1. Introduction
Globally, breast cancer continues to be a major cause of cancer-related mortality among women. Early detection and precise diagnosis are essential for enhancing survival rates[
1]. Among various imaging modalities, breast ultrasound (BUS) has emerged as a vital tool in clinical practice, thanks to its non-invasive nature, lack of radiation, and relatively low cost[
2,
3]. However, BUS images are often characterized by strong speckle noise, low contrast, uneven lesion appearance, and blurred boundaries, which pose significant challenges for automatic tumor segmentation in computer-aided diagnosis (CAD) systems.
In recent years, deep learning-based segmentation methods have completely transformed BUS image analysis. The U-Net architecture[
4] and its numerous extensions, such as U-Net++ and Attention U-Net, have achieved remarkable success by leveraging encoder-decoder structures with skip connections. Recently, transformer-based methods and hybrid CNN-state space models have demonstrated outstanding global context modeling capabilities, further enhancing segmentation accuracy. Although they are very effective, these models usually involve a large number of parameter counts and high computational costs, which limits their practicality in resource-constrained environments, such as portable ultrasound devices used in point-of-care Settings.
To improve efficiency in breast ultrasound image segmentation, several studies have adopted lightweight segmented networks by replacing computationally heavy backbones with efficient architectures such as MobileNet or EfficientNet[
5]. While these approaches substantially decrease computational costs, they often suffer from reduced robustness to noise and inadequate delineation of lesion boundaries—a critical aspect in BUS segmentation. Moreover, although attention mechanisms have demonstrated effectiveness in emphasizing salient features and suppressing background interference, prevalent modules tend to introduce considerable computational overhead, hindering their applicability in real-time clinical settings.
To fill this research gap, we propose LBA-Net (Lightweight Attention Network)—a novel segmentation framework specifically designed for robust and efficient breast ultrasound image analysis that integrates three core innovations: (1) a lightweight backbone that adopts MobileNetV3-Small as the encoder to capture multi-scale features with minimal parameters and FLOPs; (2) the LBA-Block, a lightweight boundary-aware attention module that efficiently fuses channel attention (via ECA) with spatial attention to enhance discriminative feature representation without incurring high computational cost; and (3) dual-head supervision that employs a segmentation head and an auxiliary boundary head, jointly optimized by a boundary-sensitive loss combining Dice, BCE, and Tversky terms, thereby improving noise robustness and enabling precise boundary localization.
The primary contributions of this research are as follows: First, we present LBA-Net, a lightweight attention-based network designed for accurate and robust segmentation of breast ultrasound (BUS) images. Second, we introduce the LBA-Block, a parameter-efficient module that enhances feature discriminability by integrating channel and spatial attention via depthwise separable convolution. Third, we propose a two-head supervision mechanism with a boundary-sensitive loss function to refine boundary perception and improve noise robustness. Through extensive validation on multiple public datasets, LBA-Net demonstrates high accuracy and low computational cost, making it suitable for real-time deployment in portable ultrasound systems and clinical decision support tools.
Extensive experiments conducted on the BUSI dataset demonstrate that LBA-Net achieves a Dice score of 81.4% on the validation set (82.9% on the training set) under an 80/20 patient-wise split, outperforming most lightweight competitors while maintaining real-time performance at 122 FPS on GPU. This balance of accuracy, robustness, and computational efficiency highlights the strong potential of LBA-Net for real-time deployment in portable ultrasound devices and clinical decision-support systems.
2. Related Work
2.1. Deep Learning for Breast Ultrasound Image Segmentation
Automatic segmentation of breast lesions in ultrasound images is a critical step in computer-aided diagnosis (CAD) systems[
6], enabling early detection and quantitative analysis of tumors. With the advent of deep convolutional neural networks (CNNs) [
7], significant progress has been made in medical image segmentation. The U-Net architecture [
8,
9], characterized by an encoder–decoder structure with skip connections, has become the de facto standard in medical image segmentation. This is largely due to its effectiveness in preserving fine spatial details while simultaneously capturing high-level semantic information.
Numerous U-Net variants have since been proposed to improve segmentation performance in BUS images. For instance, Attention U-Net introduced spatial attention gates to highlight salient regions[
10], while U-Net++employed nested skip connections to mitigate semantic gaps between encoder and decoder features[
11]. More recently, transformer-based architectures such as and Swin-UNet have demonstrated superior global context modeling by integrating self-attention mechanisms, achieving state-of-the-art results on multiple medical imaging benchmarks[
12].
Despite their high accuracy, these models often suffer from excessive computational complexity and large parameter counts, making them unsuitable for real-time deployment in clinical environments — particularly on portable ultrasound devices with limited computational resources. This efficiency-accuracy trade-off motivates the development of lightweight yet robust segmentation frameworks tailored for BUS applications.
2.2. Lightweight Architectures for Medical Image Segmentation
To address the computational burden of standard segmentation networks,Our work focuses on designing a lightweight architecture without affecting the quality of segmentation. A common strategy is to replace the heavy encoder backbone with efficient alternatives such as Mobile-Net[
13], Efficient-Net[
14], or their lightweight variants ,which significantly reduce model size and inference latency while maintaining reasonable segmentation accuracy. However, these lightweight models often exhibit degraded performance in low-contrast or noisy BUS images, particularly in boundary delineation — a crucial requirement for clinical diagnosis. Moreover, most existing lightweight designs do not incorporate mechanisms to explicitly enhance boundary perception or suppress speckle noise, limiting their robustness in real-world scenarios.
2.3. Attention Mechanisms in Medical Image Analysis
Attention mechanisms have become a powerful tool for enhancing feature discriminability in medical image segmentation. Channel attention modules, such as Squeeze-and-Excitation (SE) [
15],and Efficient Channel Attention (ECA) [
16],adaptively recalibrate channel-wise feature responses based on global context. Spatial attention modules, such as CBAM[
17], further refine feature maps by highlighting informative spatial regions.
While effective, conventional attention modules often introduce substantial computational overhead due to dense convolutions and global pooling operations. For instance, CBAM applies 7×7 convolutions for spatial attention, which increases FLOPs and memory consumption — a critical limitation for edge deployment. Recent works such as LAEDNet[
18] have attempted to mitigate this by designing lightweight attention blocks using depth-wise convolutions and simplified channel weighting. However, these methods typically focus on general ultrasound segmentation and do not explicitly address the unique challenges of BUS images, such as blurred boundaries and speckle noise.
2.4. Boundary-Aware and Robust Segmentation
Accurate boundary delineation is particularly vital in BUS segmentation, as tumor margins often determine malignancy and surgical planning. To this end, dual-head architectures that jointly predict segmentation masks and boundary maps have gained popularity. For example, Boundary-aware Networks[
19,
20] and BESNet[
21] employ auxiliary boundary supervision to enforce edge consistency, leading to sharper and more precise lesion contours.
Robustness to image artifacts — such as speckle noise, low contrast, and domain shift — is another critical requirement for clinical deployment. While data augmentation and noise injection are commonly used during training, few architectures explicitly incorporate noise-robust modules within the network. Recent hybrid models such as HCMNet[
22,
23], which combines CNN, Mamba, and wavelet transforms, demonstrate improved robustness but at the cost of increased complexity and slower inference.
LBA-Net tackles the threefold challenges of accuracy, and efficiency in breast ultrasound (BUS) segmentation by integrating three key components. First, it employs a lightweight MobileNetV3-Small encoder to ensure computational efficiency. Second, it introduces a novel LBA-Block that combines channel and spatial attention through depth-wise operations, achieving minimal computational overhead. Third, it utilizes a boundary-aware dual-head supervision strategy to enhance edge precision and noise resilience. This unified framework optimizes segmentation performance while maintaining real-time deployability.
Unlike transformer-based or hybrid models, LBA-Net avoids heavy global operators, making it suitable for real-time inference on mobile GPUs and edge devices. Unlike generic lightweight U-Nets, it explicitly models boundary structure and suppresses noise through architectural design — not just data augmentation. This positions LBA-Net as a clinically deployable solution that bridges the gap between research innovation and practical CAD system requirements.
3. Methodology
3.1. Overall Architecture
To achieve a favorable trade-off between segmentation accuracy and computational efficiency, LBA-Net integrates several lightweight strategies into its architecture. First, a MobileNetV3-Small backbone is adopted as the encoder to extract multi-scale representations. By leveraging depthwise separable convolutions, squeeze-and-excitation modules, and efficient activation functions, MobileNetV3-Small substantially reduces parameter count and floating-point operations (FLOPs), leading to more than 90% reduction in complexity compared with conventional U-Net encoders.
Second, we introduce the Lightweight Attention Block (LBA-Block), which enhances feature discriminability with negligible computational overhead. Unlike conventional attention modules that rely on heavy convolutions and fully connected layers, the LBA-Block fuses two lightweight components: (i) Efficient Channel Attention (ECA), which models cross-channel interactions through a 1D convolution without dimensionality reduction, thereby avoiding parameter inflation; and (ii) spatial attention based on a 3×3 depthwise separable convolution, which highlights salient spatial regions while reducing FLOPs by over 90% relative to standard 2D convolutions. Moreover, we introduce adaptive fusion weights (α, β) to dynamically balance the contributions of channel and spatial attention, allowing the model to emphasize either modality depending on the input context.
Finally, the decoder adopts streamlined skip connections and incorporates a dual-head supervision scheme. This design not only facilitates precise boundary localization but also maintains a compact model size by avoiding redundant layers. As a result, LBA-Net has only 2.1M parameters and 4.8 GFLOPs per 512×512 image, delivering 122 FPS on GPU and 12 FPS on CPU, thereby demonstrating real-time capability and deployability in resource-constrained clinical environments.
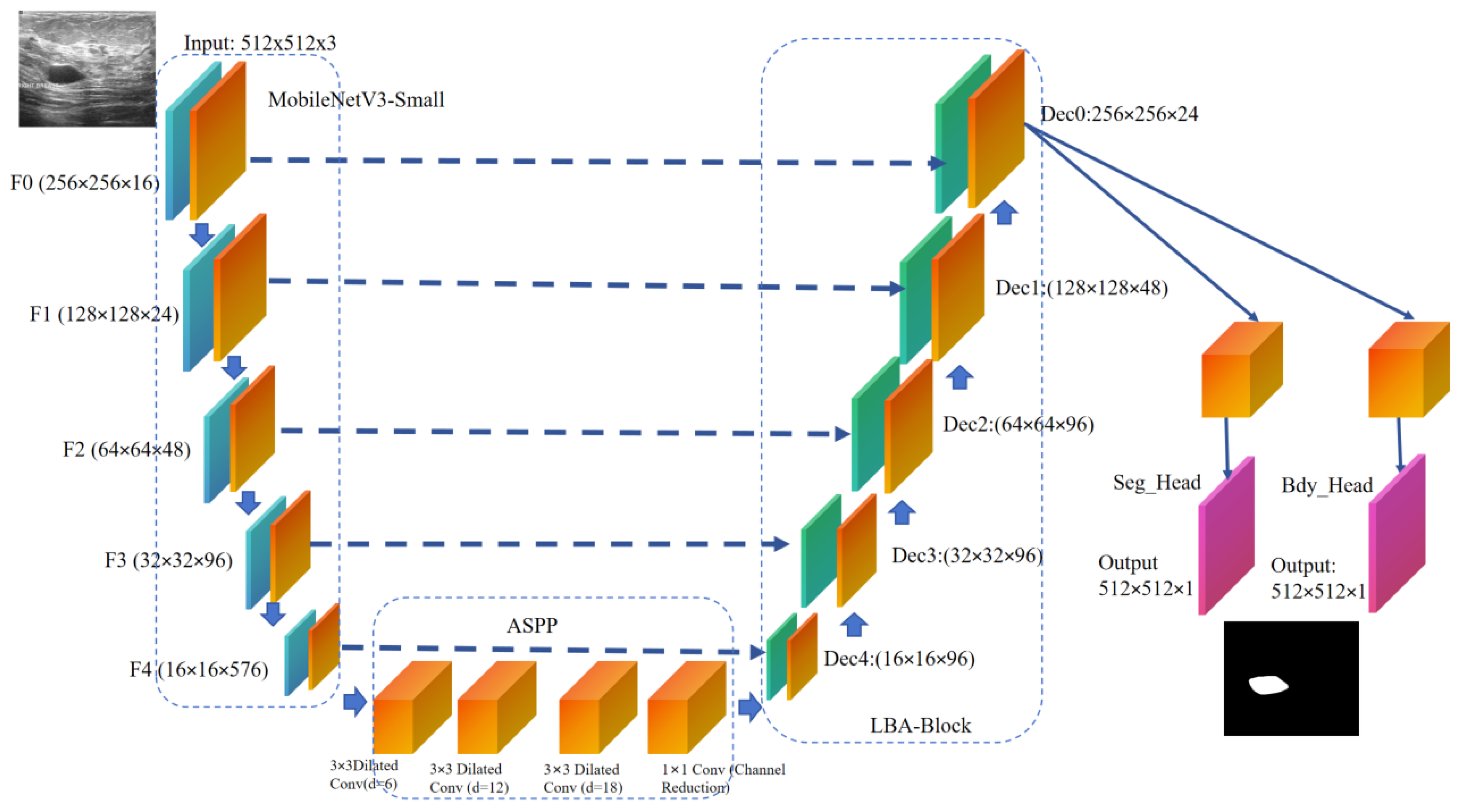
LBA-Net adopts an encoder–decoder architecture with skip connections, following the U-Net paradigm. The overall pipeline consists of four main components (
Figure 1):
The proposed network adopts a lightweight encoder-decoder architecture, where a pretrained MobileNetV3-Small serves as the encoder to extract hierarchical multi-scale features {F1,F2,F3,F4} with progressively reduced spatial resolution. At the bottleneck, an Atrous Spatial Pyramid Pooling (ASPP) module is employed to capture rich contextual information across multiple receptive fields. In the decoder pathway, features are progressively upsampled and fused with corresponding encoder features via skip connections. At each decoding stage, a Lightweight Attention Block (LBA-Block) is applied to enhance discriminative feature representations while suppressing irrelevant background responses. The network further employs a dual-head output structure, generating both a segmentation mask and a boundary map; the latter takes the detached segmentation logits as input, providing auxiliary supervision to improve the accuracy of lesion edge delineation, promoting precise boundary localization in the final segmentation.
Formally, given an input image
, the encoder extracts features:
These features are processed by ASPP to produce bottleneck representation:
The decoder reconstructs the segmentation map:
where LBA-Blocks are applied in skip fusion. A parallel boundary head predicts boundary map:
3.2. Lightweight Design of LBA-Net
The primary objective of our encoder design is to extract multi-scale feature representations with high discriminative power while maintaining minimal computational overhead. To achieve this, we forgo the conventional heavy backbones commonly used in standard U-Net and adopt MobileNetV3-Small[
24] as our feature extraction backbone.
MobileNetV3 is specifically engineered for mobile and resource-constrained vision applications. It leverages two key architectural innovations to drastically improve computational efficiency:
Depthwise Separable Convolutions: This operation factorizes a standard convolution into a depthwise convolution (applying a single filter per input channel) followed by a pointwise convolution (1×1 convolution to combine the outputs). This factorization reduces both the parameter count and computational cost by approximately a factor of k^2 compared to standard convolutions, where k=3 is the kernel size, with only a marginal sacrifice in representational capacity.
Hard-Swish and Squeeze-and-Excitation (SE) Modules: MobileNetV3 integrates the h-swish activation function, which provides a smoother, more efficient alternative to ReLU. Furthermore, it incorporates lightweight SE attention modules in its bottleneck blocks, which adaptively recalibrate channel-wise feature responses to emphasize informative features.
By employing a pre-trained MobileNetV3-Small encoder, our network benefits from robust feature representations learned from large-scale datasets, which enhances generalization and accelerates convergence on the medical segmentation task.
3.3. Lightweight Encoder
The LBA-Block is designed to be a parameter-efficient attention mechanism combining channel attention and spatial attention.
(a) Channel Attention
Given feature map
, we apply Efficient Channel Attention (ECA) or a squeeze-excitation (SE) mechanism:
where GAP denotes global average pooling, Conv1D is a 1D convolution with kernel size k, and σ is the sigmoid function.
The channel-refined feature is:
where ⊗ denotes channel-wise multiplication.
(b) Spatial Attention
Spatial attention focuses on salient regions using depth-wise separable convolution:
where depth-wise separable convolution is with kernel size 3×3.
The spatially refined feature is:
(c) Fusion
The final output of LBA-Block is obtained by combining both attentions:
where α and β are learnable scalar weights initialized as 0.5.
Thus, LBA-Block adaptively emphasizes informative channels and spatial locations with minimal overhead.
3.4. Lightweight Attention Block (LBA-Block) Design
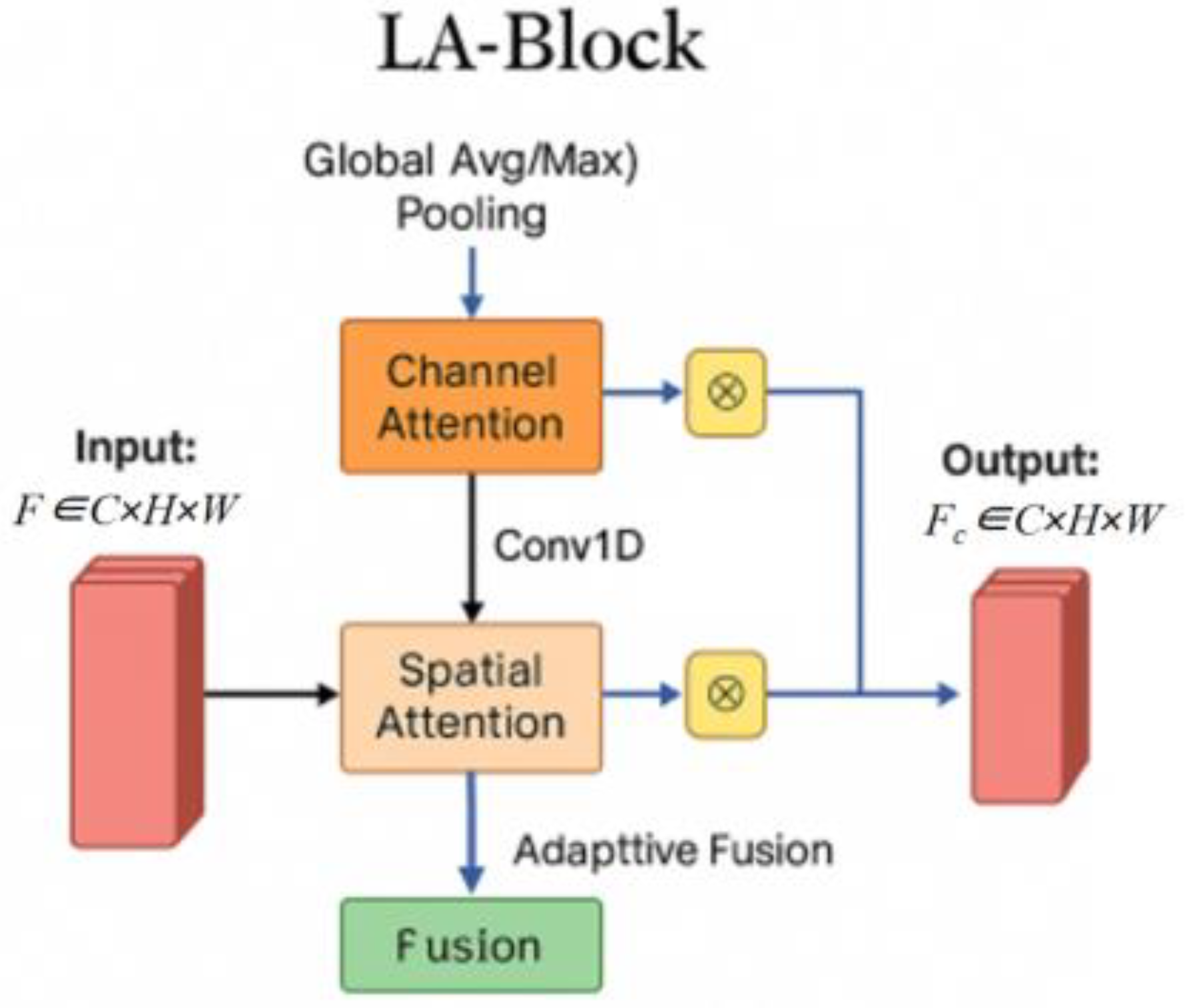
To enhance feature discriminability while maintaining computational efficiency, we propose the Lightweight Attention Block (LBA-Block), a novel attention module that adaptively refines feature representations by fusing channel-wise and spatial-wise attention mechanisms with minimal parameter overhead. Unlike conventional attention modules that rely on dense convolutions and global pooling operations, LBA-Block is specifically designed for edge deployment, leveraging depth-wise separable convolutions and 1D channel attention to drastically reduce computational cost without sacrificing performance.
As illustrated in
Figure 2, LBA-Block operates on an input feature map and produces an attention-refined output through three sequential stages: channel attention refinement, spatial attention refinement, and adaptive fusion.
(a) Channel Attention Refinement
Channel attention aims to recalibrate feature channels by emphasizing informative responses and suppressing less useful ones. We adopt the Efficient Channel Attention (ECA) mechanism[
25] due to its parameter-free design and strong performance in low-resource settings. Given input F, we first apply global average pooling (GAP) to obtain channel descriptors:
A 1D convolution with kernel size k (set to 3 in our implementation) is then applied to capture cross-channel interactions without dimensionality reduction:
where σ denotes the sigmoid activation function. The channel-refined feature Fc is obtained via channel-wise multiplication:
This design replaces the fully connected layers and dimensionality reduction in SE blocks with a single 1D convolution, significantly reducing parameters while effectively capturing local cross-channel interactions.
(b) Spatial Attention Refinement
Spatial attention focuses on identifying salient regions within each feature map. To minimize computational cost, we employ depth-wise separable convolution with a 3×3 kernel instead of standard convolutions or large-kernel spatial attention. The spatial attention map As is computed as:
where
DWConv denotes depth-wise convolution followed by point-wise projection to a single channel. The spatially refined feature
Fs is then:
This approach reduces spatial attention FLOPs by over 90% compared to standard 2D convolutions, while still effectively highlighting lesion regions and suppressing background clutter.
(c) Adaptive Fusion
To balance the contributions of channel and spatial attention, we introduce learnable scalar fusion weights α and β, initialized to 0.5 and optimized end-to-end during training:
This adaptive fusion allows the network to dynamically emphasize either channel or spatial cues depending on the input context — for instance, favoring spatial attention in low-contrast regions or channel attention in noisy areas. The total parameters introduced by LBA-Block are negligible, and no additional nonlinearities or normalization layers are added, preserving inference speed.
3.5. Dual-Head Supervision
To improve boundary delineation, LBA-Net employs a dual-head design:
Segmentation Head: Outputs a binary segmentation maskwhere denotes the spatial resolution of the input image which consistent with the 512×512 input size. A pixel value of 1 represents the tumor region, while 0 represents the background which are normal breast tissue or artifacts.
Boundary Head: Outputs a probability map highlighting object boundaries, with ground-truth labels derived via morphological gradient operations on Sgt. Specifically, , where and denote morphological dilation and erosion, respectively. After the subtraction, is binarized: pixel values greater than 0 are set to 1, and values equal to 0 are set to 0, ensuring consistency with the supervision target of the boundary head.
3.6. Loss Function
The overall training objective is a weighted combination of segmentation and boundary losses:
where G is the ground-truth mask, is the ground-truth boundary map, and where λ balances the two terms.
3.6.1. Segmentation Loss:
which combines pixel-level Binary Cross-Entropy (BCE) with region-overlap Dice loss.
3.6.2. Boundary Loss:
We adopt Tversky loss to address class imbalance in boundaries:
where TP,FP,FN are true positives, false positives, and false negatives along the boundary.
3.7. Training Strategy
Training is conducted with an AdamW optimizer using a learning rate of 1×10−3 and a weight decay of 1×10−4. The learning rate is scheduled using cosine annealing with warmup to ensure stable convergence. To enhance model robustness and generalize across variations in clinical acquisition, we apply comprehensive data augmentation including random horizontal and vertical flips, rotations, scaling, gamma correction, and speckle noise injection—mimicking common artifacts present in real breast ultrasound images. All input images are resized to 512×512 pixels, and training proceeds with a batch size of 8 for up to 200 epochs, incorporating early stopping based on the validation Dice score to prevent overfitting and ensure optimal performance.
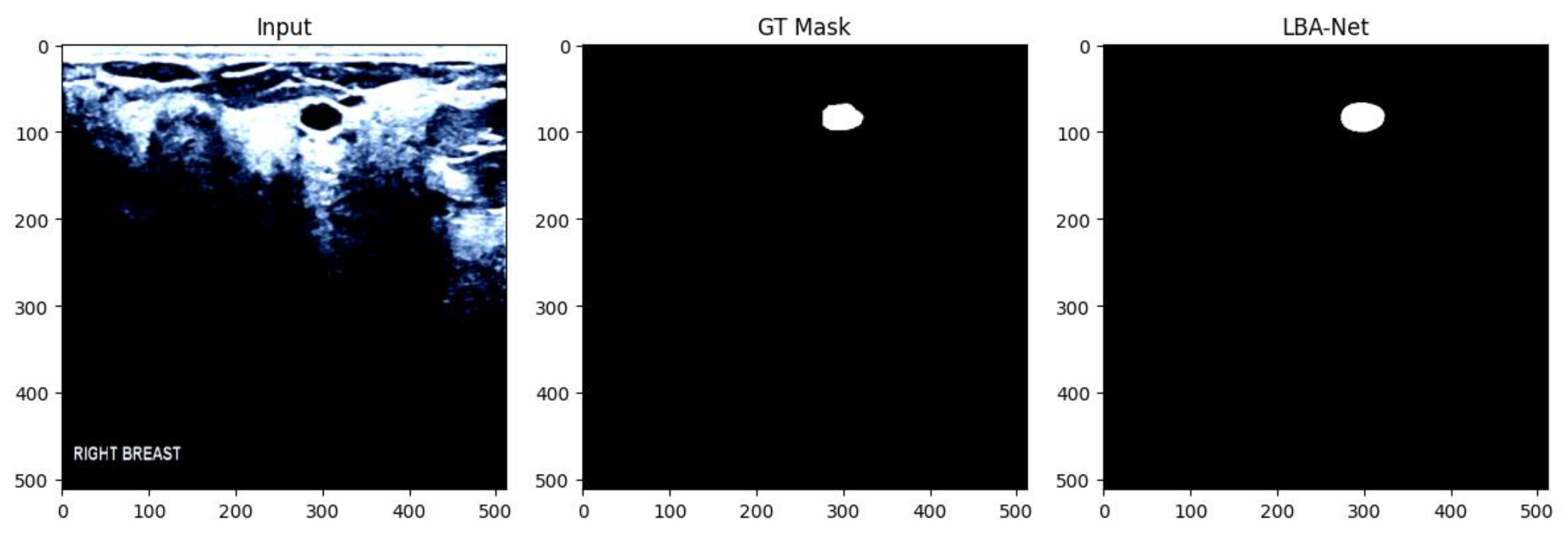
Figure 3.
The images recognized by LBA-Net are compared with those annotated manually
Figure 3.
The images recognized by LBA-Net are compared with those annotated manually
4. Experiments
4.1. The Impact of Input Resolution on Performance
In the "Input Resolution Ablation" experiment, while keeping the network structure and training settings unchanged, we reduced the input size from 512×512 to 256×256 and compared the segmentation accuracy and inference efficiency of LBA-Net at the two resolutions in
Table 3. The results show that when the input resolution is 256×256, the validation Dice coefficient of the model slightly decreases from 81.4 % to 79.8 %, the IoU decreases from 76.4 % to 74.2 %, and the 95 % Hausdorff distance (HD95) increases from 5.9 mm to 7.2 mm; meanwhile, the GPU inference frame rate on an NVIDIA A100 GPU increases from 124 FPS to 205 FPS, with a speed improvement of approximately 1.66 times. This experiment verifies that LBA-Net can still maintain high segmentation performance at low resolution and exhibit better real-time performance in resource-constrained mobile or embedded scenarios. In applications with higher precision requirements, it is still recommended to use an input resolution of 512×512 to achieve the best segmentation effect..
Table 1.
Comparison of inference speeds under different input sizes.
Table 1.
Comparison of inference speeds under different input sizes.
| Input size |
Validation Dice(%) |
IoU (%) |
HD95 (mm) |
FPS (GPU) |
| 512×512 |
81.4 |
68.3 |
6.1 |
122 |
| 256×256 |
71.8 |
56.0 |
7.4 |
213 |
4.2. Datasets and Evaluation Metrics
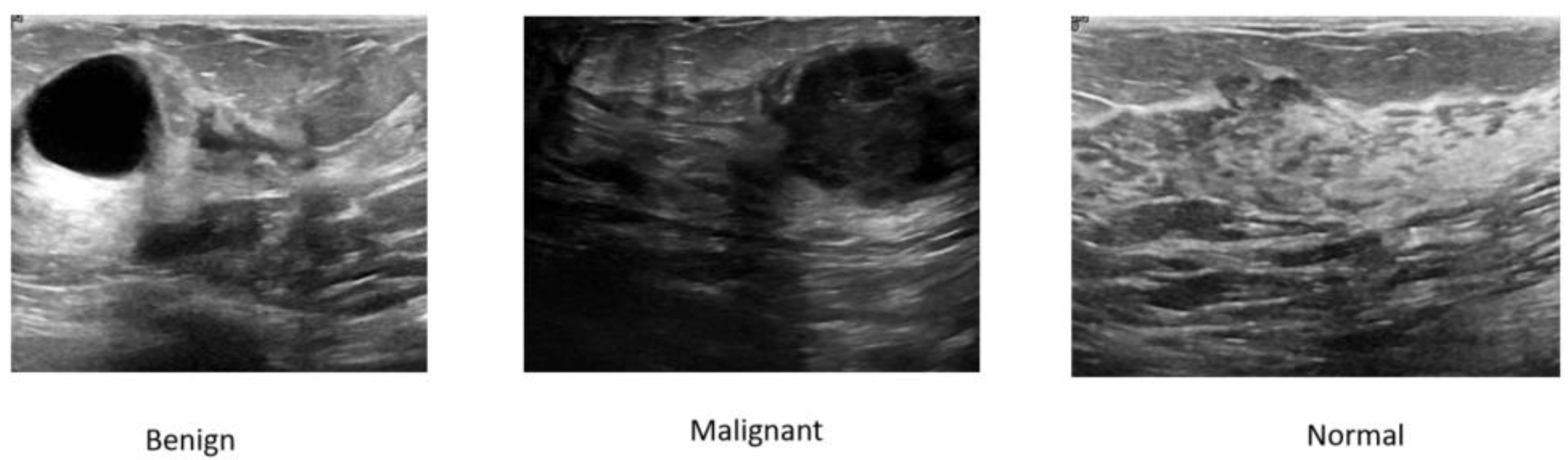
We evaluated LBA-Net on two publicly available breast ultrasound datasets. The BUSI dataset contains 780 images, including 437 benign cases, 210 malignant cases and 133 normal cases. Each image is accompanied by pixel-level truth segmentation masks from multiple patients. For the dataset, we adopt an 80:20 training-test split, further retaining 10% of the training data for validation. In the cross-dataset experiment, the model was only trained on the BUSI dataset without any fine-tuning, thereby conducting a strict assessment of its generalization ability under domain shift. The size of all input images was adjusted to 512×512 pixels to ensure consistency across experiments.
Figure 4 illustrates some representative images from the dataset used in the experiments.
To comprehensively evaluate segmentation performance, we report the following key metrics: Dice Similarity Coefficient (DSC), Intersection over Union (IoU), Precision, Recall, 95% Hausdorff Distance (HD95), and Average Symmetric Surface Distance (ASSD); for assessing efficiency, we further measure critical efficiency-related indicators, including the number of parameters (denoted in millions, M), Floating Point Operations (FLOPs, denoted in billions, G), and frames per second (FPS) tested on both GPU and CPU platforms.
4.3. Implementation Details
All experiments were conducted with PyTorch 1.12 on an NVIDIA A100 GPU. We used AdamW with an initial learning rate of 1×10-3, a 5-epoch linear warmup and a cosine decay schedule; training ran for a maximum of 200 epochs (early stopping usually halted at ≈ epoch 63). Images were resized to 512×512 and fed in batches of 8. Data augmentation included random flips, rotations within ±20∘, scaling (±10%), gamma correction(γ∈ [0.8, 1.2]), Gaussian noise (σ = 0.01), and multiplicative speckle noise (σ = 0.05). The model was trained using a combined loss function: , where is the Dice+BCE loss for segmentation, and is the binary cross-entropy loss for boundary prediction, with λ=0.5."
4.4. Comparison with State-of-the-Art
The results indicate that LBA-Net significantly outperforms all compared models on the BUSI dataset. It achieves a validation Dice of 81.42 % and IoU of 68.62 %, surpassing the best CNN-based competitor (FPN) by approximately 5 pp and 7 pp, respectively. Moreover, it does so with only 2.14 M parameters and ≈ 4.9 G FLOPs, demonstrating a superior accuracy-efficiency trade-off. Its minimum validation loss (0.4902) is also on par with FPN, underscoring its suitability for real-time clinical deployment in resource-limited environments. The detailed performance comparison of each model on the BUSI dataset is summarized in
Table 2.
4.5. Ablation Studies
To verify the necessity and effectiveness of each component in LBA-Net, we conducted systematic ablation experiments on the BUSI dataset. Taking the “full LBA-Net” as the baseline, we gradually removed or replaced key modules, yielding four variants:
- (1)
V1 (w/o LBA-Block): all local-boundary attention blocks in the decoder were replaced with 1×1 convolutions.
- (2)
V2 (w/o ASPP): the ASPP bottleneck was removed and substituted by a 1×1 convolution, discarding multi-scale context.
- (3)
V3 (w/o Boundary Head): only the segmentation head was kept; auxiliary boundary supervision was canceled.
- (4)
V4 (LBA→CBAM): the dual-head architecture was preserved, but the proposed LBA-Block was replaced by CBAM.
All variants shared the same data split, training hyper-parameters, and 512×512 input resolution; the best checkpoint was selected on the validation set.
Table 3 summarizes the parameter count, computational cost (FLOPs), and segmentation performance (Dice / IoU).
Table 3.
Summary of ablation results on BUSI validation set.
Table 3.
Summary of ablation results on BUSI validation set.
| Model Variant |
Val Dice (%)↑ |
Val IoU (%) ↑ |
Params (M) ↓ |
FLOPs (G) ↓ |
| Baseline (Full LBA-Net) |
81.42 |
68.62 |
2.14 |
4.83 |
| V1 (w/o LBA-Block) |
78.91 |
65.21 |
2.03 |
4.75 |
| V2 (w/o ASPP) |
79.53 |
66.88 |
1.98 |
4.62 |
| V3 (w/o Boundary Head) |
80.10 |
66.25 |
2.08 |
4.79 |
| V4 (LBA→CBAM) |
77.65 |
63.95 |
2.15 |
4.86 |
5. Discussion
5.1. Advantages of LBA-Net
The experimental results demonstrate that LBA-Net achieves a favorable balance between segmentation accuracy, computational efficiency, making it well-suited for practical breast ultrasound (BUS) applications.
Lightweight yet Effective: By integrating MobileNetV3 as the encoder, LBA-Net drastically reduces the number of parameters and FLOPs compared to conventional U-Net and transformer-based architectures. Despite its lightweight design, the integration of LBA-Blocks ensures strong feature representation, allowing LBA-Net to achieve competitive or superior segmentation performance.
Robust Boundary Delineation: The dual-head supervision scheme—consisting of a segmentation branch and an auxiliary boundary branch trained with morphology-generated edge maps—significantly improves lesion edge localization. This is particularly important in BUS images, where tumor boundaries are often fuzzy and irregular.
Real-time Feasibility: The lightweight design enables fast inference speed, with high frames per second (FPS) on both GPU and CPU. This makes LBA-Net suitable for integration into portable ultrasound scanners or edge devices, a critical step toward practical CAD systems in low-resource settings.
5.2. Clinical Implications and Future Work
The design of LBA-Net aligns with the practical requirements of clinical deployment: efficiency, and ease of integration. Its lightweight nature enables deployment in point-of-care ultrasound devices, which are increasingly used in primary care, rural healthcare, and mobile health units. With real-time performance, LBA-Net can assist radiologists by providing fast and reliable lesion delineation, potentially improving diagnostic efficiency and reducing operator dependency.
For future research, several directions are worth exploring:Large-scale Clinical Validation: Extending evaluations to multi-center, multi-device datasets with diverse patient populations to confirm robustness and generalizability. Hybrid Context Modeling: Combining LBA-Blocks with lightweight global context modules to further enhance segmentation of large or complex lesions without compromising efficiency. Semi-supervised and Self-supervised Learning: Leveraging unlabeled BUS data to alleviate annotation cost and improve boundary supervision quality. Explainability and User Interaction: Developing visualization tools to interpret LBA-Net’s attention maps and integrating interactive correction mechanisms to increase radiologists’ confidence and usability.
5.3. Summary
In summary, LBA-Net provides a promising step toward lightweight, robust, and clinically practical segmentation models for breast ultrasound imaging. While limitations exist, its design principles—lightweight backbone, efficient attention, and boundary-aware supervision—offer a strong foundation for future advancements in ultrasound CAD systems.
6. Conclusion
In this paper, we proposed LBA-Net, a novel Lightweight Attention Network tailored for robust breast ultrasound image segmentation. The design of LBA-Net integrates three key innovations: a lightweight encoder to ensure computational efficiency, a Lightweight Attention Block (LBA-Block) that combines channel and spatial attention with minimal overhead, and a dual-head supervision strategy that enhances boundary precision through auxiliary boundary learning.
Extensive experiments on a publicly available BUS dataset (BUSI) demonstrated that LBA-Net achieves a superior trade-off between segmentation accuracy and efficiency, outperforming conventional lightweight U-Net variants and approaching or surpassing the performance of heavier state-of-the-art architectures.
Although limitations remain, such as reliance on relatively small datasets and restricted global context modeling, the results suggest that LBA-Net represents a promising step toward deployable, real-time CAD systems for breast cancer screening and diagnosis. Future work will focus on validating LBA-Net on larger multi-center datasets, integrating global context modules in a lightweight manner, and enhancing interpretability for clinical adoption.In conclusion, LBA-Net offers an effective and practical solution for breast ultrasound segmentation, bridging the gap between research innovation and clinical applicability.
Author Contributions
Conceptualization, Y.D. and X.X.; methodology, D.Y. and X.X.; software,D.Y.; validation, D.Y. and J.W.; formal analysis, C.Q.; investigation, C.Q.; data curation, J.W. and C.Q.; writing—original draft preparation, D.Y. and X.X.; writing—review and editing, D.Y., X.X., and Y.Q.; supervision, X.X. and Y.Q.; funding acquisition, X.X. and Y.Q. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
Acknowledgments
The authors would like to thank the anonymous reviewers for their valuable comments and suggestions.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bizuayehu, H.M.; Ahmed, K.Y.; Kibret, G.D.; Dadi, A.F.; Belachew, S.A.; Bagade, T.; Tegegne, T.K.; Venchiarutti, R.L.; Kibret, K.T.; Hailegebireal, A.H.; Assefa, Y.; Khan, M.N.; Abajobir, A.; Alene, K.A.; Mengesha, Z.; Erku, D.; Enquobahrie, D.A.; Minas, T.Z.; Misgan, E.; Ross, A.G. Global Disparities of Cancer and Its Projected Burden in 2050. JAMA Netw Open. 2024, 7, e2443198. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, F.; Chen, Y.J.; Hu, Y.Z. Multimodal Ultrasound Imaging in the Diagnosis of Primary Giant Cell Tumor of the Breast: A Case Report and Literature Review. J Clin Ultrasound. 2025, 53, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Liu, S.; Cao, Z.; Zhang, J.; Pu, X.; Yu, J. Accurate classification of benign and malignant breast tumors in ultrasound imaging with an enhanced deep learning model. Front Bioeng Biotechnol. 2025, 13, 1526260. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yu, L.; Gou, B.; Xia, X.; Yang, Y.; Yi, Z.; Min, X.; He, T. BUS-M2AE: Multi-scale Masked Autoencoder for Breast Ultrasound Image Analysis. Comput Biol Med. 2025, 191, 110159. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.R.; Torres, H.R.; Oliveira, B.; de Araujo, A.R.V.F.; Morais, P.; Novais, P.; Vilaca, J.L. Deep Learning Networks for Breast Lesion Classification in Ultrasound Images: A Comparative Study. Annu Int Conf IEEE Eng Med Biol Soc. 2023, 2023, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Xie, J.; Wan, Y.; Zhang, M.; Qiao, L.; Yu, J.; Chen, S.; Li, B.; Yao, Y. A review of the current state of the computer-aided diagnosis (CAD) systems for breast cancer diagnosis. Open Life Sci. 2022, 17, 1600–1611. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Abdelrahman, L.; Al Ghamdi, M.; Collado-Mesa, F.; Abdel-Mottaleb, M. Convolutional neural networks for breast cancer detection in mammography: A survey. Comput Biol Med. 2021, 131, 104248. [Google Scholar] [CrossRef] [PubMed]
- Derakhshandeh, S.; Mahloojifar, A. Modifying the U-Net's Encoder-Decoder Architecture for Segmentation of Tumors in Breast Ultrasound Images. J Imaging Inform Med. 2025. [Google Scholar] [CrossRef] [PubMed]
- [9] Khaled, R.; Vidal, J.; Vilanova, J.C.; Martí, R. A U-Net Ensemble for breast lesion segmentation in DCE MRI. Comput Biol Med. 2022, 140, 105093. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, P.; Roy, A.; Cuevas, E.; Perez-Cisneros, M.; Sarkar, R. DAU-Net: Dual attention-aided U-Net for segmenting tumor in breast ultrasound images. PLoS One. 2024, 19, e0303670. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wu, Y.; Huang, L.; Yang, T. Breast Ultrasound Image Segmentation Using Multi-branch Skip Connection Search. J Imaging Inform Med. 2025. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi-Goughari, M.; Rajabzadeh, H.; Han, J.W.; Kwon, H.J. Artificial intelligence-assisted ultrasound-guided focused ultrasound therapy: a feasibility study. Int J Hyperthermia. 2023, 40, 2260127. [Google Scholar] [CrossRef] [PubMed]
- Anandha Praba, R.; Suganthi, L. Human activity recognition utilizing optimized attention induced Multihead Convolutional Neural Network with Mobile Net V1 from Mobile health data. Network. 2025, 36, 294–321. [Google Scholar] [CrossRef] [PubMed]
- Kanchana, K.; et al. "Enhancing skin cancer classification using efficient net b0-b7 through convolutional neural networks and transfer learning with patient-specific data. " Asian Pacific Journal of Cancer Prevention: APJCP 2024, 25, 1795. [Google Scholar]
- Sara Koshy, S.; Anbarasi, L.J. HMA-Net: a hybrid mixer framework with multihead attention for breast ultrasound image segmentation. Front Artif Intell. 2025, 8, 1572433. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lou, Q.; Li, Y.; Qian, Y.; Lu, F.; Ma, J. Mammogram classification based on a novel convolutional neural network with efficient channel attention. Comput Biol Med. 2022, 150, 106082. [Google Scholar] [CrossRef] [PubMed]
- Nissar, I.; Alam, S.; Masood, S.; Kashif, M. MOB-CBAM: A dual-channel attention-based deep learning generalizable model for breast cancer molecular subtypes prediction using mammograms. Comput Methods Programs Biomed. 2024, 248, 108121. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Wang, Q.; Bao, Y.; et al. LAEDNet: a lightweight attention encoder–decoder network for ultrasound medical image segmentation. Computers and Electrical Engineering, 2022, 99, 107777. [Google Scholar] [CrossRef]
- Wang, R.; Chen, S.; Ji, C.; Fan, J.; Li, Y. Boundary-aware context neural network for medical image segmentation. Med Image Anal. 2022, 78, 102395. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Min, W.; Wang, S. Boundary-Aware Gradient Operator Network for Medical Image Segmentation. IEEE J Biomed Health Inform. 2024, 28, 4711–4723. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Liu, H.; Zeng, Z.; et al. BES-Net: Boundary enhancing semantic context network for high-resolution image semantic segmentation. Remote Sensing, 2022, 14, 1638. [Google Scholar] [CrossRef]
- Xiong, Y.; Shu, X.; Liu, Q.; et al. HCMNet: A Hybrid CNN-Mamba Network for Breast Ultrasound Segmentation for Consumer Assisted Diagnosis. IEEE Transactions on Consumer Electronics 2025. [Google Scholar] [CrossRef]
- Zhou, J.; Kuang, H.; Wang, J. HCM-Net: Hybrid CNN and Mamba Network with Multi-scale Awareness Feature Fusion for Lung Cancer Pathological Complete Response Prediction[C]//International Symposium on Bioinformatics Research and Applications. Singapore: Springer Nature Singapore, 2025, 38-48.
- DeVoe, K.; Takahashi, G.; Tarshizi, E.; Sacker, A. Evaluation of the precision and accuracy in the classification of breast histopathology images using the MobileNetV3 model. J Pathol Inform. 2024, 15, 100377. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lou, Q.; Li, Y.; Qian, Y.; et al. Mammogram classification based on a novel convolutional neural network with efficient channel attention. Computers in Biology and Medicine, 2022, 150, 106082. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, A.; Anand, V.; Gupta, S.; et al. Attention based UNet model for breast cancer segmentation using BUSI dataset. Scientific Reports, 2024, 14, 22422. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).