Supplemental Figure S2) The tag cleavage results in a “tag scar” which leaves behind an additional 8 amino acids on the N-terminus (GPLGSPEF). This tag scar is consistent between WT and N53D.
Next, protein was dialyzed against an EDTA-rich buffer (50mM Tris, 200 mM NaCl, 100 mM EDTA, pH 8.0) to remove any contaminating non-native divalent metal ions. Second, the solution was dialyzed against an acidic buffer solution to encourage unfolding of apo-SOD1 (100 mM sodium acetate, 200 mM NaCl, pH 3.8). Third, the solution was dialyzed against a refolding buffer (25 mM MOPS, 200 mM NaCl, pH 6.9) into which metal cofactor addition described in the following section could be performed while maintaining near-neutral pH with minimal precipitate formation. After the addition of metal cofactors, the solution was dialyzed against Tris buffer (25 mM Tris, 200 mM NaCl, pH 7.4) to remove unreacted metals, and then finally against PBS (11.8 mM PO4, 137 mM NaCl, 4.5 mM KCl, pH 7.4) before dimeric Cu/Zn SOD1 was purified by size exclusion chromatography.
Size Exclusion Chromatography (SEC)
Dimeric SOD1 was purified by SEC using an AKTA Explorer FPLC system (Amersham Biosciences). Protein samples were first centrifuged at 10,000 x g for 5 minutes before being loaded into the system in 1-2 mL injections. Protein was run over a Superdex™ 75 Increase 10/300 GL column (Cytiva cat # 29-1487-22) at a flowrate of 0.75 mL/min at room temperature and 1 mL fractions were collected. Dimer fractions for each biological replicate of each Cu/Zn addition order variant were separately pooled, and each dimer pool was then concentrated to approximately 30 µM using 10 kDa spin columns (Vivaspin®20, Cytiva). Spin column membranes were first treated with sterile filtered 20% glycerol to minimize protein binding. Peak integration tools in Unicorn 5.31 software were used to integrate the dimer (12 mL) and monomer peaks (20 mL), to calculate a ratio of monomeric to dimeric SOD1. The relative quantities of dimer and monomer were also used to estimate the ∆∆G of dimerization for each variant using the following equation: where for the holoprotein standard (KWTZn) and the SOD1 variant (Kx). Absorbance at 280 nm SEC chromatograms from at least 3 independent growths of WT and N53D SOD1 were used to calculate the monomer:dimer ratios.
SEC purified SOD1 was subjected to intact mass spectrometry to confirm sequence identity. Mass spectrometry also confirmed that the intramonomer disulfide was present in both species (-2 Da from expected mass, cysteine disulfide bond between AA65-AA154, corresponding to C57 and C146 in the native sequence shifted by the 8 amino acid tag scar). (Supplemental
Figure S3)
Intact Protein Analysis by Tandem Mass Spectrometry
Purified recombinant WT and N53D SOD1 was quantified by spectrophotometer absorbance at 280 nm. Sample was diluted to 10 pmol/μL in Solvent A (0.1% formic acid in Fisher Optima grade water) and analyzed using a Thermo Scientific Ultimate 3000 RSLCnano ultra-high performance liquid chromatography (UPLC) system coupled to a high-resolution Thermo Scientific Eclipse Tribrid Orbitrap mass spectrometer.
Chromatography was performed using a nanoEase M/Z Protein BEH C4 column (1.7 μm, 75 μm x 100 mm, Waters Corporation) heated to 50°C with a reversed-phase UPLC gradient of 4-30% Solvent B (0.1% formic acid in Fisher Optima LC/MS grade acetonitrile) over a 50 minute gradient at 300 nL/min flow, followed by a ramp of 30-90% Solvent B over 10 minutes, for a 60 minute total gradient. Proteins were eluted directly into the Eclipse using positive mode nanoflow electrospray ionization with a capillary voltage of 2200 V. MS1 scans were acquired at 120,000 resolution, with an AGC target of 4e
5, maximum ion time set to Auto, RF lens setting of 30%, and a scan range of 300-2000 m/z. Data-dependent MS2 scans were acquired in the Orbitrap at a resolution of 60,000, for any ions over 2.5e
4 intensity threshold, with AGC target set to 8e
5, maximum ion time set to Auto, mass range set to Normal, scan range set to Auto, isolation window set to 2 m/z, and a fixed cycle time of 3 sec. ETD fragmentation was applied with a 20 ms reaction time, ETD reagent target of 3e
5, and 12% supplemental activation energy applied. Dynamic exclusion was set to exclude for 15 sec after 1 observation, for charge states 8-40. Peak deconvolution was performed in UniDec [
26], with the following parameters: charge range 9-20, mass range 2,000 – 50,000, mass sampled every 1 Da, 5 Da peak detection range, 0.1 peak detection threshold.
Circular Dichroism (CD)
Circular dichroism experiments were performed on a Jasco J-810 polarimeter to measure and monitor changes to the secondary structure of SOD1 variants in response to heating. Samples were diluted in PBS to ~14 µM and subjected to a thermal melt from 5-95°C. CD experiments were carried out on 2 independent growths of WT and N53D SOD1 (2 biological replicates) in duplicate (2 experimental replicates). The exact concentration of each sample from A280nm was used to convert the CD spectra to molar ellipticity and the resulting 4 CD runs for each species were averaged to create the spectrums shown.
Heated Dynamic Light Scattering (DLS)
As a complementary approach to determine differences in structural integrity and aggregation propensity, the hydrodynamic radius of purified SOD1 was measured by dynamic light scattering using a Horiba nanoPartica SZ-100V2 Series particle analyzer. After centrifugation at 10,000 x g for 5 minutes followed by filtration through 0.45 micron syringe-tip filters, samples of ~30 µM SOD1 were measured at a 90° angle at temperatures spanning 30°C to 90°C in 10°C intervals, and the Z-average hydrodynamic radius of particles in solution was averaged across viable scans. Samples were held at each temperature for 10 minutes before each set of DLS measurements, with each measurement consisting of 10 separate scans collected over a 90 second period.
Each 10-scan data set was recalculated using a channel range determined to be most suitable for the specific autocorrelation data. Channel ranges were determined to be suitable first by ensuring the autocorrelation decay was sufficiently captured within the range (i.e. the decay curve occurred before the final point of the channel range) and, where this criteria applied to multiple tested channel ranges, the calculation that yielded the lesser standard deviation of Z-average radii was selected. Prior to finally determining the appropriate channel range, outlier scans were identified by several approaches in the following order: first, scans that yielded non-viable autocorrelation data, such as those showing immediate decay after a single point, were eliminated from the sets. Next, scans yielding a Z-average radius beyond two standard deviations from the average of the set were eliminated. Finally, data with a polydispersity index beyond 10 were eliminated from the set. After the identification and elimination of scan outliers, 3 to 5 experimental replicates of each variant were analyzed, each including protein prepared from 2 to 3 independent growths.
Cu2+/Zn2+ Quantification
To determine the abundance of metal cofactors present in the SOD1 variants, divalent copper and zinc ion concentrations were simultaneously quantified with a colorimetric 4-(2-pyridylazo) resorcinol (PAR) chelate complex assay [
27]. The solutions described below were prepared in MilliQ water that was first treated with Chelex®100 resin to remove any contaminating divalent metal ions. A stock solution of 10 mM PAR was prepared by dissolving PAR in MilliQ water with a small quantity of 1 M NaOH to facilitate total dissolution. The 4X PAR assay working solution (400 µM PAR, 200 mM HEPES, pH 8.0) was then prepared by mixing 1.6 mL of 10 mM PAR with 35 mL of HEPES buffer, adjusting pH with 1 M NaOH and/or HCl, and then diluting to a final volume of 40 mL. All PAR solutions were protected from light by wrapping tubes and plates in aluminum foil and were stored at 4°C. Solutions of 1 M ZnCl
2 and CuCl
2 were prepared by dissolving the appropriate amount of each solid in MilliQ water, and 250 µM working solutions of each were then prepared by sequential dilutions. A solution of 8 M guanidine HCl (GuHCl) was prepared in warm MilliQ water and also subjected to CHELEX resin to remove metal contaminants.
In a clear 96-well plate (FisherSci cat # 12565501), duplicate or triplicate samples of each SOD1 variant were first denatured in 7 M GuHCl to release bound metal cofactors into solution; 25 µL of approximately 30 µM SOD1 were thoroughly mixed with 168.75 µL of deionized 8 M GuHCl. After incubating for 1 hour at room temperature, 56.25 µL of 4X PAR working solution was added to each well for a final assay volume of 250 µL and final sample concentration of 0.1X. Standard curves for Cu2+ and Zn2+ were established spanning a range of either 0-17.5 or 0-10 µM by diluting the appropriate volume of the 250 µM metal standards into PBS for a sample volume of 25 µL with a concentration of 10X the target standard concentration. For standards, a 1X PAR solution was prepared as 3:1 mixture of GuHCl and 4X PAR. After the addition of 225 µL of 1X PAR to each standard well (i.e. 168.75 µL of 8 M GuHCl and 56.25 µL of 4X PAR per well), all wells consisted of 0.1X initial concentration of samples or standards, 5.4 M GuHCl, 90 µM PAR, and 45 mM HEPES.
Absorbance spectra spanning 300-600 nm were then measured in 5 nm increments using a VarioSkan Lux™ microplate reader (Thermo Scientific) and SkanIt software (v7.0.2). (
Supplemental Figure S4) Spectra for standards and protein samples were then processed in R to establish a standard series for PAR:Cu
2+ and PAR
2:Zn
2+ complexes. With these standards, spectral decomposition of the multicomponent solutions of PAR and unfolded SOD1 were performed by linear least squares fit analysis to quantify the weights of the constituent spectra of PAR, PAR:Cu
2+, and PAR
2:Zn
2+ complexes. The resulting metal concentrations contributing to the composite PAR/SOD1 spectra were then divided by the concentration of SOD1 monomers to determine the relative quantities of each cofactor per binding site in each variant. The PAR assay was performed for each of 3 independent growths of each SOD1 variant.
In silico preparation
The reference PDB structure 1HL5 was used and a GST-tag scar was added using cyclic coordinate descent loop modeling in the Rosetta macromolecular modeling software such that the resulting primary structure matched that of the recombinant SOD1 used in this study [
28]. Cu
2+ was selectively removed from all variants in PyMOL to match the measured over incorporation of Zn
2+. Mutation at N53 was performed using PyMOL. Metal-coordinating histidine residues and an aspartic acid residue were modified based on previous literature [
29].
Molecular Dynamics (MD) and Alchemical Morphing
Energy minimization was carried out using GROMACS. Energy minimized structures were then equilibrated at constant volume and temperature for 200 ps. Following that, structures were equilibrated in three 20 ps NPT simulations using all-atom position restraints of 500 kJ, 250 kJ, and 125 kJ. NPT equilibration was then carried out for another 20 ps first with backbone restraints only then with C-alpha restraints only. MD simulations were carried out with GROMACS. The AMBER
ff99SB*-ILDN [
30,
31] force field and TIP3P water model were used. A triclinic box was used with 1.5 Å spacing between the box and the solute. Joung Zn
+ and Cl
- ions were added to the box at a concentration of 150 mM [
32]. Production simulations were run at constant temperature and pressure, with the V-rescale thermostat and the C-rescale barostat holding temperature and pressure constant, respectively. All bonds were constrained using the LINCS algorithm. Free-energy perturbation was performed using the PMX software package and in-house scripts [
33,
34].
3. Results
3.1. SOD1 Deamidation Identified in WT iPSC Derived Neuronal Cultures
We are interested in why there is cell type specificity of SOD1 associated ALS symptoms (degeneration of motor neurons) when SOD1 is ubiquitously expressed throughout the body. We hypothesized there could be cell type specific PTMs on SOD1 that disrupt SOD1 structural integrity, thus causing misfolding only in motor neurons. To identify such cell specific PTMs, we first generated different cell types from single iPSC lines such that the samples have the same genomic background and only the cell identity varied. Next, we immunoprecipitated SOD1 from each cell type and subjected the sample to tandem mass spectrometry (MS/MS) to identify and locate SOD1 PTMs. We used one WT iPSC line (1016a, Harvard Stem Cell Core) to make motor neurons and astrocytes, and, to understand our hypothesis in a SOD1-associated ALS model, we also used an A4V SOD1 patient-derived iPSC line (39b, Harvard Stem Cell Core) to make motor neurons.
Low SOD1 protein yields caused low read depth and therefore low abundant PTMs were unable to be identified. However, several deamidations were found and are summarized in
Table 1. Deamidation at N19 was consistent in both motor neurons and astrocytes but absent in stem cells, which may be of interest in future studies, although this site may be easily deamidated through conditions in the sample preparation. Rare deamidations (1 out of 9 samples) were discovered at Q15, N26, and N53 exclusively in our WT motor neuron preparations. While it was a rare occurrence, the position of N53 near the structurally stabilizing zinc binding loop, intramonomer disulfide bond, and the dimer interface made it particularly interesting for us to examine how this deamidation could affect structural integrity. (
Figure 1) Further, N53 is predicted to have an extremely low likelihood of spontaneous deamidation (less than 20% N53 deamidated after 700 days) making its detection in our motor neuron cultures stand out. [
21]
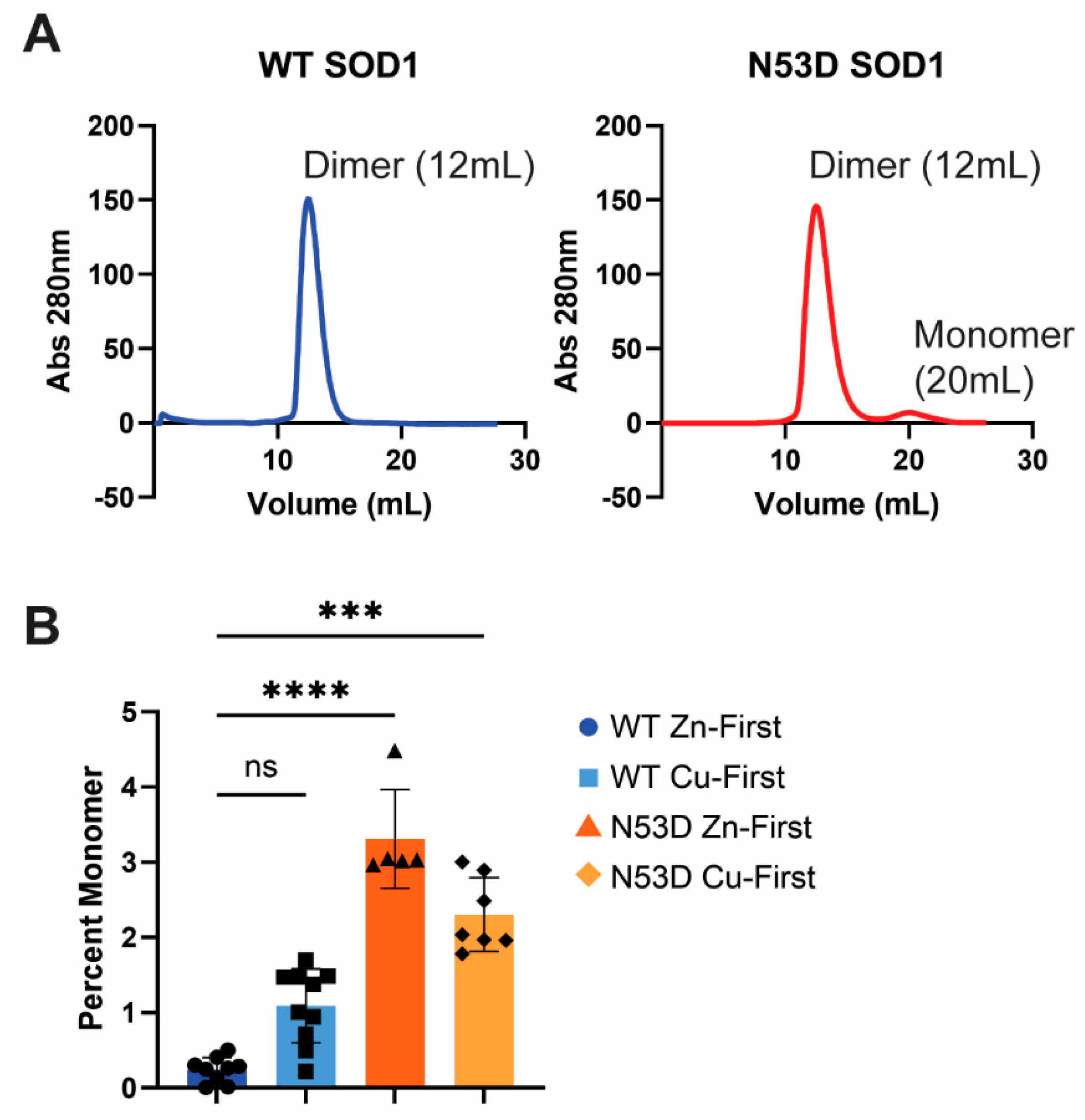
3.2. The Relative Abundance of Monomeric SOD1 is Increased by the N53D Mutation
The relative abundance of dimeric and monomeric SOD1 was determined by the integrated area of the corresponding peaks in SEC A
280 chromatograms; dimeric SOD1 abundance was measured as the integral of the peak at 12 mL, monomeric SOD1 abundance was measured as the integral of the peak at 20 mL, and the ratio of these integrals was then calculated for each purification and averaged across purifications of each biological replicate. (
Figure 2) In all cases, the dimer population dominated the monomer population. As described in the methods, SOD1 samples were prepared with different orders of metal cofactor addition, either with Zn
2+ addition preceding Cu
2+ addition (Zn-first) or Cu
2+ addition preceding Zn
2+ addition (Cu-first) during maturation. The relative abundance of monomers in the Zn-first WT holo-SOD1 control was the lowest with a 0.002 monomer/dimer ratio, while both N53D species had significantly more monomer. N53D SOD1 Zn-first had the most monomer with a 0.037 monomer/dimer ratio, approximately 18.3 times that of WT. WT and N53D Cu-first variants had monomer/dimer ratios of 0.011 and 0.020, respectively. Using these ratios, the ΔΔG of dimerization at 25°C were estimated to be approximately +2.02 kcal/mol, +3.45 kcal/mol, and +2.71 kcal/mol for WT Cu-first, N53D Zn-first, and N53D Cu-first, respectively.
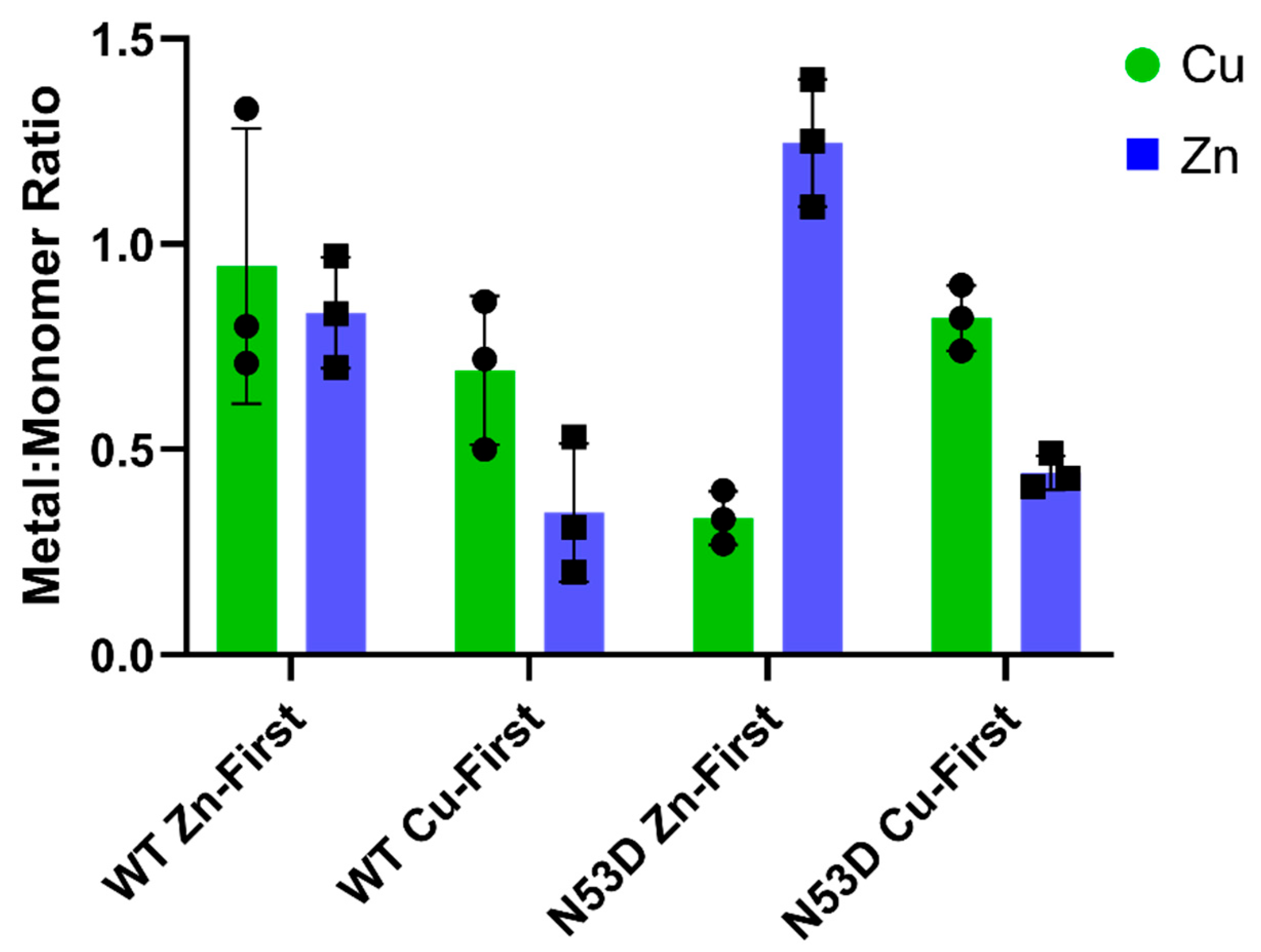
3.3. N53D SOD1 Exhibits Aberrant Metal Binding
In the native holo-SOD1 dimer, each monomer coordinates one copper and one zinc cation. To determine the influence of N53 deamidation on SOD1 metal binding properties, a dimeric WT SOD1 control with a 1:1:1 Cu/Zn/monomer ratio was needed for comparison. Purified dimeric WT SOD1 Zn-first was measured by PAR assay to have approximately 1:1 Cu/Zn ratio and by using the 280 nm extinction coefficient ɛ(A
280) = 5400 M
-1cm
-1, molar ratios of 0.94 Cu
2+ and 0.83 Zn
2+ per monomer were measured. (
Figure 3) The near 1:1 metal ratio and high total metal/monomer ratios were considered sufficient validation of WT SOD1 Zn-first dimer as a native holo-SOD1 control. In contrast to the WT control, dimeric N53D SOD1 Zn-first was measured by PAR assay to have 0.33 Cu
2+ and 1.25 Zn
2+ per monomer, giving a Cu/Zn ratio of approximately 1:4. Notably, this was the only variant consistently measured to have both more zinc cofactors than copper and more zinc cofactors than zinc binding sites. In Cu-first proteins, the relative abundance of copper and zinc were consistent between WT and N53D, while both had a reduced total abundance of both cofactors per monomer; the Cu/Zn ratios were roughly 2:1 with 0.69 Cu
2+ and 0.35 Zn
2+ per monomer in WT, and 0.81 Cu
2+ and 0.44 Zn
2+ per monomer in N53D SOD1. This data further confirms the importance of following the in vivo maturation metal incorporation order (i.e. zinc-first).
3.4. Aggregates of N53D SOD1 Zn-First Form at a Lower Temperature
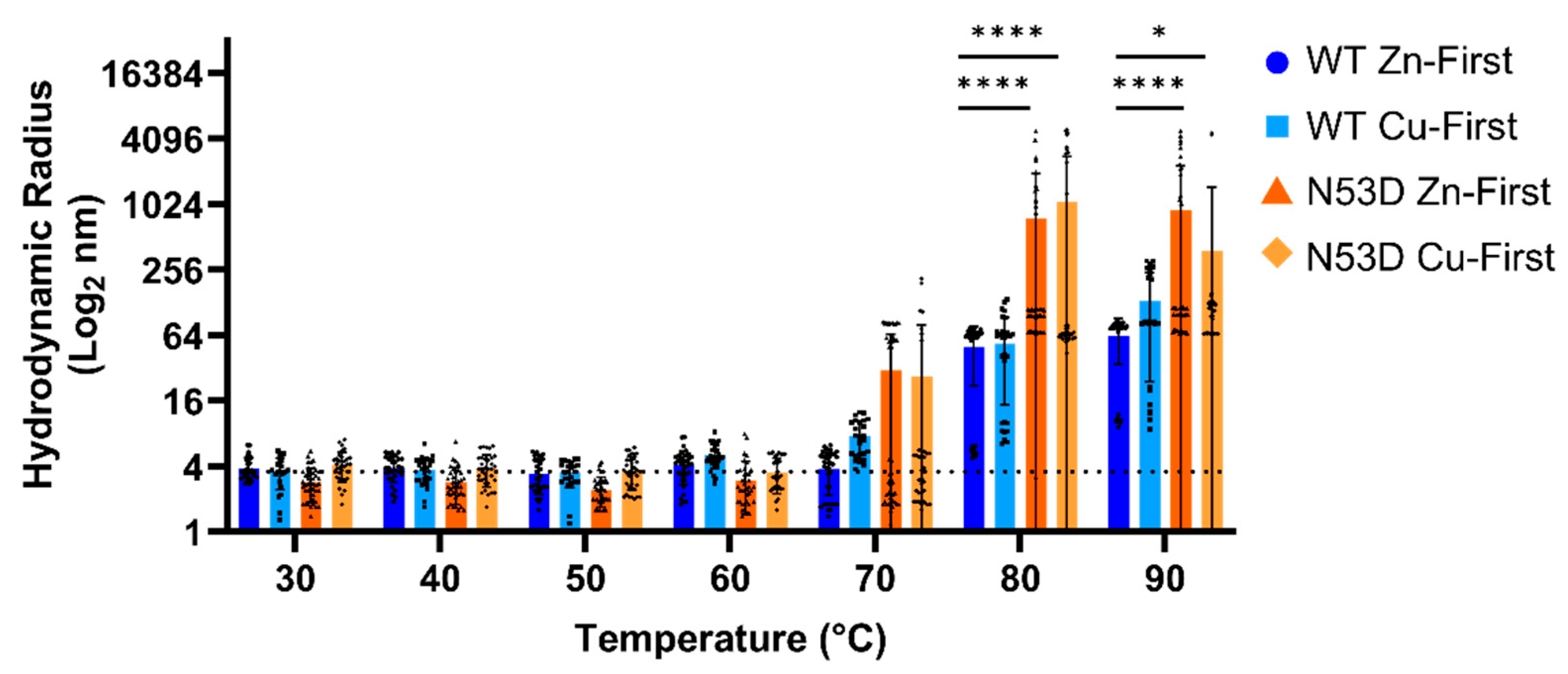
In theory, destabilized variants of SOD1 will more readily lose structural integrity with increasing thermal energy, allowing templated misfolding and the formation of unstructured aggregable monomers. In heated DLS experiments, at 30°C the Z-average radius of all tested variants started with an average radius of roughly 3.6 nm and then increased with increasing temperature to differing degrees. (
Figure 4) However, both the Zn-first and Cu-first N53D started to increase in radii at 70°C as evidenced by the increased standard deviation of the data (N53D Zn-first 30.75 ± 34.98 nm, N53D Cu-first 26.72 ± 53.67 nm) at this temperature compared to Zn-first and Cu-first WT (WT Zn-first 3.79 ± 1.60 nm, WT Cu-first 7.66 ± 2.87 nm). WT SOD1 began to notably increase in size at 80°C (WT Zn-first 49.97 ± 27.78 nm, WT Cu-first 54.17 ± 39.28 nm). All SOD1 species had large aggregates at 80-90°C (i.e. Z-average radius >10 times the radius at 30°C, ~36 nm) with N53D aggregates having significantly larger radii at high temperatures. One replicate of both Zn-first and Cu-first WT SOD1 never achieved such a steep increase throughout the heating process, replicating the reported 95°C melting temperature [
4]. Additionally, we used a 5,000 nm radius cut off for our DLS calculations as DLS is inherently biased towards larger particles, thereby skewing z-average radii. This cutoff prevented the inclusion of some scans of both N53D Cu-first and N53D Zn-first at 90°C as radii exceeding 5,000 nm were calculated. Since DLS measurements assume spherical particle morphology, calculations fail to accurately account for highly elongated species. Because of this, the >5,000 nm radius data was interpreted as an artifact of the presence of large fibrillar aggregates formed by N53D SOD1. This means that the reported large average radii of N53D aggregates at 90°C are an underestimation in some replicates.
3.5. Secondary Structures of Cu- and Zn-First Variants of WT and N53D SOD1 Are Similar Before Heating
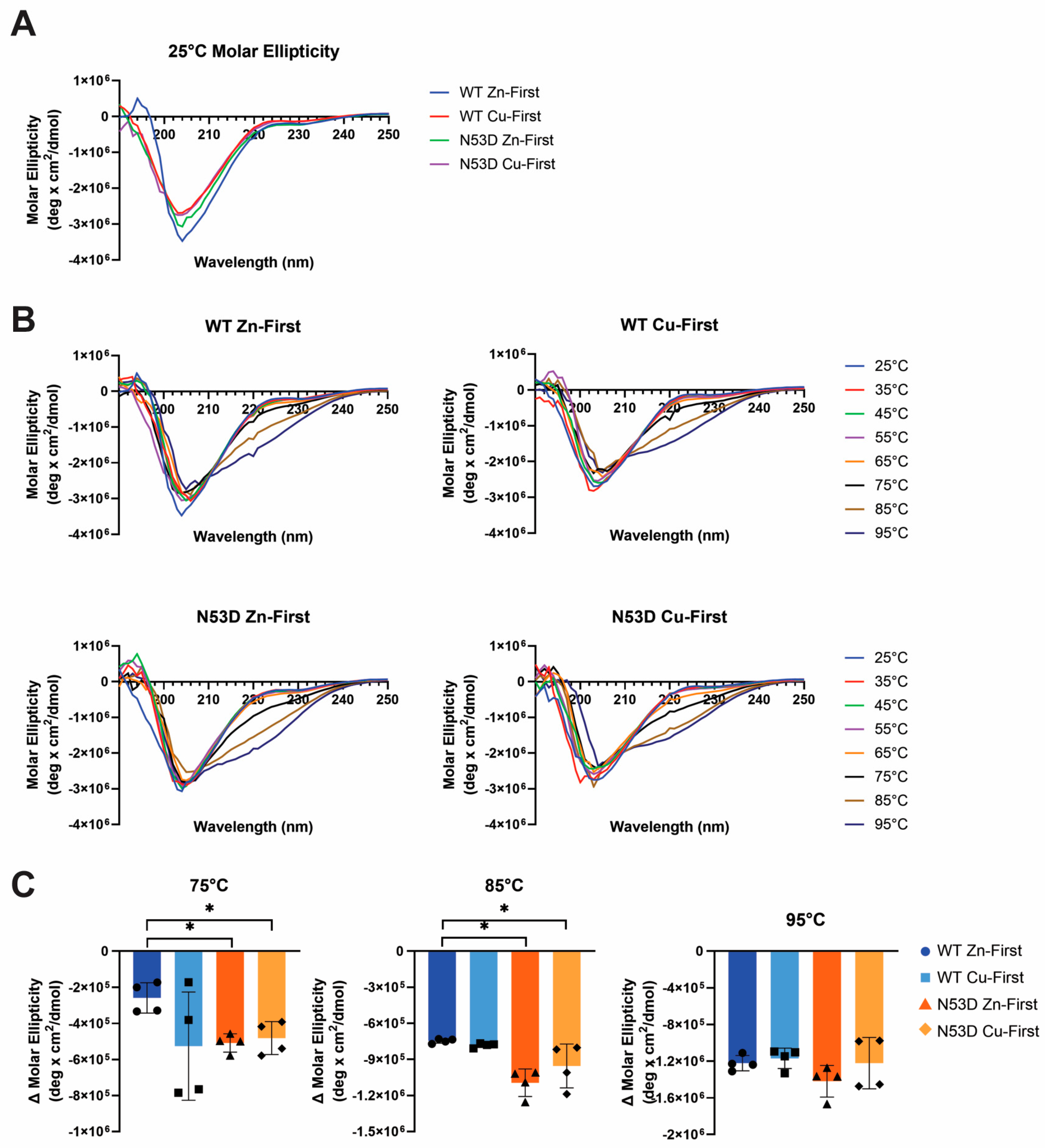
The overpopulation of Zn
2+ and reduced Cu
2+ per monomer in N53D SOD1 Zn-first measured by the PAR assay suggested divergent metal cofactor coordination, which would be expected to change the orientation of metal binding amino acids to account for the distinct geometries of the d9 Cu
2+ and d10 Zn
2+ ions. Additionally, the change in charge from Asn53 to Asp53 had the potential to affect folding. However, CD analysis at 25°C indicated little variation in secondary structure between WT and N53D SOD1 Zn-first variants. (
Figure 5A) Pure alpha helix gives a double minima at approximately 208 nm and 222 nm while purely beta-sheet gives a single minima at 216-218 nm and a positive maximum around 195-198 nm [
35]. One distinction in the room temperature CD is that our native control species, WT Zn-first, was the only species with a positive peak at 195 nm, which suggests beta sheet; the major secondary element of SOD1. (
Figure 1) The control species also has the most negative 208 nm peak, suggesting that our control species has the most alpha helical structure; a minor secondary component of SOD1. At 25°C, the other 3 species, all with aberrant metal content, group together across the spectra range. Once heated, the secondary structure expectedly changes, although with no striking differences between species. (
Figure 5B) With heating, all samples increased in negative ellipticity around 220nm and we therefore chose to compare species at this wavelength across temperatures. (
Figure 5C) At this specific wavelength, we begin to see some significant changes between the native control and the N53D mutants and WT Cu-first at higher temperatures (75°C and 85°C). At 95°C, all species seem to be denatured to a similar degree. At the lower temperatures (≤ 65°C), significant differences between the native control and the other SOD1 species were not observed.
3.6. Molecular Dynamics Suggests Zn2+ Insertion Does Not Stabilize N53D SOD1
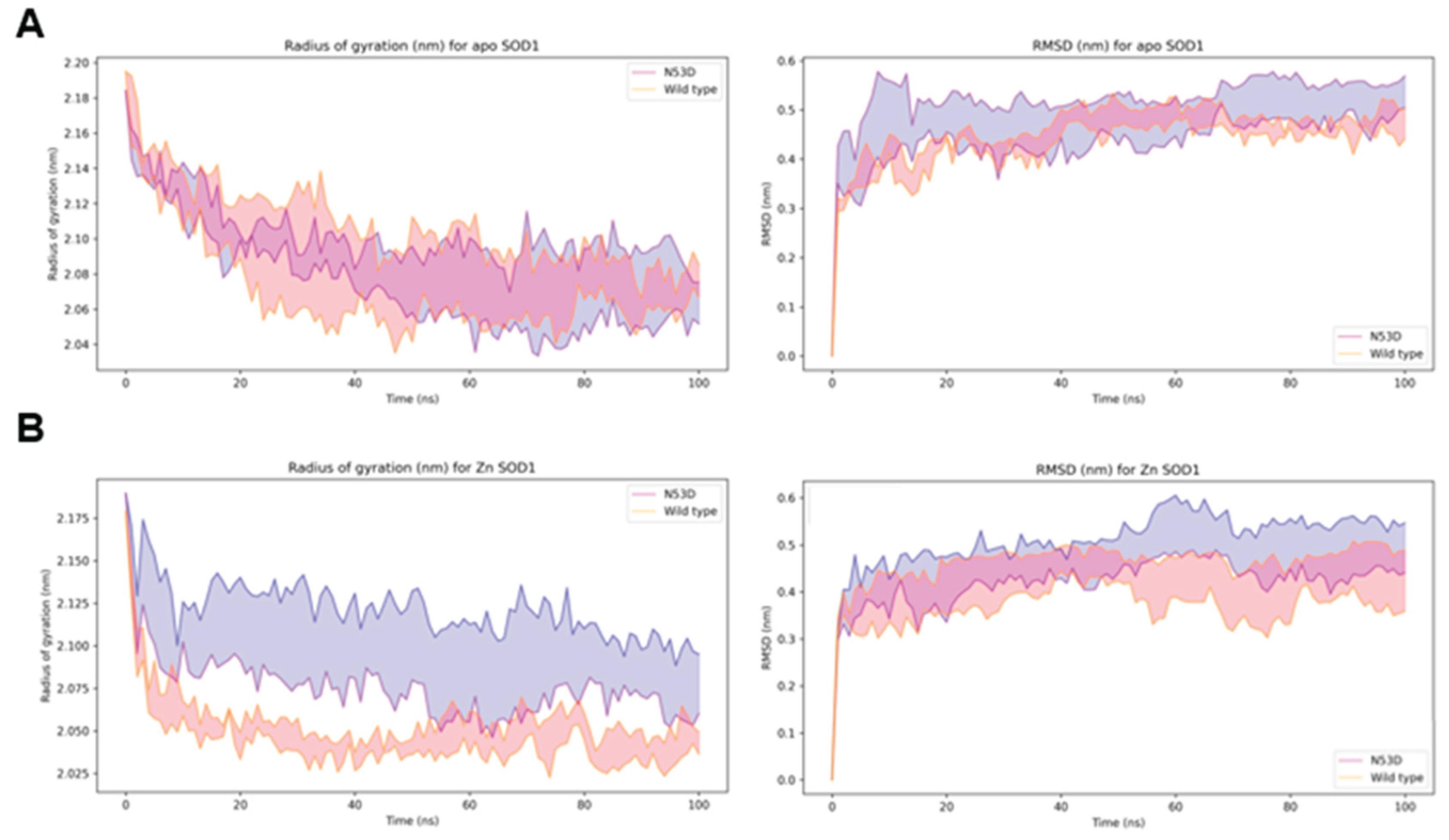
We carried out molecular dynamics (MD) simulations in GROMACS on WT and N53D SOD1 to investigate how the N53D mutation and increased Zn
2+ binding may disrupt the structure and cause the increase in aggregation potential we measured. First, we measured how flexible/tightly held together the energy minimized homodimer structures were by measuring their radius of gyration over a 100 ns simulation. When metals are removed (apo), the WT and N53D have similar average radii and root mean square deviation (RMSD) from the starting structure. (
Figure 6) The native coordination of Zn
2+ into the SOD1 structure is known to increase stability[
5]. When simulated with Zn
2+ in the homodimers, the radius of the WT homodimer quickly decreases from the starting structure and becomes much tighter between runs (decreased width of the shaded region indicating mean ± SEM), indicating a more compact and less flexible structure. However the addition of Zn
2+ does not have the same effect on the N53D homodimer and the radius of gyration stays broad between runs over the simulation, with the greater variation suggesting a more flexible structure than the WT homodimer.
3.7. Alchemical Morphing Simulations Indicates N53D-WT Heterodimer Is More Stable than the WT-WT Homodimer
Heterodimerization between WT and mutant SOD1 increases SOD1 aggregation and decreased SOD1 function which has been implicated in leading to more severe ALS symptoms.[
16,
36] Because we measure an increased amount of monomer in the N53D mutant by SEC, we wondered if this could lead to an increased amount of WT-N53D heterodimer. To investigate this, we calculated the ΔΔG of hetero-dimerization using an alchemical morphing simulation strategy we previously developed for mutant SOD1.[
34] Briefly, in these simulations one of the monomers in the WT dimer is morphed over time from N53 to D53 (forward trajectory) and then back to N53 (reverse trajectory) and work values from these morphs are used to calculated ΔG values. The difference between the ΔG values of the forward and reverse trajectories gives the ΔΔG. We found the ΔΔG of dimerization of WT-N53D heterodimer (-3.0±0.8 kJ/mol) suggesting that the heterodimer is more stable (
Supplemental Figure 5).
4. Discussion
As expected, the holoprotein control, WT SOD1 Zn-first, had the lowest relative abundance of monomers among all variants and a metal cofactor incorporation most in line with that of the native holo-SOD1 dimer. In the non-physiological case of Cu-first metal cofactor addition, a similar non-native metal cofactor abundance was observed in both WT and N53D SOD1 samples with similar relative deficiency of Zn2+ cofactors. Both zinc deficient Cu-first variants had a decreased preference for dimerization compared to the Zn-first WT control; monomer/dimer ratio of WT SOD1 Cu-first was ~5.5 times that of WT SOD1 Zn-first, while N53D SOD1 Cu-first was ~9.85 times that of Zn-first and ~1.79 times that of WT SOD1 Cu-first. In the case of N53D SOD1 Zn-first, a unique profile of aberrantly high of Zn2+ and low Cu2+ incorporation was observed, and this distinct metal cofactor profile corresponded with a distinct decrease in dimer preference with a monomer/dimer ratio ~18.3 times that of the control. Alchemical morphing simulations indicated that the N53D-WT heterodimer is more stable than the WT-WT homodimer suggesting that our measured increase in N53D monomer could lead to increased heterodimer formation in vivo and increased disease severity.
In these experiments, zinc addition preceding copper addition consistently yielded a greater abundance of coordinated Zn2+ in N53D versus WT SOD1, with the molar quantity of Zn2+ exceeding the predicted molar quantity of zinc binding sites. Additionally, copper addition preceding zinc addition yielded a similarly reduced relative abundance of coordinated Zn2+ in both N53D and WT SOD1. Considered together, these data indicate that metal binding is altered in N53 deamidated SOD1, possibly via an increased affinity for non-native Zn2+ ion coordination to the copper binding site precluding Cu2+ incorporation. Alternatively, in vivo, mature holo-SOD1 may become deamidated at N53 and lose affinity for Cu and bind another Zn co-factor as no chaperone is needed for Zn insertion.
Taken together, the data from SEC and PAR experiments indicate that N53 deamidation both generally decreases dimer stability and facilitates a non-native coordination of additional zinc cofactors, further reducing dimer stability. Corroborating this, our room temperature CD spectra show decreased alpha helicity and beta sheet signatures for variants with non-native metal coordination, namely WT Cu-first and both N53D Cu-first and Zn-first, indicating the importance of native metal incorporation for folding. Importantly, the in vitro model for N53 deamidation in this study was limited in that the N53D mutation mimicking the PTM of interest was present before metal cofactor addition. In contrast, N53 deamidation in vivo is predicted to occur very slowly, most likely after native cofactor incorporation is complete [
21]. However, CD results showing a significant reduction in the ellipticity at 220 nm in N53D variants versus control align with the spectra of metal-free SOD1 in previous studies, suggesting a greater tendency for the mutant to release metal cofactors [
9]. In the case that SOD1 in vivo more readily releases metal cofactors and achieves metal pocket vacancies following N53 deamidation, especially if stabilizing dimerization is reduced as it is in vitro, then the passive uptake of zinc into N53 deamidated SOD1 species could culminate in the observed aberrant metal coordination despite the PTM occurring after the proper maturation of WT SOD1. Previous studies have indeed suggested that aberrant Zn/Zn coordinated SOD1 species have an increased propensity to misfold, and considering that N53 deamidation was identified specifically in cultured motor neurons, the observation that this PTM may introduce a bias towards for Zn/Zn coordination could be relevant to ALS pathophysiology [
37].
Increased misfolding due to N53 deamidation was also captured in heated DLS experiments. Although differences between the control Zn-first WT SOD1 and N53D variants lacked statistical significance at 70°C, it was notable that an increase in particle size was reflected in the higher cumulative average radius and broader standard deviation in N53D variants. This indicator of aggregation at 70°C was exclusively observed in N53D variants, more frequently so in Zn-first N53D SOD1. Further, at 80°C and 90°C both metal variants of N53D SOD1 were found to have significantly greater radii than the Zn-first WT control, which had the lowest cumulative average radius of all variants as expected. The formation of heat-induced aggregates in N53D variants at lower temperatures than Zn-first WT control and the significantly greater size of aggregates at 80°C and 90°C indicate that N53 deamidation increases SOD1 aggregability, possibly introducing or enhancing aggregation pathways that facilitate the generation of significantly larger aggregates.
Unstructured monomers readily serve as a feedstock for aggregate growth and their greater relative population suggests dimer destabilization, which may also facilitate the templated misfolding of dimeric N53D SOD1. Correlations between heated DLS results and relative monomer populations in SEC were apparent; the variant with the least monomer population, Zn-first WT SOD1, achieved the lowest maximum hydrodynamic radius with heating and had no notable propensity to aggregate at 70°C, while the variant with the highest monomer population, Zn-first N53D SOD1, achieved the highest hydrodynamic radius with heating and showed a propensity to aggregate at 70°C. This suggests that aberrant Zn/Zn coordination and dimer destabilization facilitates aggregation in N53D SOD1.
Our secondary structure analysis via CD spectroscopy confirms that N53D SOD1 is less stable at higher temperatures (75-85°C). Notably, at 95°C none of the proteins are fully denatured; consistent with the formation of stable aggregates rather than fully melted protein. While stability and metal content analysis would predict gross structural changes, the CD spectra of Zn-first WT and the N53D variants were similar at 25°C and for much of the melting process. Other studies have corroborated that while Zn/Zn coordinated SOD1 has greater aggregation propensity, the crystal structure of Zn/Zn SOD1 was previously determined to align with that of the native Cu/Zn protein outside of the Cu site [
37,
38].
5. Conclusions
Through mass spectrometry analysis across multiple cell types, we have identified deamidation sites that may be cell type specific and may contribute to SOD1 destabilization and, consequently, ALS pathology. Future experiments that would provide further validation of disease relevance and cell type specificity would include additional IP/MS experimental replicates from tested lines, ideally with increased read depth to bolster statistical power, and the evaluation of more iPSC derived cell lines and ultimately human tissue is necessary. From the preliminary data, the most compelling PTM site for investigation was deamidation at N53, both because of the apparent disease cell type specificity of the PTM and because N53 is proximal to critical stabilizing motifs including the dimer interface, metal binding loop, and disulfide loop, giving modifications at this site notable potential to disrupt SOD1 structure. As we hypothesized, N53 deamidation reduces structural stability and increases relative monomer populations. Unexpectedly, instead of a loss of Zn binding, N53D SOD1 showed an increased affinity for Zn beyond the molar abundance of Zn binding sites, suggesting a structural shift allowing the Cu binding site to instead bind Zn. This tendency for N53 deamidated SOD1 to aberrantly bind additional Zn
2+ could be expected in vivo after the release of metal cofactors following N53 deamidation, and the associated non-native SOD1 species could be of interest in the study of sALS pathophysiology. Previous studies have suggested that aberrant Zn/Zn coordinated SOD1 species have an increased propensity to misfold and this tendency was observed in the case of the aberrantly Zn overpopulated N53D SOD1 Zn-first variant [
37]. Furthermore, previous studies suggest increased misfolding and toxicity in heterodimeric SOD1 species, the formation of which is both predicted to be favorable in simulations of N53D SOD1 and expected to be enhanced by increased monomer populations, as was observed in SEC of N53D SOD1 [
36]. The influence of this PTM on the properties of fALS mutant SOD1 has yet to be similarly studied, and determining whether N53 deamidation may exacerbate the instability, aggregability, and toxicity of such mutants may also be of interest in future studies.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figure S1: iPSC-derived cells; Figure S2: SDS-PAGE of recombinant GST SOD1 processing; Figure S3: Intact protein mass spectrometry confirms the N53D mutation in recombinant SOD1; Figure S4: Representative spectra for PAR assay; Figure S5: Alchemical morphing simulations of heterodimerization.
Author Contributions
Conceptualization, A.L.O. and C.S..; methodology, A.L.O., C.S. E.Z., M.M., O.R., P.S.; software, C.S., E.Z., O.R.; validation, C.S., E.Z., O.R.; formal analysis, A.L.O., E.Z., O.R.; investigation, E.Z., M.M., O.R., P.S.; resources, A.L.O.; data curation, C.S., E.Z., O.R.; writing—original draft preparation, E.Z.; writing—review and editing, A.L.O, C.S., E.Z.; visualization, A.L.O, C.S., E.Z., O.R.; supervision, A.L.O., C.S.; project administration, A.L.O.; funding acquisition, A.L.O. All authors have read and agreed to the published version of the manuscript.
Funding
The authors would like to acknowledge the NIH S10 High-End Instrumentation Award 1S10-OD028445-01A1, which supported this work by providing funds to acquire the Orbitrap Eclipse Tribrid mass spectrometer housed in the University of Connecticut Proteomics & Metabolomics Facility.
Institutional Review Board Statement
All research has a “non-human subject” designation from Wesleyan University’s Institutional Review Board and approval from Wesleyan’s Embryonic Stem Cell Research Oversight committee.
Informed Consent Statement
Our research uses de-identified, established human induced pluripotent stem cell (iPSC) lines. The creators of the lines, Harvard University (Harvard Stem Cell Institute) maintain the signed donor consent forms.
Data Availability Statement
The original contributions presented in this study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Acknowledgments
We gratefully acknowledge the quantitative proteomics analysis conducted by Dr. Jen Liddle and Dr. Jeremy Balsbaugh, of the Proteomics & Metabolomics Facility, a component of the Center for Open Research Resources and Equipment at the University of Connecticut. Graphical Abstract created in BioRender. Zanderigo, E. (2025)
https://BioRender.com/99ku1bh
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| fALS |
Familial ALS |
| SOD1 |
Superoxide Dismutase |
| sALS |
Sporadic ALS |
| PTMs |
Post translational modifications |
| ALS |
Amyotrophic lateral sclerosis |
| WT |
Wild-type |
| WTL |
Wild-type like |
| MBR |
Metal-binding region |
| N53D |
Asparagine at position 53 mutated to aspartic acid |
| iPSC |
Induced pluripotent stem cells |
| CD |
Circular dichroism |
| DLS |
Dynamic light scattering |
| PAR |
4-(2-pyridylazo) resorcinol |
| MD |
Molecular dynamics |
| MS/MS |
Tandem mass spectrometry |
| SEC |
Size exclusion chromatography |
| ns |
Not significant |
References
- Rosen, D.R.; Siddique, T.; Patterson, D.; Figlewicz, D.A.; Sapp, P.; Hentati, A.; Donaldson, D.; Goto, J.; O'Regan, J.P.; Deng, H.-X.; Rahmani, Z.; Krizus, A.; McKenna-Yasek, D.; Cayabyab, A.; Gaston, S.M.; Berger, R.; Tanzi, R.E.; Halperin, J.J.; Herzfeldt, B.; Van den Bergh, R.; Hung, W.-Y.; Bird, T.; Deng, G.; Mulder, D.W.; Smyth, C.; Laing, N.G.; Soriano, E.; Pericak–Vance, M.A.; Haines, J.; Rouleau, G.A.; Gusella, J.S.; Horvitz, H.R.; Brown, R.H. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 1993, 362, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Broom, H.R.; Rumfeldt, J.A.; Meiering, E.M. Many roads lead to Rome? Multiple modes of Cu,Zn superoxide dismutase destabilization, misfolding and aggregation in amyotrophic lateral sclerosis. Essays Biochem 2014, 56, 149–165. [Google Scholar] [CrossRef]
- Tokuda, E.; Takei Y-i Ohara, S.; Fujiwara, N.; Hozumi, I.; Furukawa, Y. Wild-type Cu/Zn-superoxide dismutase is misfolded in cerebrospinal fluid of sporadic amyotrophic lateral sclerosis. Molecular Neurodegeneration 2019, 14, 42. [Google Scholar] [CrossRef] [PubMed]
- Trist, B.G.; Hilton, J.B.; Hare, D.J.; Crouch, P.J.; Double, K.L. Superoxide Dismutase 1 in Health and Disease: How a Frontline Antioxidant Becomes Neurotoxic. Angewandte Chemie International Edition. [CrossRef]
- Wright, G.S.A.; Antonyuk, S.V.; Hasnain, S.S. The biophysics of superoxide dismutase-1 and amyotrophic lateral sclerosis. Q Rev Biophys 2019, 52, e12. [Google Scholar] [CrossRef] [PubMed]
- Huai, J.; Zhang, Z. Structural Properties and Interaction Partners of Familial ALS-Associated SOD1 Mutants. Front Neurol 2019, 10, 527–527. [Google Scholar] [CrossRef]
- Valentine, J.S.; Doucette, P.A.; Zittin Potter, S. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu Rev Biochem 2005, 74, 563–593. [Google Scholar] [CrossRef]
- Wang, Q.; Johnson, J.L.; Agar, N.Y.; Agar, J.N. Protein aggregation and protein instability govern familial amyotrophic lateral sclerosis patient survival. PLoS Biol 2008, 6, e170. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Durazo, A.; Girotto, S.; Gralla, E.B.; Martinelli, M.; Valentine, J.S.; Vieru, M.; Whitelegge, J.P. Metal-free superoxide dismutase forms soluble oligomers under physiological conditions: a possible general mechanism for familial ALS. Proc Natl Acad Sci U S A 2007, 104, 11263–11267. [Google Scholar] [CrossRef]
- Reaume, A.G.; Elliott, J.L.; Hoffman, E.K.; Kowall, N.W.; Ferrante, R.J.; Siwek, D.F.; Wilcox, H.M.; Flood, D.G.; Beal, M.F.; Brown, R.H., Jr.; Scott, R.W.; Snider, W.D. Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat Genet 1996, 13, 43–47. [Google Scholar] [CrossRef]
- Saccon, R.A.; Bunton-Stasyshyn, R.K.A.; Fisher, E.M.C.; Fratta, P. Is SOD1 loss of function involved in amyotrophic lateral sclerosis? Brain 2013, 136, 2342–2358. [Google Scholar] [CrossRef]
- Zarei, S.; Carr, K.; Reiley, L.; Diaz, K.; Guerra, O.; Altamirano, P.F.; Pagani, W.; Lodin, D.; Orozco, G.; Chinea, A. A comprehensive review of amyotrophic lateral sclerosis. Surg Neurol Int 2015, 6, 171. [Google Scholar] [CrossRef]
- Sangwan, S.; Zhao, A.; Adams, K.L.; Jayson, C.K.; Sawaya, M.R.; Guenther, E.L.; Pan, A.C.; Ngo, J.; Moore, D.M.; Soriaga, A.B.; Do, T.D.; Goldschmidt, L.; Nelson, R.; Bowers, M.T.; Koehler, C.M.; Shaw, D.E.; Novitch, B.G.; Eisenberg, D.S. Atomic structure of a toxic, oligomeric segment of SOD1 linked to amyotrophic lateral sclerosis (ALS). Proc Natl Acad Sci U S A 2017, 114, 8770–8775. [Google Scholar] [CrossRef]
- Redler, R.L.; Fee, L.; Fay, J.M.; Caplow, M.; Dokholyan, N.V. Non-native soluble oligomers of Cu/Zn superoxide dismutase (SOD1) contain a conformational epitope linked to cytotoxicity in amyotrophic lateral sclerosis (ALS). Biochemistry 2014, 53, 2423–2432. [Google Scholar] [CrossRef]
- Proctor, E.A.; Fee, L.; Tao, Y.; Redler, R.L.; Fay, J.M.; Zhang, Y.; Lv, Z.; Mercer, I.P.; Deshmukh, M.; Lyubchenko, Y.L.; Dokholyan, N.V. Nonnative SOD1 trimer is toxic to motor neurons in a model of amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A 2016, 113, 614–619. [Google Scholar] [CrossRef]
- Zhu, C.; Beck, M.V.; Griffith, J.D.; Deshmukh, M.; Dokholyan, N.V. Large SOD1 aggregates, unlike trimeric SOD1, do not impact cell viability in a model of amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A 2018, 115, 4661–4665. [Google Scholar] [CrossRef] [PubMed]
- Gill, C.; Phelan, J.P.; Hatzipetros, T.; Kidd, J.D.; Tassinari, V.R.; Levine, B.; Wang, M.Z.; Moreno, A.; Thompson, K.; Maier, M.; Grimm, J.; Gill, A.; Vieira, F.G. SOD1-positive aggregate accumulation in the CNS predicts slower disease progression and increased longevity in a mutant SOD1 mouse model of ALS. Sci Rep 2019, 9, 6724. [Google Scholar] [CrossRef] [PubMed]
- Bosco, D.A.; Morfini, G.; Karabacak, N.M.; Song, Y.; Gros-Louis, F.; Pasinelli, P.; Goolsby, H.; Fontaine, B.A.; Lemay, N.; McKenna-Yasek, D.; Frosch, M.P.; Agar, J.N.; Julien, J.P.; Brady, S.T.; Brown, R.H., Jr. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat Neurosci 2010, 13, 1396–1403. [Google Scholar] [CrossRef] [PubMed]
- Sekhar, A.; Rumfeldt, J.A.O.; Broom, H.R.; Doyle, C.M.; Sobering, R.E.; Meiering, E.M.; Kay, L.E. Probing the free energy landscapes of ALS disease mutants of SOD1 by NMR spectroscopy. Proc Natl Acad Sci U S A 2016, 113, E6939–e6945. [Google Scholar] [CrossRef]
- Crown, A.; McAlary, L.; Fagerli, E.; Brown, H.; Yerbury, J.J.; Galaleldeen, A.; Cashman, N.R.; Borchelt, D.R.; Ayers, J.I. Tryptophan residue 32 in human Cu-Zn superoxide dismutase modulates prion-like propagation and strain selection. PLoS One 2020, 15, e0227655. [Google Scholar] [CrossRef]
- Shi, Y.; Rhodes, N.R.; Abdolvahabi, A.; Kohn, T.; Cook, N.P.; Marti, A.A.; Shaw, B.F. Deamidation of asparagine to aspartate destabilizes Cu, Zn superoxide dismutase, accelerates fibrillization, and mirrors ALS-linked mutations. J Am Chem Soc 2013, 135, 15897–15908. [Google Scholar] [CrossRef]
- Trist, B.G.; Genoud, S.; Roudeau, S.; Rookyard, A.; Abdeen, A.; Cottam, V.; Hare, D.J.; White, M.; Altvater, J.; Fifita, J.A.; Hogan, A.; Grima, N.; Blair, I.P.; Kysenius, K.; Crouch, P.J.; Carmona, A.; Rufin, Y.; Claverol, S.; Van Malderen, S.; Falkenberg, G.; Paterson, D.J.; Smith, B.; Troakes, C.; Vance, C.; Shaw, C.E.; Al-Sarraj, S.; Cordwell, S.; Halliday, G.; Ortega, R.; Double, K.L. Altered SOD1 maturation and post-translational modification in amyotrophic lateral sclerosis spinal cord. Brain 2022, 145, 3108–3130. [Google Scholar] [CrossRef] [PubMed]
- Rigamonti, A.; Repetti, G.G.; Sun, C.; Price, F.D.; Reny, D.C.; Rapino, F.; Weisinger, K.; Benkler, C.; Peterson, Q.P.; Davidow, L.S.; Hansson, E.M.; Rubin, L.L. Large-Scale Production of Mature Neurons from Human Pluripotent Stem Cells in a Three-Dimensional Suspension Culture System. Stem Cell Reports 2016, 6, 993–1008. [Google Scholar] [CrossRef]
- Rapino, F.; Natoli, T.; Limone, F.; O'Connor, E.; Blank, J.; Tegtmeyer, M.; Chen, W.; Norabuena, E.; Narula, J.; Hazelbaker, D.; Angelini, G.; Barrett, L.; O'Neil, A.; et al. Small-molecule screen reveals pathways that regulate C4 secretion in stem cell-derived astrocytes. Stem Cell Reports 2023, 18, 237–253. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology, 1367. [Google Scholar] [CrossRef]
- Marty, M.T.; Baldwin, A.J.; Marklund, E.G.; Hochberg, G.K.A.; Benesch, J.L.P.; Robinson, C.V. Bayesian Deconvolution of Mass and Ion Mobility Spectra: From Binary Interactions to Polydisperse Ensembles. Analytical Chemistry 2015, 87, 4370–4376. [Google Scholar] [CrossRef] [PubMed]
- Doyle, C.M.; Naser, D.; Bauman, H.A.; Rumfeldt, J.A.O.; Meiering, E.M. Spectrophotometric method for simultaneous measurement of zinc and copper in metalloproteins using 4-(2-pyridylazo)resorcinol. Anal Biochem 2019, 579, 44–56. [Google Scholar] [CrossRef]
- Leaver-Fay, A.; Tyka, M.; Lewis, S.M.; Lange, O.F.; Thompson, J.; Jacak, R.; Kaufman, K.W.; Renfrew, P.D.; Smith, C.A.; Sheffler, W.; Davis, I.W.; Cooper, S.; Treuille, A.; Mandell, D.J.; Richter, F.; Ban, Y.-E.A.; Fleishman, S.J.; Corn, J.E.; Kim, D.E.; Lyskov, S.; Berrondo, M.; Mentzer, S. ; Popović; Z; Havranek, J.J.; Karanicolas, J.; Das, R.; Meiler, J.; Kortemme, T.; Gray, J.J.; Kuhlman, B.; Baker, D.; Bradley, P. (2011) Chapter nineteen - Rosetta3: An Object-Oriented Software Suite for the Simulation and Design of Macromolecules, In Methods Enzymol (Johnson, M. L., and Brand, L., Eds.), pp 545-574, Academic Press.
- Branco, R.J.F.; Fernandes, P.A.; Ramos, M.J. Molecular Dynamics Simulations of the Enzyme Cu, Zn Superoxide Dismutase. The Journal of Physical Chemistry B 2006, 110, 16754–16762. [Google Scholar] [CrossRef]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins: Structure, Function, and Bioinformatics, 1950. [Google Scholar] [CrossRef]
- Aliev, A.E.; Kulke, M.; Khaneja, H.S.; Chudasama, V.; Sheppard, T.D.; Lanigan, R.M. Motional timescale predictions by molecular dynamics simulations: Case study using proline and hydroxyproline sidechain dynamics. Proteins: Structure, Function, and Bioinformatics. [CrossRef]
- Joung, I.S.; Cheatham, T.E., III. Determination of Alkali and Halide Monovalent Ion Parameters for Use in Explicitly Solvated Biomolecular Simulations. The Journal of Physical Chemistry B 2008, 112, 9020–9041. [Google Scholar] [CrossRef]
- Gapsys, V.; Michielssens, S.; Seeliger, D.; de Groot, B.L. pmx: Automated protein structure and topology generation for alchemical perturbations. Journal of Computational Chemistry 2015, 36, 348–354. [Google Scholar] [CrossRef]
- Wells, N.G.M.; Tillinghast, G.A.; O'Neil, A.L.; Smith, C.A. Free energy calculations of ALS-causing SOD1 mutants reveal common perturbations to stability and dynamics along the maturation pathway. Protein Science 2021, 30, 1804–1817. [Google Scholar] [CrossRef]
- Rogers, D.M.; Jasim, S.B.; Dyer, N.T.; Auvray, F.; Réfrégiers, M.; Hirst, J.D. Electronic Circular Dichroism Spectroscopy of Proteins. Chem 2019, 5, 2751–2774. [Google Scholar] [CrossRef]
- Brasil Ad, A.; de Carvalho, M.D.C.; Gerhardt, E.; Queiroz, D.D.; Pereira, M.D.; Outeiro, T.F.; Eleutherio, E.C.A. Characterization of the activity, aggregation, and toxicity of heterodimers of WT and ALS-associated mutant Sod1. Proceedings of the National Academy of Sciences 2019, 116, 25991–26000. [Google Scholar] [CrossRef]
- Leal, S.S.; Gomes, C.M. Calcium dysregulation links ALS defective proteins and motor neuron selective vulnerability. Frontiers in Cellular Neuroscience, 2015. [Google Scholar]
- Strange, R.W.; Antonyuk, S.V.; Hough, M.A.; Doucette, P.A.; Valentine, J.S.; Hasnain, S.S. Variable metallation of human superoxide dismutase: atomic resolution crystal structures of Cu-Zn, Zn-Zn and as-isolated wild-type enzymes. J Mol Biol 2006, 356, 1152–1162. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).