1. Introduction
Nuclear instance segmentation is a fundamental task in computational pathology, underpinning a range of downstream applications such as cell classification, tissue phenotyping, and tumor microenvironment analysis [
1]. Accurate delineation of individual nuclei is critical for automated diagnosis and quantitative analysis in histopathological images, which are often challenged by dense cellular arrangements, overlapping nuclei, and diverse morphological variations [
2].
Conventional image processing techniques have shown limited robustness in these complex settings due to their sensitivity to occlusion and shape variability. In contrast, deep learning approaches—particularly convolutional neural network CNN-based models like U-Net [
3] and nnU-Net [
4]—have greatly advanced performance by learning semantic features directly from data. Among these, HoVer-Net [
5] has emerged as a state-of-the-art multi-task architecture capable of simultaneously predicting nuclear pixel maps (NP), horizontal–vertical distance maps (HoVer), and nuclear type maps (TP). This design has established a strong benchmark for nuclear instance segmentation and classification, particularly on large-scale datasets such as PanNuke [
6].
However, despite its effectiveness, HoVer-Net is built upon a residual CNN encoder, which may constrain its ability to capture long-range dependencies and global context. Recent developments in Transformer-based models—such as the Vision Transformer (ViT) [
7], Swin Transformer [
8], and hybrid models like CellViT [
9]—have demonstrated powerful capabilities in modeling spatial relationships, which are especially important for the heterogeneous patterns seen in histopathology images [
10]. Nevertheless, the incorporation of Transformer backbones into multi-task segmentation frameworks like HoVer-Net remains relatively underexplored.
In this study, I present a systematic investigation into enhancing the HoVer-Net architecture through the integration of advanced modules and modern design principles. My key contributions are as follows:
I investigate the integration of Squeeze-and-Excitation (SE) blocks [
11] into the original CNN-based encoder of HoVerNet, aiming to enhance channel-wise feature recalibration. This modification leads to measurable improvements in segmentation accuracy.
I explore the replacement of HoVerNet’s residual CNN encoder with Transformer-based modules, including Vision Transformer (ViT) and Swin Transformer. A unified architecture is proposed to preserve compatibility with HoVerNet’s three-branch output structure.
I design an enhanced decoder architecture that leverages dense connections and dropout regularization, facilitating improved information flow and mitigating overfitting risks during training.
I conduct comparative experiments against the current state-of-the-art model, CellViT, and perform ablation studies to assess the individual contributions of each architectural component.
All experiments are conducted on the PanNuke dataset, following consistent training protocols—such as fixed epoch count, learning rate, pretrained models, optimizer, and train-validation splits—to ensure fair evaluation. While not all proposed models surpass the original HoVerNet in every metric, the findings emphasize the value of attention mechanisms and architectural enhancements in advancing segmentation and classification performance.
2. Materials and Methods
2.1. Dataset
All experiments in this study were conducted on the publicly available PanNuke dataset, a large-scale benchmark specifically curated for nuclei instance segmentation and classification tasks. PanNuke contains pixel-wise annotated histopathology image patches spanning 19 distinct tissue types. Each nucleus within the dataset is labeled according to one of five predefined categories: neoplastic, inflammatory, connective, dead, and epithelial [
6].
The dataset was chosen for its comprehensive and high-quality annotations, as well as its established usage in prior research. These characteristics make PanNuke particularly suitable for evaluating multi-class nuclear segmentation methods and enable consistent comparisons with other state-of-the-art approaches.
2.2. Preprocessing and Augmentation
To ensure fair and consistent evaluation across all models, I adopted the preprocessing and augmentation procedures from the original HoVerNet pipeline. Input histopathology images were first normalized and resized, followed by padding to align with architectural constraints and to maintain spatial consistency across samples. Random shuffling of training data was applied to mitigate sampling bias and enhance the models’ generalization performance [
12].
The PanNuke dataset was partitioned into training, validation, and testing subsets using a ratio of 2:6:2, ensuring a balanced distribution of tissue types and cell classes across splits.
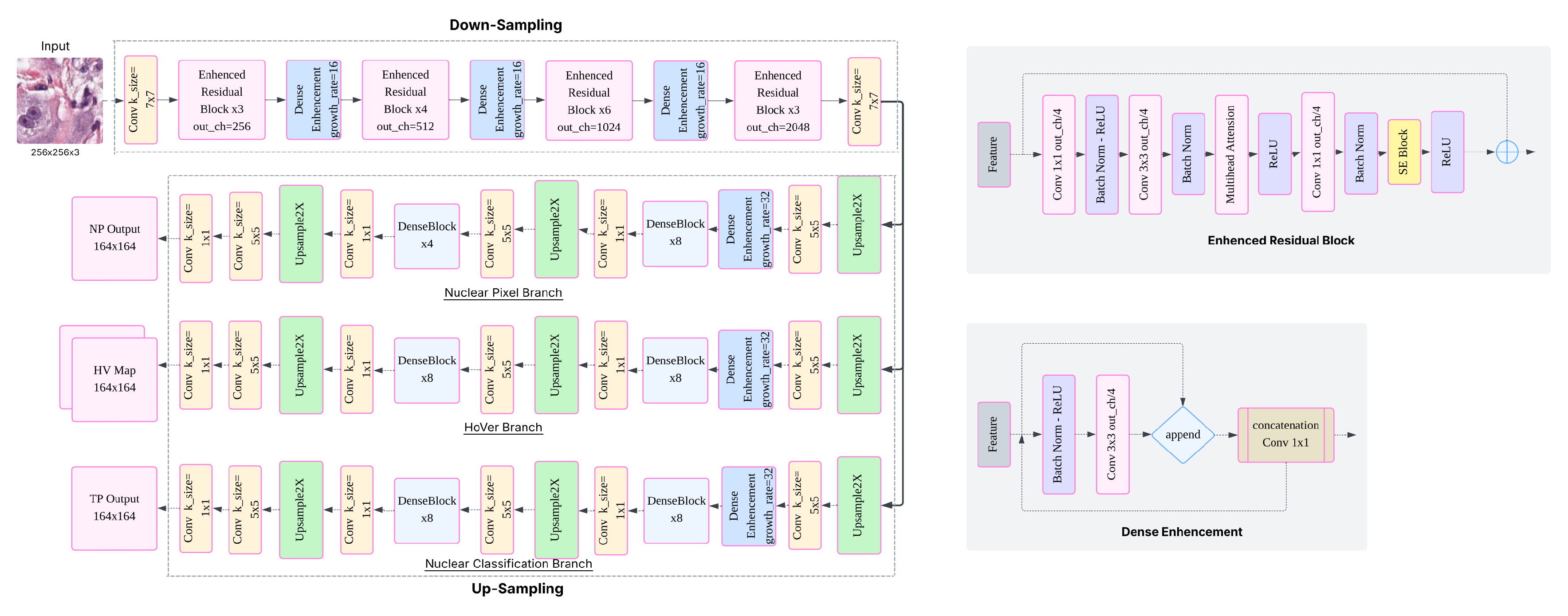
2.3. Model Variants Based on HoVerNet Architecture
To investigate the effects of architectural modifications on nuclei instance segmentation and classification, we developed six variants of the original HoVerNet framework. Each variant retains HoVerNet’s distinctive three-head output structure—
np_map,
hv_map, and
tp_map—which enables joint nuclear segmentation and classification [
5]. The modifications focus on enhancing the encoder and decoder designs, and are detailed as follows
(the pretrained model can be found in Appendix A and training code in Appendix B):
HoVerNet + SE (HoverNetEnhanced): This variant augments the encoder with Squeeze-and-Excitation (SE) blocks, which are integrated within the residual units. SE blocks perform adaptive channel-wise recalibration, emphasizing informative features while suppressing less useful ones, thereby enhancing representational capacity [
11].
HoVerNet + Multi-head Attention (Multihead-HoverNet): In this model, multi-head self-attention (MHSA) modules [
13] are embedded into the encoder to capture long-range dependencies and global contextual cues. Inspired by prior analysis of attention heads [
14], this design seeks to improve performance on complex spatial structures.
HoVerNet + SE + MHSA + Enhanced Decoder (MSDHV-Net): This is the most comprehensive CNN-based modification. It combines both SE and MHSA modules in the encoder and introduces a redesigned decoder featuring deeper DenseBlock structures, additional skip connections, and convolutional refinement layers. This design aims to facilitate robust feature propagation and enhanced spatial resolution restoration [
15].
HoVerNet + ViT Encoder (HoverViTNet): Here, the conventional CNN encoder is replaced with a Vision Transformer (ViT) [
7], enabling the model to extract patch-wise global representations using self-attention. These transformer-derived features are decoded via a CNN-based decoder, allowing comparative evaluation of attention-driven global context modeling [
16].
HoVerNet + Custom SwinViT Encoder (HoVerIT): This architecture integrates a custom Swin Transformer encoder into the HoVerNet pipeline. The hierarchical design and shifted window self-attention in SwinViT capture both local and global dependencies more effectively than vanilla ViT [
17]. The idea draws inspiration from the Swin-UNETR architecture [
18], while maintaining compatibility with HoVerNet’s three-branch outputs.
HoVerNet + SwinUNETR from MONAI (HoverSwinNet): In this variant, the encoder is directly replaced with the SwinUNETR backbone from MONAI [
18]. Transformer-derived multi-scale features are routed through HoVerNet’s original three-branch decoders, serving as a strong baseline to assess the integration feasibility and performance of prebuilt transformer encoders.
2.4. Training Configuration
All models were trained using the Adam optimizer [
19] with a fixed learning rate of 1e
-4. Pre-trained ResNet-50 weights [
20] were used to initialize the encoder when applicable. Although alternative optimizers such as AdamW [
21] were evaluated, they did not produce noticeable improvements in performance. Attempts to modify the learning rate led to unstable training dynamics, including instances of gradient explosion, thereby justifying the choice of a conservative fixed schedule.
Each model was trained for 80 epochs, based on empirical observations that validation performance typically saturated between epochs 50 and 60
(See Figure A1). A batch size of 16 was used to optimize memory utilization on an NVIDIA L20 GPU with 44 GB VRAM.
The loss functions followed the original HoVerNet formulation: binary cross-entropy (BCE) combined with Dice loss for nuclear pixel (np_map) and nuclear type (tp_map) predictions, and a joint loss of mean squared error (MSE) and mean squared gradient error (MSGE) for the horizontal-vertical distance (hv_map) regression task.
2.5. Evaluation Metrics
To comprehensively evaluate model performance, we employed several quantitative metrics: DICE coefficient, Panoptic Quality (PQ), Precision, F1-score, and Recall. These metrics were computed separately for each predicted cell type, enabling a fine-grained analysis of segmentation and classification performance. This multi-metric evaluation framework facilitated rigorous comparisons across different architectural variants and offered insight into the effectiveness and limitations of each proposed enhancement [
22].
3. Results
3.1. Baseline Performance
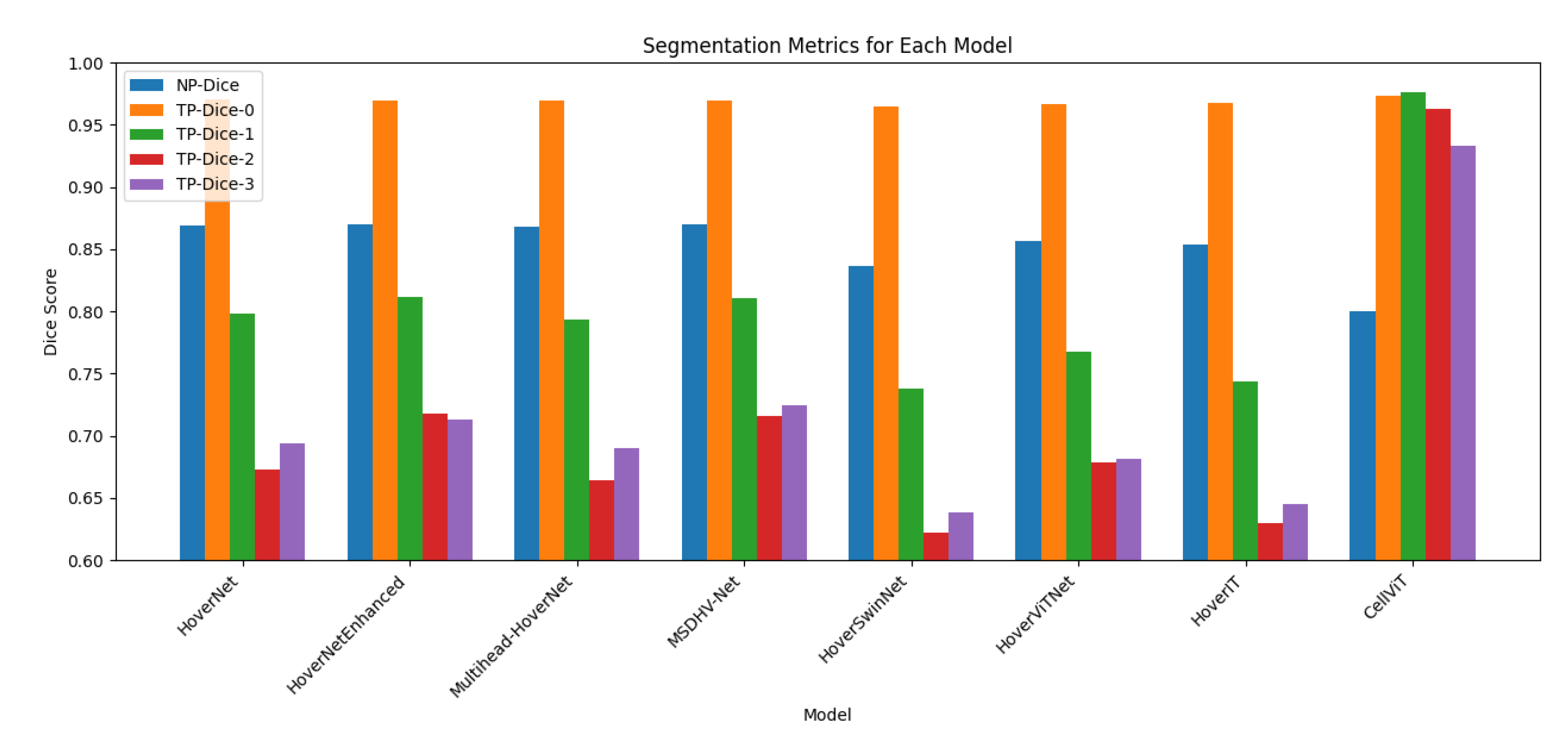
To establish a reliable benchmark, I trained the original HoVer-Net architecture on the PanNuke dataset using the standardized training pipeline described earlier. The baseline model achieved an NP-Dice score of 0.8686, indicating strong segmentation accuracy for nuclear regions. For type prediction (TP), Dice scores were 0.9703, 0.7978, 0.6728, and 0.6939 across the four nuclear subtypes, reflecting varying levels of classification performance.
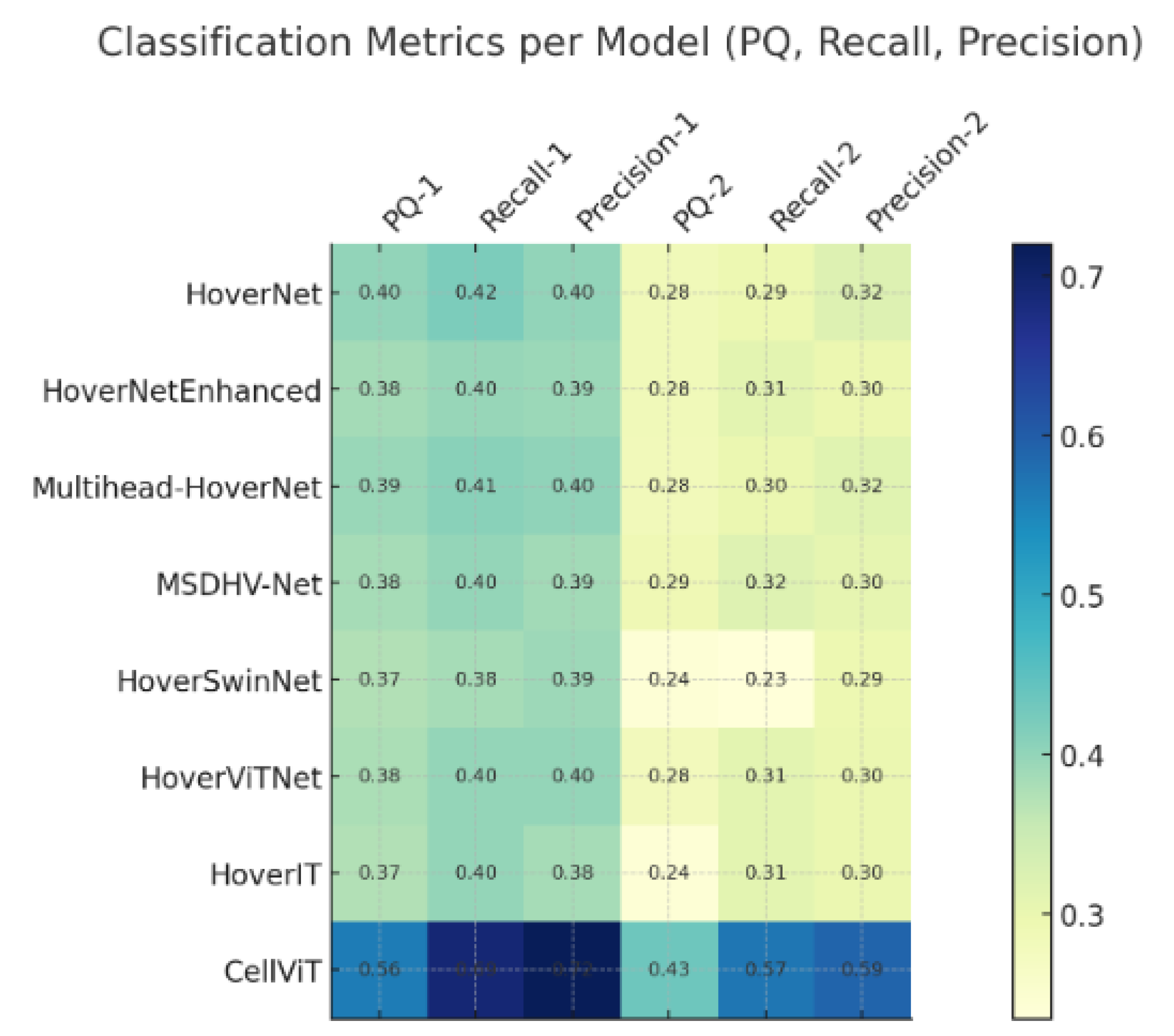
In terms of instance-level evaluation, Panoptic Quality (PQ) scores were 0.4005 for neoplastic, 0.2830 for inflammatory, and 0.4526 for other cell types. Additionally, standard classification metrics such as F1-score, Precision, and Recall were computed to provide a comprehensive view of the model’s strengths and weaknesses. These results serve as a foundational reference for comparing the performance of all proposed architectural variants in subsequent experiments.
3.2. CNN-Based Architectural Enhancements
Three key modifications were explored within the traditional CNN-based design framework: the incorporation of Squeeze-and-Excitation (SE) blocks [
11], the integration of multi-head self-attention modules [
13], and the use of an enhanced DenseBlock-based decoder. The most effective configuration, referred to as
MSDHV-Net, combined all three enhancements.
Compared to the original HoVerNet, MSDHV-Net achieved consistent performance gains across multiple metrics. Specifically, the TP-Dice score for inflammatory nuclei increased from 0.6728 to 0.7162, while the TP-Dice score for the “other” cell category improved from 0.6939 to 0.7241. The NP-Dice and overall classification Dice scores also exhibited an average improvement of approximately 2%
(See Table 1 and Table 2 for more details).
However, despite these enhancements, performance on neoplastic cells remained challenging. Both the PQ and F1 scores for this class experienced a slight decline of around 2% relative to the baseline, suggesting that additional strategies may be required to address the morphological variability and contextual ambiguity characteristic of neoplastic nuclei.
3.3. Transformer-Based Architectural Variants
The effectiveness of replacing the original CNN encoder with Transformer-based architectures was also evaluated. Three model variants were explored:
HoverSwinNet, which directly employed the SwinViT encoder from the MONAI SwinUNETR implementation [
18];
HoVerIT, which integrated a customized Swin Transformer encoder into the original HoVerNet three-branch framework; and
HoverViTNet, which substituted the encoder with a vanilla Vision Transformer (ViT) [
7].
All Transformer-based models underperformed compared to the CNN-based baseline. Among them, HoverViTNet demonstrated slightly better results than HoverSwinNet, but overall, the Transformer variants exhibited a consistent degradation of 3–5% across Dice, Panoptic Quality (PQ), and classification-related metrics
(See Table 1 and Table 2 for more details). These results suggest that while Transformer encoders offer strong theoretical advantages in capturing long-range dependencies, their practical integration into multi-task nuclei segmentation frameworks remains a non-trivial challenge that warrants further investigation.
3.4. Summary of Comparative Performance
Among all proposed architectures,
MSDHV-Net demonstrated the most consistent and meaningful improvements
(See Figure 1 and Figure 2 for comparision), particularly for inflammatory and other nuclear types. The transformer-based models, while conceptually promising, require further tuning or hybridization to outperform well-optimized CNN backbones on the PanNuke dataset.
3.5. Formatting of Mathematical Components
Binary Cross-Entropy (BCE):
Mean Squared Error (MSE):
4. Discussion
This study aimed to investigate architectural enhancements to the HoVer-Net framework for simultaneous nuclei segmentation and classification [
5]. My findings demonstrate that thoughtful modifications within the CNN paradigm—specifically the integration of Squeeze-and-Excitation (SE) blocks, multi-head attention, and an enhanced DenseBlock decoder—can lead to measurable gains across multiple evaluation metrics. The best-performing model, MSDHV-Net
(See Figure 3 for architecture), showed improved TP-Dice scores for Neoplastic, inflammatory and other nuclei types, indicating enhanced discriminative power in challenging classification scenarios.
These improvements are consistent with prior research emphasizing the importance of adaptive channel recalibration (as in SE blocks) and attention mechanisms in deep feature extraction for biomedical imaging tasks [
11]. The enhanced decoder appears to better capture spatial dependencies and refine instance boundaries, especially in complex tissue environments.
In contrast, my exploration of Vision Transformer (ViT and SwinViT) encoders—although theoretically appealing due to their global receptive field—did not yield superior performance. HoverSwinNet and HoVerIT
(See Figure 4 for architecture) underperformed compared to both the baseline and CNN-enhanced models. One likely reason is that transformer-based models may require significantly larger training data or more domain-specific pretraining to outperform CNNs in medical imaging, for example the SAM-ViT pretrained model used in CellVit [
9]. Another limitation lies in integrating transformer encoders into multi-task frameworks like HoVer-Net, which may require carefully aligned intermediate feature representations.
Figure 4.
Overview of the HoVerIT architecture.
Figure 4.
Overview of the HoVerIT architecture.
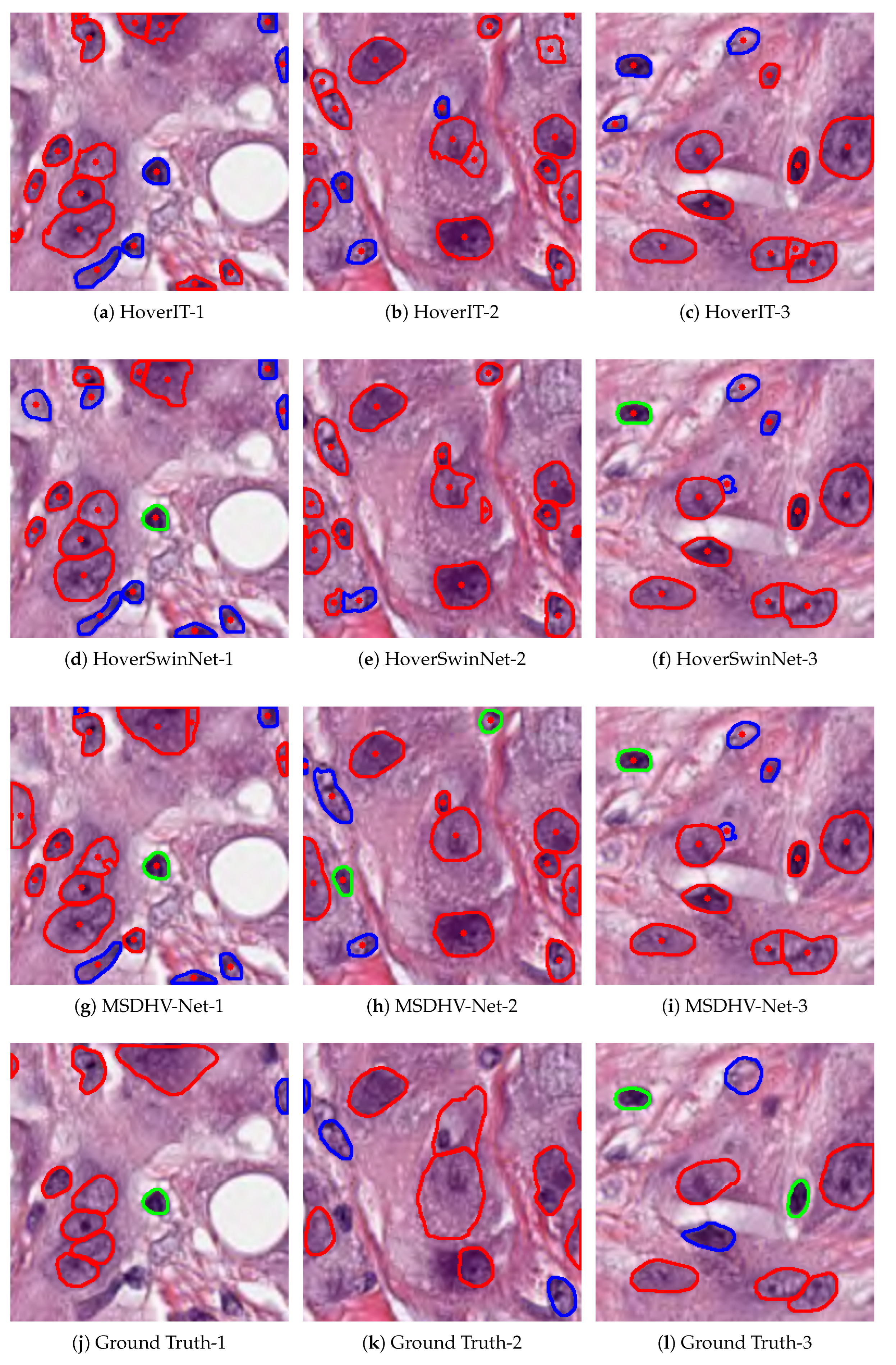
Figure 5.
Segmentation and classification results of three different models on multiple test patches.
Figure 5.
Segmentation and classification results of three different models on multiple test patches.
Interestingly, all models struggled to improve classification metrics for neoplastic nuclei. This suggests either intrinsic ambiguity in their visual features or insufficient discriminatory signal in current feature representations. Future work could explore class-specific loss weighting or incorporate cell microenvironment context to enhance neoplastic classification.
Looking forward, promising directions include hybrid CNN–transformer architectures, domain-adaptive pretraining strategies, and exploring self-supervised representation learning to leverage unlabeled histopathology data. Additionally, integrating spatially aware attention mechanisms and refining decoder design may further enhance both instance segmentation and fine-grained classification capabilities.
Overall, this work highlights the value of selectively integrating modern deep learning components into established architectures, balancing innovation with task-specific constraints in computational pathology.
5. Conclusions
In this work, I systematically investigated architectural enhancements to the HoVer-Net framework [
5] for nuclei instance segmentation and classification on the PanNuke dataset. Through the integration of SE blocks [
11], multi-head attention, and a more expressive decoder, I developed MSDHV-Net, which consistently outperformed the original HoVer-Net in both segmentation and classification tasks—particularly for inflammatory and other nuclei types.
In contrast, transformer-based variants such as HoverSwinNet and HoVerIT did not demonstrate improved performance, suggesting that CNN-based backbones remain more robust and effective under the current dataset and training conditions. These results underscore the value of carefully engineered improvements to established CNN architectures, while also highlighting that successful transformer integration—such as those attempted in CellViT with SAM-ViT pretraining [
9]—requires further domain adaptation and architectural refinement.
Overall, these findings provide practical insights for designing more accurate and efficient nuclei analysis models and lay a foundation for future work exploring hybrid CNN-transformer architectures and self-supervised learning approaches in computational pathology [
21].
Author Contributions
Conceptualization, Shizhuo Qu; methodology, Shizhuo Qu; software, Shizhuo Qu; validation, Shizhuo Qu; formal analysis, Shizhuo Qu; investigation, Shizhuo Qu; resources, Shizhuo Qu; data curation, Shizhuo Qu; writing—original draft preparation, Shizhuo Qu; writing—review and editing, Shizhuo Qu; visualization, Shizhuo Qu; supervision, Shizhuo Qu; project administration, Shizhuo Qu. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The dataset used in this study is publicly available. All experiments were conducted on the PanNuke dataset, which provides annotated histopathological images for nuclei instance segmentation and classification. The dataset can be accessed at
https://warwick.ac.uk/fac/cross_fac/tia/data/pannuke. No new datasets were generated during the current study.
Acknowledgments
The author would like to express sincere gratitude to Jiang Yu from Yangtze River Delta Guozhi (Shanghai) Intelligent Medical Technology Co., Ltd., for entrusting the author with the responsibility of conducting the nuclei segmentation and classification component in the group project AI Multi-model System Prediction of Treatment Response to PD-1 Combined Chemotherapy in Advanced Gastric Cancer. This opportunity provided both inspiration and direction for the experimental work presented in this paper. Special thanks are also extended to Zhou Yin for technical support in server maintenance and PanNuke dataset preprocessing, and to Zhang Zihao (MSc, King’s College London) for generously providing fundamental tutorials and guidance in computer vision for medical imaging. Their support has been instrumental in the successful completion of this study.
Conflicts of Interest
The author declares no conflicts of interest. This study was conducted during a cooperative research internship at Yangtze River Delta Guozhi (Shanghai) Intelligent Medical Technology Co., Ltd. While the company provided the research direction and access to computing resources, it had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| MDPI |
Multidisciplinary Digital Publishing Institute |
| SE |
Squeeze-and-Excitation |
| CNN |
Convolutional Neural Network |
| ViT |
Vision Transformer |
Appendix A. Additional Materials and Resources
To promote reproducibility and transparency, additional materials are provided as follows:
Appendix B. Model Structure and Implementation Code
These resources are intended solely for academic and non-commercial use.
Appendix C. TensorBoard Training Logs and Visualizations
Figure A1.
Segmentation and classification results of different models on multiple validation metrics.
Figure A1.
Segmentation and classification results of different models on multiple validation metrics.
References
- Sirinukunwattana, K.; Snead, D.; Epstein, D.; Aftab, Z.; Mujeeb, I.; Tsang, Y.W.; Cree, I.; Rajpoot, N. Novel digital signatures of tissue phenotypes for predicting distant metastasis in colorectal cancer. Scientific reports 2018, 8, 13692. [Google Scholar] [CrossRef] [PubMed]
- Javed, S.; Fraz, M.M.; Epstein, D.; Snead, D.; Rajpoot, N.M. Cellular community detection for tissue phenotyping in histology images. In Computational Pathology and Ophthalmic Medical Image Analysis; Springer: Cham, Switzerland, 2018; pp. 120–129. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A Self-Configuring Method for Deep Learning-Based Biomedical Image Segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Graham, S.; Vu, Q.D.; Raza, S.E.; Azam, A.; Tsang, Y.W.; Kwak, J.T.; Rajpoot, N. Hover-net: Simultaneous segmentation and classification of nuclei in multi-tissue histology images. Medical Image Analysis 2019, 58, 101563. [Google Scholar] [CrossRef] [PubMed]
- Gamper, J.; Koohbanani, N.A.; Graham, S.; Jahanifar, M.; Khurram, S.A.; Azam, A.; Hewitt, K.; Rajpoot, N. PanNuke Dataset Extension, Insights and Baselines. arXiv 2020, arXiv:2003.10778. https://arxiv.org/abs/2003.10778. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T. An Image Is Worth 16×16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. https://arxiv.org/abs/2010.11929. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; ...; Guo, B. Swin Transformer: Hierarchical Vision Transformer Using Shifted Windows. In Proc. IEEE/CVF Int. Conf. Comput. Vis.; 2021; pp. 10012–10022. [CrossRef]
- Hörst, F.; Rempe, M.; Heine, L.; Seibold, C.; Keyl, J.; Baldini, G.; Kleesiek, J. CellViT: Vision Transformers for Precise Cell Segmentation and Classification. Medical Image Analysis 2024, 94, 103143. [Google Scholar] [CrossRef]
- H. Xu et al., "Vision Transformers for Computational Histopathology," IEEE Reviews in Biomedical Engineering, vol. 17, pp. 63–79, 2024. [CrossRef]
- J. Hu, L. Shen, and G. Sun, "Squeeze-and-excitation networks," in Proc. IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR), 2018, pp. 7132–7141. [CrossRef]
- Zhang, Z. Enhancing Distributed Machine Learning through Data Shuffling: Techniques, Challenges, and Implications. In ITM Web of Conferences 2025, 73, 03018. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is All You Need. Advances in Neural Information Processing Systems 2017, 30. [Google Scholar] [CrossRef]
- Voita, E.; Talbot, D.; Moiseev, F.; Sennrich, R.; Titov, I. Analyzing multi-head self-attention: Specialized heads do the heavy lifting, the rest can be pruned. arXiv 2020, arXiv:1905.09418. https://aclanthology.org/P19-1580/. [Google Scholar]
- Mao, X.; Shen, C.; Yang, Y.B. Image Restoration Using Very Deep Convolutional Encoder-Decoder Networks with Symmetric Skip Connections. Advances in Neural Information Processing Systems 2016, 29, 2802–2810. [Google Scholar] [CrossRef]
- Wang, Z.; Li, T.; Zheng, J.Q.; Huang, B. When CNN Meet with ViT: Towards Semi-Supervised Learning for Multi-Class Medical Image Semantic Segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Cham, Switzerland, October 2022; Springer Nature Switzerland: Cham, 2022; pp. 424–441. [Google Scholar] [CrossRef]
- Yang, J.; Li, C.; Zhang, P.; Dai, X.; Xiao, B.; Yuan, L.; Gao, J. Focal Self-Attention for Local-Global Interactions in Vision Transformers. arXiv preprint 2021, arXiv:2107.00641. [Google Scholar] [CrossRef]
- Hatamizadeh, A.; Nath, V.; Tang, Y.; Yang, D.; Roth, H.R.; Xu, D. Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images. In International MICCAI Brainlesion Workshop; Springer: Cham, Switzerland, 2021; pp. 272–284. [Google Scholar] [CrossRef]
- D. P. Kingma and J. Ba, "Adam: A method for stochastic optimization," arXiv preprint arXiv:1412.6980, 2014. https://doi.org/10.48550/arXiv.1412.6980. [CrossRef]
- K. He, X. Zhang, S. Ren, and J. Sun, "Deep residual learning for image recognition," in Proc. IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR), 2016, pp. 770–778. https://www.cv-foundation.org/openaccess/content_cvpr_2016/papers/He_Deep_Residual_Learning_CVPR_2016_paper.pdf.
- I. Loshchilov and F. Hutter, "Decoupled weight decay regularization," arXiv preprint arXiv:1711.05101, 2017. https://www.semanticscholar.org/paper/Decoupled-Weight-Decay-Regularization-Loshchilov-Hutter/d07284a6811f1b2745d91bdb06b040b57f226882.
- A. Kirillov, K. He, R. Girshick, C. Rother, and P. Dollár, "Panoptic segmentation," in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit. (CVPR), 2019, pp. 9404–9413. https://openaccess.thecvf.com/content_CVPR_2019/papers/Kirillov_Panoptic_Segmentation_CVPR_2019_paper.pdf.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).