Submitted:
16 September 2025
Posted:
17 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. An Overview of the Artificial Intelligence (AI): Categories and Evolving Techniques
1.2. Current Challenges in Veterinary AI Adoption
1.3. AI Applications in Veterinary Medicine
1.4. Some Potential Roles of AI/ML in Studying Viral Diseases in the Context of the One Health Concept
1.5. Accelerating the Vaccines and Antiviral Therapy Development Using the Enhanced ML/AI Tools
2. Historical and Conceptual Background of Vaccines and the Integration of AI in Modern Vaccine Development
2.1. Types of the Currently Known Vaccines
2.1.1. First Generation Vaccines:
2.1.2. Second Generation Vaccines
2.1.3. Third Generation Vaccines
2.2. Integration of ML/AI and Bioinformatics in Vaccine Design and Evaluation
3. AI-Driven Approaches in Vaccine Design and Development
3.1. Comparative Analysis of AI/ML and the Traditional Approaches in Vaccine Design and Development
3.2. Identification of the Key Viral Proteins/Immunogens for Vaccine Design
3.3. The Procedure of Vaccine Design and Optimization Using the AI/ML Tools
3.4. Roles of AI/ML in the Design and Development of the Next Generation Viral Vaccines
3.5. Transforming the Preclinical and Clinical Vaccine Development Through the AI-Driven Simulations and Trial Optimization
4. Pipelines of Vaccine Design Using Enhanced AI/ML Tools
4.1. Preprocessing and Curation of Viral Protein Sequences for the AI/ML-Based Vaccine Design for Some Common Viral Diseases of Poultry
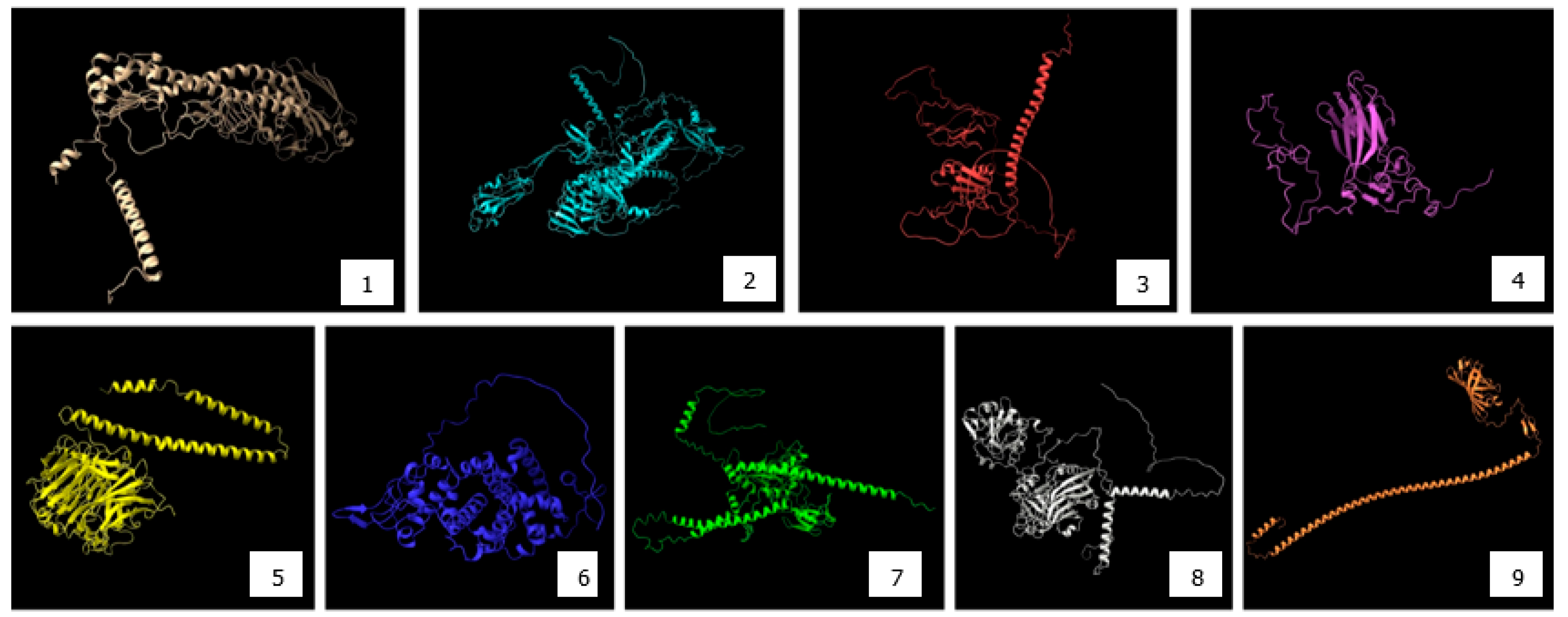
4.2. Epitope Mapping and Antigen-Predicting Tools
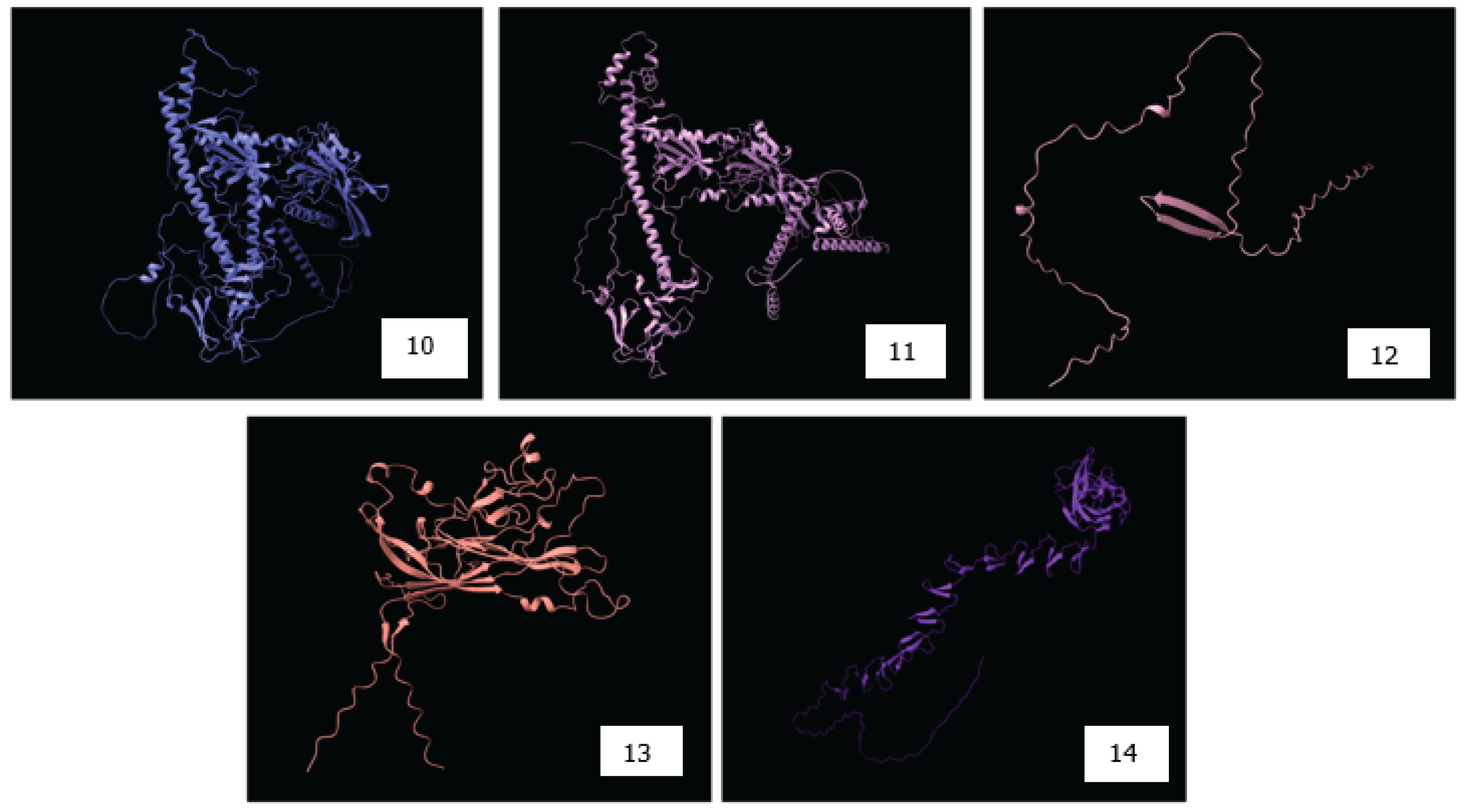
4.3. Structural Modeling and Validation of Viral Proteins Using AlphaFold2 for Accurate Epitope Localization and Vaccine Design
4.4. Approaches to the Design and Optimization of Some Multi-Epitope-Based DNA Vaccine Constructs Against Some Common Avian Viruses
4.5. The in Silico Assessment of the Immunogenicity, Allergenicity, Toxicity, and Structural Stability of the Multi-Epitope DNA Vaccine Constructs
4.6. In Silico Immune Simulation
5. Challenges and Limitations of the Use of AI/ML in Vaccine Design
5.1. Data Quality and Availability
5.2. Some ethical and Regulatory Challenges in the AI/ML-Driven Vaccine Development
5.3. The Availability of the Infrastructure and Sustainability Challenges in Using AI/ML in Vaccine Development
6. Some Future Directions in the Application of AI/ML in Avian Viral Vaccines Design and Development
6.1. Integration of AI/ML and Other Bioinformatics Tools to Enhance the Vaccine Design and Development
6.2. Integration of AI/ML in the Personalized Vaccine Development
6.3. The Potential Roles of the AI Tools and Their Applications in the Emergency Preparedness Plans for the Next Pandemic
7. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| ML | Machine learning |
| DNA | Deoxyribonucleic acid |
| RNA | Ribonucleic acid |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus-2 |
| COVID-19 | Coronavirus disease-2019 |
References
- Lu, H. , Generative AI for vaccine misbelief correction: Insights from targeting extraversion and pseudoscientific beliefs. Vaccine, 2025. 54: p. 127018.
- Feuerriegel, S. , et al., Generative AI. Business & Information Systems Engineering, 2024. 66(1): p. 111-126.
- Topol, E.J. , High-performance medicine: the convergence of human and artificial intelligence. Nature Medicine, 2019. 25(1): p. 44-56.
- Appleby, R.B. and P.S. Basran, Artificial intelligence in veterinary medicine. J Am Vet Med Assoc, 2022. 260(8): p. 819-824.
- Duraisamy, N. , et al., Machine learning tools used for mapping some immunogenic epitopes within the major structural proteins of the bovine coronavirus (BCoV) and for the in silico design of the multiepitope-based vaccines. Front Vet Sci, 2024. 11: p. 1468890.
- Duraisamy, N. , et al., A Pan-H5N1 Multiepitope DNA Vaccine Construct Targeting Some Key Proteins of the Clade 2.3.4.4b Using AI-Assisted Epitope Mapping and Molecular Docking. Viruses, 2025. 17(9): p. 1152.
- Khan, M.Y. , et al., Repurposing of Some Nucleoside Analogs Targeting Some Key Proteins of the Avian H5N1 Clade 2.3.4.4b to Combat the Circulating HPAI in Birds: An In Silico Approach. Viruses, 2025. 17(7).
- Khan, M.Y. , et al., Identification of potential inhibitors of the main protease from feline infectious peritonitis virus using molecular docking and dynamic simulation approaches. PeerJ, 2025. 13: p. e19744.
- Khan, M.Y. , et al., Leveraging Artificial Intelligence and Gene Expression Analysis to Identify Some Potential Bovine Coronavirus (BCoV) Receptors and Host Cell Enzymes Potentially Involved in the Viral Replication and Tissue Tropism. Int J Mol Sci, 2025. 26(3).
- Krishna, K. , et al., Generating SOAP Notes from Doctor-Patient Conversations Using Modular Summarization Techniques. Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (Volume 1: Long Papers), 2021/8.
- Guo, W. , et al., Innovative applications of artificial intelligence in zoonotic disease management. Science in One Health, 2023. 2: p. 100045.
- Sharma, S., P. D. Yadav, and S. Cherian, Comprehensive immunoinformatics and bioinformatics strategies for designing a multi-epitope based vaccine targeting structural proteins of Nipah virus. Front Immunol, 2025. 16: p. 1535322.
- Naveed, M. , et al., Immunoinformatics Approach to Design Multi-Epitope-Based Vaccine against Machupo Virus Taking Viral Nucleocapsid as a Potential Candidate. Vaccines (Basel), 2022. 10(10).
- Dashti, F. , et al., A computational approach to design a multiepitope vaccine against H5N1 virus. Virol J, 2024. 21(1): p. 67.
- Saihar, A. , et al., From bytes to bites: In-silico creation of a novel multi-epitope vaccine against Murray Valley Encephalitis Virus. Microb Pathog, 2025. 198: p. 107171.
- de Sena, B.V. , et al., Extreme lymphocytosis in a dog with T-zone lymphoma. Open Vet J, 2023. 13(12): p. 1760-1768.
- Zaher, M.R. , et al., A novel immunoinformatic approach for design and evaluation of heptavalent multiepitope foot-and-mouth disease virus vaccine. BMC Vet Res, 2025. 21(1): p. 152.
- Zhu, Y. , et al., Construction of an attenuated goatpox virus AV41 strain by deleting the TK gene and ORF8-18. Antiviral Res, 2018. 157: p. 111-119.
- Izhari, M.A. , et al., Design and Validation of a Multi-Epitope mRNA Vaccine Construct Against Human Monkeypox Virus (hMPXV) by Annotating Protein of Intracellular Mature Virus (IMV) Form of hMPXV. Biomedicines, 2025. 13(6).
- Nielsen, P.H. and C.M. Singleton, Parasitic bacteria control foam formation. Nat Microbiol, 2021. 6(6): p. 701-702.
- Sanetra, A.M. , et al., Electrophysiological complexity in the rat dorsomedial hypothalamus and its susceptibility to daily rhythms and high-fat diet. Eur J Neurosci, 2022. 56(4): p. 4363-4377.
- Kar, P.P. , et al., Design of a multi-epitope protein as a subunit vaccine against lumpy skin disease using an immunoinformatics approach. Sci Rep, 2022. 12(1): p. 19411.
- Tataje-Lavanda, L. , et al., Identification and evaluation in-vitro of conserved peptides with high affinity to MHC-I as potential protective epitopes for Newcastle disease virus vaccines. BMC Vet Res, 2023. 19(1): p. 196.
- Elshafei, S.O., N. A. Mahmoud, and Y.A. Almofti, Immunoinformatics, molecular docking and dynamics simulation approaches unveil a multi epitope-based potent peptide vaccine candidate against avian leukosis virus. Sci Rep, 2024. 14(1): p. 2870.
- Chen, L.M., F. Jiang, and H. Hu, A Case of Spontaneous Ovarian Artery Haemorrhage in a Post-Menopausal Woman with Congenital Solitary Kidney. J Coll Physicians Surg Pak, 2022. 32(12): p. SS137-SS139.
- Paul, B. , et al., Immunoinformatics for Novel Multi-Epitope Vaccine Development in Canine Parvovirus Infections. Biomedicines, 2023. 11(8).
- Simbulan, A.M. , et al., Immunoinformatics-guided approach for designing a pan-proteome multi-epitope subunit vaccine against African swine fever virus. Sci Rep, 2024. 14(1): p. 1354.
- Sharma, A.D. , et al., Immunoinformatics-driven design of a multi-epitope vaccine targeting neonatal rotavirus with focus on outer capsid proteins VP4 and VP7 and non structural proteins NSP2 and NSP5. Sci Rep, 2025. 15(1): p. 11879.
- Pang, F., Q. Long, and S. Liang, Designing a multi-epitope subunit vaccine against Orf virus using molecular docking and molecular dynamics. Virulence, 2024. 15(1): p. 2398171.
- Long, Q. , et al., Design of a multi-epitope vaccine against goatpox virus using an immunoinformatics approach. Front Cell Infect Microbiol, 2023. 13: p. 1309096.
- Roy, R.R. , et al., Identification of B-cell epitopes of Indian Zika virus strains using immunoinformatics. Front Immunol, 2025. 16: p. 1534737.
- Naskar, S. , et al., Super epitope dengue vaccine instigated serotype independent immune protection in-silico. Vaccine, 2024. 42(18): p. 3857-3873.
- Fatima, I. , et al., Designing of a multi-epitopes-based peptide vaccine against rift valley fever virus and its validation through integrated computational approaches. Comput Biol Med, 2022. 141: p. 105151.
- Katayama, M. , et al., Antigenic commonality and divergence of hemagglutinin-esterase-fusion protein among influenza D virus lineages revealed using epitope mapping. J Virol, 2024. 98(3): p. e0190823.
- Mahdeen, A.A. , et al., Designing novel multiepitope mRNA vaccine targeting Hendra virus (HeV): An integrative approach utilizing immunoinformatics, reverse vaccinology, and molecular dynamics simulation. PLoS One, 2024. 19(10): p. e0312239.
- El Arab, R.A. , et al., Artificial intelligence in vaccine research and development: an umbrella review. Front Immunol, 2025. 16: p. 1567116.
- Waheed, A., I. H. Aljundi, and U. Baig, Recovery of Dissolved Hydrogen Sulfide from Various Wastewater Streams Using Membranes and Other Relevant Techniques: A Review. Membranes (Basel), 2023. 13(7).
- Venkateswaran, D. , et al., Designing a multi-epitope vaccine against African swine fever virus using immunoinformatics approach. Sci Rep, 2025. 15(1): p. 16044.
- Kolla, H.B. , et al., Immuno-informatics study identifies conserved T cell epitopes in non-structural proteins of Bluetongue virus serotypes: formulation of a computationally optimized next-generation broad-spectrum multi-epitope vaccine. Front Immunol, 2024. 15: p. 1424307.
- Hou, W. , et al., Developing a multi-epitope vaccine candidate to combat porcine epidemic diarrhea virus and porcine deltacoronavirus co-infection by employing an immunoinformatics approach. Front Microbiol, 2023. 14: p. 1295678.
- Ullah, S. , et al., In Silico Designed Multi-Epitope Vaccine Based on the Conserved Fragments in Viral Proteins for Broad-Spectrum Protection Against Porcine Reproductive and Respiratory Syndrome Virus. Vet Sci, 2025. 12(6).
- Adhikari, U.K. and M.M. Rahman, Overlapping CD8+ and CD4+ T-cell epitopes identification for the progression of epitope-based peptide vaccine from nucleocapsid and glycoprotein of emerging Rift Valley fever virus using immunoinformatics approach. Infect Genet Evol, 2017. 56: p. 75-91.
- Sinha, P.R. , et al., In Silico Development of a Multi-Epitope Subunit Vaccine against Bluetongue Virus in Ovis aries Using Immunoinformatics. Pathogens, 2024. 13(11).
- Sira, E. , et al., Design of a Multiepitope Pan-Proteomic mRNA Vaccine Construct Against African Swine Fever Virus: A Reverse Vaccinology Approach. Vet Med Int, 2025. 2025: p. 2638167.
- Lei, X. , et al., A Universal Multi-Epitope Vaccine Design Against Porcine Reproductive and Respiratory Syndrome Virus via Bioinformatics and Immunoinformatics Approaches. Vet Sci, 2024. 11(12).
- Barazesh, M. , et al., Bioinformatics analysis to design a multi-epitope mRNA vaccine against S. agalactiae exploiting pathogenic proteins. Sci Rep, 2024. 14(1): p. 28294.
- Martinez, G.S. , et al., PoxiPred: An Artificial-Intelligence-Based Method for the Prediction of Potential Antigens and Epitopes to Accelerate Vaccine Development Efforts against Poxviruses. Biology (Basel), 2024. 13(2).
- Olawade, D.B. , et al., Leveraging artificial intelligence in vaccine development: A narrative review. J Microbiol Methods, 2024. 224: p. 106998.
- Akinsulie, O.C. , et al., The potential application of artificial intelligence in veterinary clinical practice and biomedical research. Frontiers in Veterinary Science, 2024. Volume 11 - 2024.
- Izmailyan, R. , et al., Discovery of new antiviral agents through artificial intelligence: In vitro and in vivo results. Antiviral Res, 2024. 222: p. 105818.
- Zhavoronkov, A. , et al., Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat Biotechnol, 2019. 37(9): p. 1038-1040.
- MacLachlan, J.N.a.D. , Edward J., Antiviral Immunity and Virus Vaccines, in Fenner’s Veterinary Virology, F. Edition, Editor. 2017. p. 79-104.
- Tahamtan, A. , et al., An Overview of History, Evolution, and Manufacturing of Various Generations of Vaccines. Journal of Archives in Military Medicine, 2017. In Press.
- Aida, V. , et al., Novel Vaccine Technologies in Veterinary Medicine: A Herald to Human Medicine Vaccines. Frontiers in Veterinary Science, 2021. Volume 8 - 2021.
- Bishop, D.H.L. , The release into the environment of genetically engineered viruses, vaccines and viral pesticides. Trends in Ecology & Evolution, 1988. 3(4): p. S12-S15.
- Nguyen-Hoai, T. , et al., HER2/neu DNA vaccination by intradermal gene delivery in a mouse tumor model: Gene gun is superior to jet injector in inducing CTL responses and protective immunity. Oncoimmunology, 2012. 1(9): p. 1537-1545.
- Kannan, S., K. Subbaram, and M. Faiyazuddin, Chapter 17 - Artificial intelligence in vaccine development: Significance and challenges ahead, in A Handbook of Artificial Intelligence in Drug Delivery, A. Philip, et al., Editors. 2023, Academic Press. p. 467-486.
- Kaushik, R., R. Kant, and M. Christodoulides, Artificial intelligence in accelerating vaccine development - current and future perspectives. Frontiers in Bacteriology, 2023. Volume 2 - 2023.
- Gorki, V. and B. Medhi, Use of artificial intelligence in vaccine development against pathogens: Challenges and future directions. Indian J Pharmacol, 2024. 56(2): p. 77-79.
- Shafi, M. , et al., The role of artificial intelligence in detecting avian influenza virus outbreaks: A review. Open Vet J, 2025. 15(5): p. 1880-1894.
- Ong, E. , et al., Vaxign-ML: supervised machine learning reverse vaccinology model for improved prediction of bacterial protective antigens. Bioinformatics, 2020. 36(10): p. 3185-3191.
- Ali, L. , et al., In silico design of multi-epitope vaccines against the hantaviruses by integrated structural vaccinology and molecular modeling approaches. PLoS One, 2024. 19(7): p. e0305417.
- Srivastava, S. , et al., Deep learning and artificial intelligence in epitope prediction: A novel approach for vaccine design. Frontiers in Immunology, 2021.
- Wickramasinghe, I.N. , et al., The avian coronavirus spike protein. Virus Res, 2014. 194: p. 37-48.
- Dortmans, J. , et al., Newcastle disease virus outbreaks: Vaccine mismatch or inadequate application? Veterinary microbiology, 2012. 160: p. 17-22.
- Tripathy, D.N.a.R., W. M, Pox, in Diseases of Poultry, Y.M. Saif, Editor. 2000, Blackwell. p. 291-307.
- Sun, H. , et al., Molecular characterization of chicken infectious anaemia virus (CIAV) in China during 2020–2021. Avian Pathology, 2023. 52(2): p. 119-127.
- Kelley, L.A. , et al., The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols, 2015. 10(6): p. 845-858.
- UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res, 2023. 51(D1): p. D523-d531.
- Doytchinova, I.A. and D.R. Flower, VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics, 2007. 8: p. 4.
- Jumper, J. , et al., Highly accurate protein structure prediction with AlphaFold. Nature, 2021. 596(7873): p. 583-589.
- Varadi, M. , et al., AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res, 2022. 50(D1): p. D439-d444.
- Zost, S.J. , et al., Immunodominance and Antigenic Variation of Influenza Virus Hemagglutinin: Implications for Design of Universal Vaccine Immunogens. J Infect Dis, 2019. 219(Suppl_1): p. S38-S45.
- Saha, S. and G.P. Raghava, Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins, 2006. 65(1): p. 40-8.
- Vita, R. , et al., The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res, 2019. 47(D1): p. D339-d343.
- Larsen, M.V. , et al., Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics, 2007. 8: p. 424.
- Nielsen, M. , et al., Improved prediction of MHC class I and class II epitopes using a novel Gibbs sampling approach. Bioinformatics, 2004. 20(9): p. 1388-97.
- Rapin, N. , et al., Computational Immunology Meets Bioinformatics: The Use of Prediction Tools for Molecular Binding in the Simulation of the Immune System. PLOS ONE, 2010. 5(4): p. e9862.
- Dimitrov, I. , et al., AllerTOP v.2--a server for in silico prediction of allergens. J Mol Model, 2014. 20(6): p. 2278.
- Paul, S. , et al., HLA class I alleles are associated with peptide-binding repertoires of different size, affinity, and immunogenicity. J Immunol, 2013. 191(12): p. 5831-9.
- Li, K. , et al., Genetic, antigenic and pathogenic characterization of four infectious bursal disease virus isolates from China suggests continued evolution of very virulent viruses. Infect Genet Evol, 2015. 30: p. 120-127.
- Tartaglia, J., S. Pincus, and E. Paoletti, Poxvirus-based vectors as vaccine candidates. Crit Rev Immunol, 1990. 10(1): p. 13-30.
- Zhavoronkov, A. , Artificial Intelligence for Drug Discovery, Biomarker Development, and Generation of Novel Chemistry. Mol Pharm, 2018. 15(10): p. 4311-4313.
- Lv, H. , et al., Application of artificial intelligence and machine learning for COVID-19 drug discovery and vaccine design. Brief Bioinform, 2021. 22(6).
- Cellina, M. , et al., Digital Twins: The New Frontier for Personalized Medicine? Applied Sciences, 2023. 13(13): p. 7940.
- Jreich, R. , et al., Evaluating the robustness of an AI pathfinder application on eligibility criteria in multiple myeloma trials using real-world data and historical trials. J Comp Eff Res, 2024. 13(7): p. e230164.
- Benson, D.A. , et al., GenBank. Nucleic Acids Research, 2017. 46(D1): p. D41-D47.
- Li, W. and A. Godzik, Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics, 2006. 22(13): p. 1658-1659.
- Sievers, F. , et al., Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology, 2011. 7(1): p. 539.
- Edgar, R.C. , MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 2004. 32(5): p. 1792-1797.
- Thompson, J.D., D. G. Higgins, and T.J. Gibson, CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 1994. 22(22): p. 4673-4680.
- Falcon, A. , et al., Development of a Fully Protective Pandemic Avian Influenza Subunit Vaccine in Insect Pupae. Viruses, 2024. 16(6).
- Yang, T. , et al., Multivalent DNA vaccine enhanced protection efficacy against infectious bronchitis virus in chickens. J Vet Med Sci, 2009. 71(12): p. 1585-90.
- Govindarajan, D., A. S. Yunus, and S.K. Samal, Complete sequence of the G glycoprotein gene of avian metapneumovirus subgroup C and identification of a divergent domain in the predicted protein. J Gen Virol, 2004. 85(Pt 12): p. 3671-3675.
- Wei, L. , et al., The VP1 protein of avian encephalomyelitis virus is a major host-protective immunogen that serves as diagnostic potential. J Virol Methods, 2008. 149(1): p. 56-62.
- Firouzamandi, M. , et al., Improved immunogenicity of Newcastle disease virus inactivated vaccine following DNA vaccination using Newcastle disease virus hemagglutinin-neuraminidase and fusion protein genes. J Vet Sci, 2016. 17(1): p. 21-6.
- Olbert, M. , et al., Viral vector vaccines expressing nucleoprotein and phosphoprotein genes of avian bornaviruses ameliorate homologous challenge infections in cockatiels and common canaries. Sci Rep, 2016. 6: p. 36840.
- Lei, T. , et al., Gp85 protein encapsulated by alginate-chitosan composite microspheres induced strong immunogenicity against avian leukosis virus in chicken. Front Vet Sci, 2024. 11: p. 1374923.
- Kant, A. , et al., Classification of Dutch and German avian reoviruses by sequencing the sigma C protein. Vet Res, 2003. 34(2): p. 203-12.
- Ali, S.A., Y. A. Almofti, and K.A. Abd-Elrahman, Immunoinformatics Approach for Multiepitopes Vaccine Prediction against Glycoprotein B of Avian Infectious Laryngotracheitis Virus. Adv Bioinformatics, 2019. 2019: p. 1270485.
- Fazel, F. , et al., Efficacy and tolerability of an mRNA vaccine expressing gB and pp38 antigens of Marek’s disease virus in chickens. Virology, 2024. 590: p. 109970.
- Fang, L. , et al., Efficacy of CpG-ODN and Freund’s immune adjuvants on antibody responses induced by chicken infectious anemia virus VP1, VP2, and VP3 subunit proteins. Poult Sci, 2019. 98(3): p. 1121-1126.
- Mato, T. , et al., Recombinant subunit vaccine elicits protection against goose haemorrhagic nephritis and enteritis. Avian Pathol, 2009. 38(3): p. 233-7.
- Wang, B. , et al., An inactivated novel chimeric FAdV-4 containing fiber of FAdV-8b provides full protection against hepatitis-hydropericardium syndrome and inclusion body hepatitis. Vet Res, 2022. 53(1): p. 75.
- Kringelum, J.V. , et al., Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS Comput Biol, 2012. 8(12): p. e1002829.
- Haste Andersen, P., M. Nielsen, and O. Lund, Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci, 2006. 15(11): p. 2558-67.
- Tunyasuvunakool, K. , et al., Highly accurate protein structure prediction for the human proteome. Nature, 2021. 596(7873): p. 590-596.
| Virus | Host | AI method | Reference |
|---|---|---|---|
| Nipah Virus | Animal Reservoirs | Epitope mapping, structural validation | [12] |
| Machupo Virus | Animal Reservoirs | Epitope prediction, vaccine construct design | [13] |
| H5N1 Influenza | Birds | B-/T-cell epitope pipeline + docking | [14] |
| MVEV | Birds/Mammals | Envelope epitope mapping, structural validation | [15] |
| Feline Infectious Peritonitis Virus (FIPV) | Cats | Epitope prediction, docking | [8,16] |
| BCoV | Cattle | ML epitope mapping + AlphaFold2 | [5] |
| Foot-and-Mouth Disease Virus (FMDV) | Cattle | Immunoinformatics, structural modeling | [17] |
| Foot-and-Mouth Disease Virus (FMDV) | Cattle | Epitope fusion with TLR agonist | [18] |
| FMDV | Cattle | Multi-serotype epitope design, stability/autogenetics analysis | [19] |
| FMDV | Cattle | Serotype mapping, structural validation | [17] |
| BEFV | Cattle | Multi-epitope subunit | [20] |
| Bovine Leukemia Virus (BLV) | Cattle | Epitope prediction, MD, immune simulation | [21] |
| Lumpy Skin Disease Virus (LSDV) | Cattle | Epitope prediction, TLR docking, MD simulation | [22] |
| Newcastle Disease Virus (NDV) | Chickens | ANN epitope affinity prediction, docking | [23] |
| Avian Leukosis Virus (ALV) | Chickens | Peptide design, TLR7 docking | [24] |
| Infectious Bursal Disease Virus (IBDV) | Chickens | Immunoinformatic (VP2/VP3 proteins) | [25] |
| CPV-2 | Dogs | Immunoinformatics + docking | [26] |
| Canine Circovirus (CanineCV) | Dogs | Immunoinformatics, in vivo validation | [27] |
| Rota virus | Elephants | Epitope prediction, structural validation, and immune simulation | [28] |
| Goatpox Virus (GTPV) | Goats | Epitope mapping, TLR docking, immune sim | [29] |
| Orf Virus | Goats/Sheep | Immunoinformatics, epitope selection | [30] |
| Zika | Humans | Epitope prediction + docking | [31] |
| Dengue (multi-serotype) | Humans | Structural modeling + conserved targeting | [32] |
| Rift Valley Fever Virus (RVFV) | Livestock | Epitope prediction, docking, and molecular dynamics | [33] |
| Influenza D Virus (IDV) | Livestock | Epitope mapping, allergenicity & toxicity checks | [34] |
| Hendra Virus (HeV) | Livestock | Epitope prediction, immune simulation, TLR docking | [35] |
| HeV | Livestock | Epitope screening, TLR docking | [36] |
| Marburg Virus (MARV) | Non-human primates/Humans | Reverse vaccinology, epitope mapping, TLR docking | [37] |
| ASFV | Pigs | Immunoinformatics + docking + simulation | [38] |
| Porcine Epidemic Diarrhea Virus (PEDV) | Pigs | Epitope selection, TLR docking, immune sim | [39] |
| Porcine Epidemic Diarrhea Virus (PEDV) | Pigs | Epitope selection, immune simulation | [40] |
| PRRSV | Pigs | Conserved epitope selection, immune modeling | [41] |
| Avian Influenza A (H5N1) | Poultry | Reverse vaccinology, epitope mapping | [6,14] |
| RVFV | Ruminants | ML peptide prediction + immune simulation | [33] |
| RVFV (M-protein) | Ruminants | Reverse vaccinology + docking | [42] |
| Bluetongue Virus (BTV) | Sheep | Epitope mapping, TLR docking | [43] |
| ASFV | Swine | Reverse vaccinology + dynamics | [38] |
| African Swine Fever Virus (ASFV) | Swine | Reverse vaccinology, SLA docking, MD simulations | [44] |
| Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) | Swine | Epitope mining, antigenicity/allergenicity | [45] |
| ASFV | Swine | Reverse vaccinology, SLA docking, MD simulations | [46] |
| PRRSV | Swine | Global epitope mining, antigenicity/allergenicity profiling | [41] |
| ASFV | Swine | SLA docking, MD simulation | [38] |
| Poxviruses | various | Proteome-wide AI epitope predictor | [47] |
| Serial numbers |
RNA Viruses | Target Protein | Protein Function | Protein Length (aa) | Structural Coverage by Alpha Fold 2 | Accession Numbers of the reference NCBI sequences for each virus | References |
|---|---|---|---|---|---|---|---|
| 1. | Avian Influenza | HA | Binds to host cell receptors (sialic acids) | 1760 | 81 | NC_007362.1 | [92] |
| 2. | Infectious Bronchitis Virus (IBV) | Spike | Inserts into the ligand on the surface of the host cell receptor, opening the cell wall | 3461 | 58 | NC_048213.1 | [93] |
| 3. | Avian Metapneumovirus | G protein | Attachment to the host cell surface receptor allows viral entry into the host | 1175 | 0 | NC_039231.1 | [94] |
| 4. | Avian Encephalomyelitis | VP1 | Host protective immunogen | 270 | 63 | AFM73888.1 - VP1 partial | [95] |
| 5. | Newcastle Disease | HN | Bind to host cell receptors | 1715 | 81.4 | NC_039223.1 | [96] |
| 6. | Avian Bornavirus | N and P protein | Packing viral RNA, essential for polymerase activity, shuttles RNP into and out of the nucleus; a cofactor of bornavirus polymerase | 1121, 605 | 64.2 | NC_039189.1 | [97] |
| 7. | Avian Leukosis | G protein | Mediates viral attachment to the cell surface | 2111 | 53.3 | MT179556.1 | [98] |
| 8. | Avian Reovirus | Sigma C | binding to the host cell surface | 326 | 71.4 | AAK18188.1 | [99] |
| DNA Viruses | Target Protein | Protein Function | Protein Length (bp) | Structural Coverage by Alpha Fold 2 | Accession Numbers | References | |
| 9. | Infectious Laryngotracheitis Virus | B-glycoprotein spike | viral attachment to the host cell surface to form a heterodimer | 2651 | 62.7 | NC_006623.1 | [100] |
| 10. | Marek’s Disease (Gallid Herpesvirus-2) | B-glycoprotein spike | viral attachment to the host cell surface to form a heterodimer | 2597 | 62.8 | NC_002229.3 | [101] |
| 11. | Chicken Infectious Anemia Virus | VP3 | induces apoptosis in chicken lymphocytes | 388 | 29.8 | AF199501.1 | [102] |
| 12. | Avian Polyoma Virus | VP1 | Capsid protein that binds to host cell receptors for infection | 1031 | 87.2 | PP057981.1 | [103] |
| 13. | Fowl Adenovirus | Fiber Genes | responsible for hemagglutination | 1386 | 81.3 | DQ864436.1 | [104] |
| Types of Epitopes | Recognized by | Immune function | AI prediction tool used | Reference |
|---|---|---|---|---|
| Linear B Cell Epitopes | B-cell Receptors (BCRs) | Induce antibody production; direct neutralization of extracellular virus | BepiPred 2.0, ABC Pred | https://services.healthtech.dtu.dk/services/BepiPred-2.0/ (accessed on 14 September 2025) |
| Conformational B-cell Epitopes | BCRs (3D -dependent) | Target protein folding-dependent antigenic sites | DiscoTope, Ellipro | https://services.healthtech.dtu.dk/services/DiscoTope-3.0/ (accessed on 14 September 2025) |
| CD8+ T cell Epitopes | Cytotoxic T Lymphocytes (CTLs) | Kill infected cells by recognizing MHC-I-bound peptides | NetMHCpan | https://services.healthtech.dtu.dk/services/NetMHCpan-4.1/ (accessed on 14 September 2025) |
| CD4+ T cell Epitopes | Helper T Lymphocytes (Th cells) | Aid in inactivating B cells and CTLs via cytokine release | NetMHCIIpan | https://services.healthtech.dtu.dk/services/NetMHCIIpan-4.0/ (accessed on 14 September 2025) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





