1. Introduction
Semi-supervised clustering (SSC) has emerged as an important paradigm in modern machine learning, as it leverages auxiliary information to improve clustering quality [1,2]. SSC enables learning from both limited labeled data and abundant unlabeled data, and is particularly useful in scenarios involving noisy or complexly distributed data. Pedrycz [3] demonstrated that, for practical applications which often require multiple intermediate ways of discovering structure in a dataset, performance can be significantly improved by using prior information; even a small proportion of labeled samples can markedly enhance clustering results.
Semi-supervised fuzzy clustering (SSFC) extends fuzzy clustering by incorporating semi-supervised learning. SSFC integrates the flexibility of fuzzy logic with the guidance of partial supervision [4,5,6]. Unlike hard clustering, which assigns each data point to exactly one cluster, fuzzy clustering allows each point to belong to multiple clusters with varying degrees of membership. This feature is especially suitable for problems with unclear cluster boundaries or noisy data.
Within the SSFC framework, the selection of high-quality seed instances plays a decisive role in clustering performance [7]. Well-chosen seeds can reduce labeling costs, enhance cluster separability, and stabilize convergence, whereas poorly chosen seeds may propagate errors, degrade clustering accuracy, and increase reliance on expert intervention. Despite its importance, seed selection remains a challenging and underexplored problem, requiring methods that are both theoretically sound and practically efficient. Thanks to its ability to effectively exploit a small amount of prior knowledge to guide the learning process, SSFC has attracted significant attention from the research community and has been widely applied in domains such as pattern recognition, information processing, and healthcare [2,6,8,9,10,11,12,13].
The difficulty of selecting good seeds arises from three main factors: (i) the non-trivial task of identifying informative patterns within uncertain cluster boundaries; (ii) the presence of noisy and imbalanced data, which can distort the clustering process; and (iii) the limited labeling budget, which necessitates efficient and adaptive query strategies.
Several studies have attempted to improve SSC through different strategies, including graph-based propagation, constraint expansion, and probabilistic seed selection [7,13]. While these methods have achieved certain successes, they still share common limitations: reliance on randomly chosen seeds or predefined constraints, lack of mechanisms to actively evaluate seed reliability, and insufficient robustness when dealing with noisy or overlapping clusters.
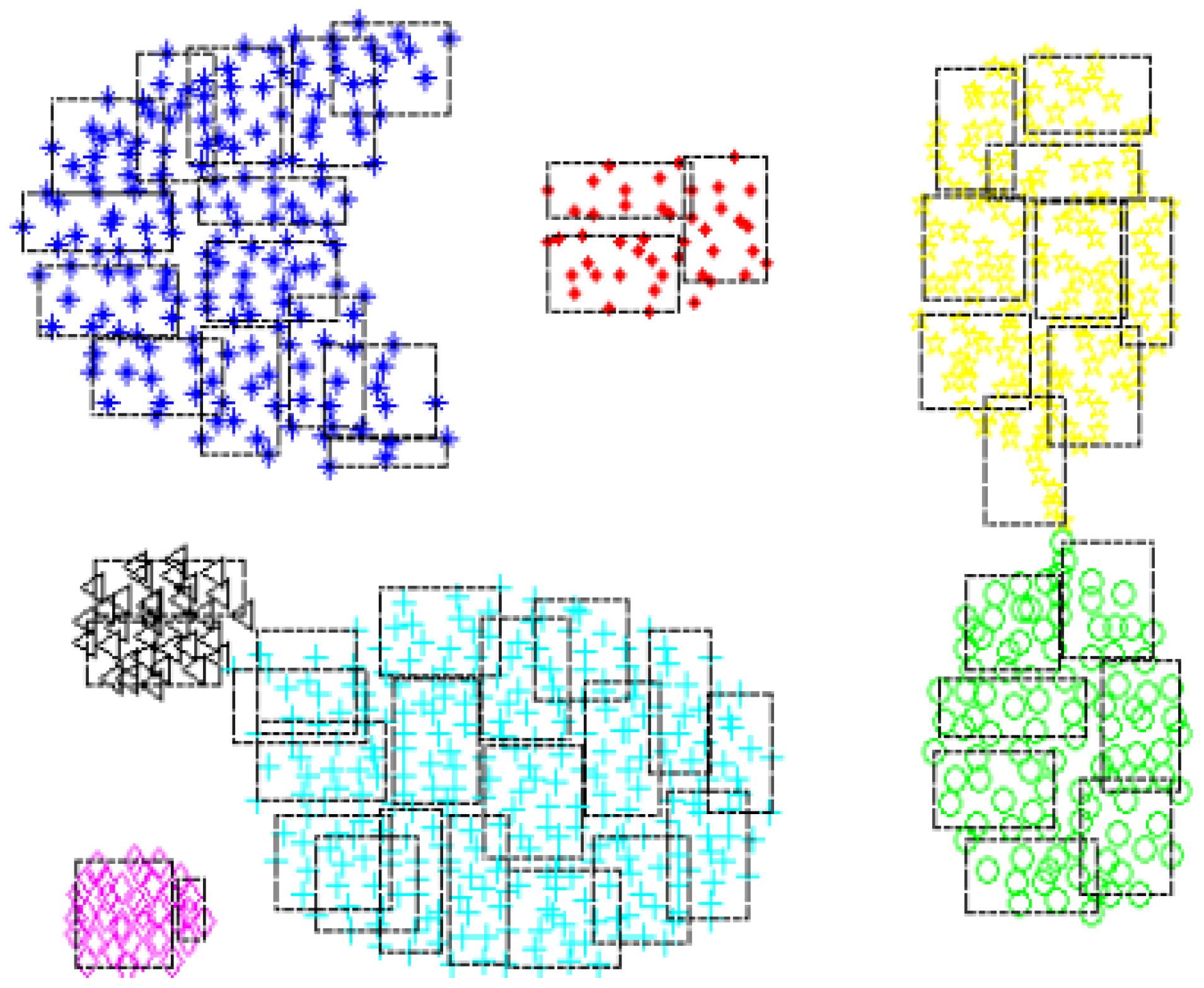
Fuzzy Min–Max Neural Networks (FMNN) have recently garnered increased attention in semi-supervised learning research, thanks to their ability to make effective use of limited labeled data. FMNN functions by dividing the data space into HXs. The visualization in
Figure 1 illustrates the distribution of 2D HXs belonging to seven clusters on the Aggregation dataset [16], as partitioned by FMNN.
Based on FMNN, the models such as GFMNN (General FMNN) [17], SS-FMM [2], SCFMN (SSC in FMNN) [11], MSCFMN (Modified SCFMN) [6], and WHFMN (Weighted Hyperboxes in FMNN) [18] have demonstrated the extensibility of FMNN in SSC tasks. However, approaches based on FMNN variants, although demonstrating flexibility in partitioning data through HXs, remain constrained by their dependence on seed quality and the absence of active querying strategies. These limitations highlight an important research gap: how to effectively select critical data points for labeling in order to reduce dependence on labeled data while maintaining clustering accuracy.
Although FMNN-based approaches have certain limitations, they remain a promising research direction with considerable potential and applicability [6]. In this study, we introduce a SSC model built upon FMNN and enhanced with an active seed selection strategy, referred to as FA-Seed. Rather than selecting seeds randomly, the model prioritizes data points that are expected to yield the most informative labels. This design aims to maximize information gain from each expert query while simultaneously minimizing the overall labeling effort.
The proposed model is applied to benchmark datasets to demonstrate its effectiveness in handling clustering problems characterized by fuzzy boundaries, class imbalance, and scarce labeled data. The main contributions of this work include:
Proposing an active seed selection strategy based on FMNN;
Evaluating seed quality under limited label availability;
Providing label suggestions to experts along with an optimized validation mechanism;
Assessing performance on benchmark datasets.
The remainder of this paper is organized as follows: Section 2 presents the background and related work. Section 3 describes the proposed model in detail. Section 4 reports the experimental results. Finally, Section 5 discusses the conclusions and future research directions.
2. Related Works
In [7], the research team developed SSGC (Semi-Supervised Graph-based Clustering), an active learning algorithm that identifies seed instances using the k-nearest neighbor and min–max algorithms. Its key idea is to construct a weighted k-NN graph based on shared neighbors and then partition it into connected components under a cut condition. Although its label propagation mechanism—combining an adaptive cut threshold () with graph connectivity—is efficient and flexible for varying cluster shapes, seed selection in SSGC remains random, making performance sensitive to initial labels.
Beyond graph-based approaches, Xin Sun et al. [13] proposed a constraint-driven framework consisting of CS-PDS (Constraints Self-learning from Partial Discriminant Spaces) and FC2 (Finding Clusters on Constraints). CS-PDS iteratively expands initial constraints using an Expectation–Maximization cycle, while FC2 performs clustering on the enriched constraint set. This approach achieves strong performance on small-scale data but relies heavily on the representativeness of initial constraints and lacks an explicit active query mechanism.
A probabilistic alternative was introduced by Bajpai et al. [12], who employed Determinantal Point Processes (DPP) to optimize seed diversity. Unlike SSGC, DPP selects seeds that are both representative and broadly distributed, which are then used as K-Means centroids, reducing convergence to poor local optima. However, this approach incurs high computational cost () and struggles with noisy or overlapping clusters due to the absence of propagation or denoising mechanisms.
Another important line of research builds on FMNN, which leverages HXs to represent uncertainty and handle limited supervision. Among these approaches, MSCFMN stands out by refining HX construction and overlap handling. In MSCFMN, the number of HXs is fixed to the number of clusters; they expand under a threshold , with contractions applied to resolve overlaps. While such refinements improve accuracy over earlier FMNN-based models, they still suffer from rigid partitioning of the feature space, the absence of active seed selection, and high sensitivity to the threshold parameter .
Our proposed FA-Seed extends MSCFMN by retaining its HX-based partitioning mechanism while introducing adaptive seed selection, seed-quality scoring, and few-shot generalization. These enhancements address the limitations of MSCFMN and enable FA-Seed to achieve superior clustering performance under limited supervision.
3. Proposed Method: FA-Seed
3.1. The Idea of Proposed Model
The effectiveness of SSC depends strongly on the quality of selected seeds. Poor seeds can propagate errors, while reliable seeds reduce labeling costs and improve clustering accuracy. However, selecting high-quality seeds is challenging in practice due to noisy data, overlapping clusters, and limited supervision. To address this, the proposed FA-Seed model extends the MSCFMN framework [6] by integrating HX partitioning, active querying, and few-shot generalization.
Built on the MSCFMN, FA-Seed represents clusters as HXs, naturally handling uncertainty and overlapping regions. Unlike random or fixed initialization, the model adaptively scores candidate seeds using density, centrality, and uncertainty, and selectively queries the most informative samples. Verified seeds then guide controlled label propagation through the HX structure, ensuring consistency and reducing error propagation.
Unlike MSCFMN, which automatically assigns a single labeled point to each HX based on data distribution characteristics, FA-Seed incorporates an active learning mechanism to purposefully evaluate and select potential seed candidates. Rather than relying on default labeling, the model actively queries and verifies informative data points with experts. Additionally, FA-Seed integrates various criteria such as centroid deviation, data density, and uncertainty to filter and assess seed quality, thereby enhancing the model’s generalization ability and clustering accuracy under limited labeled data conditions.
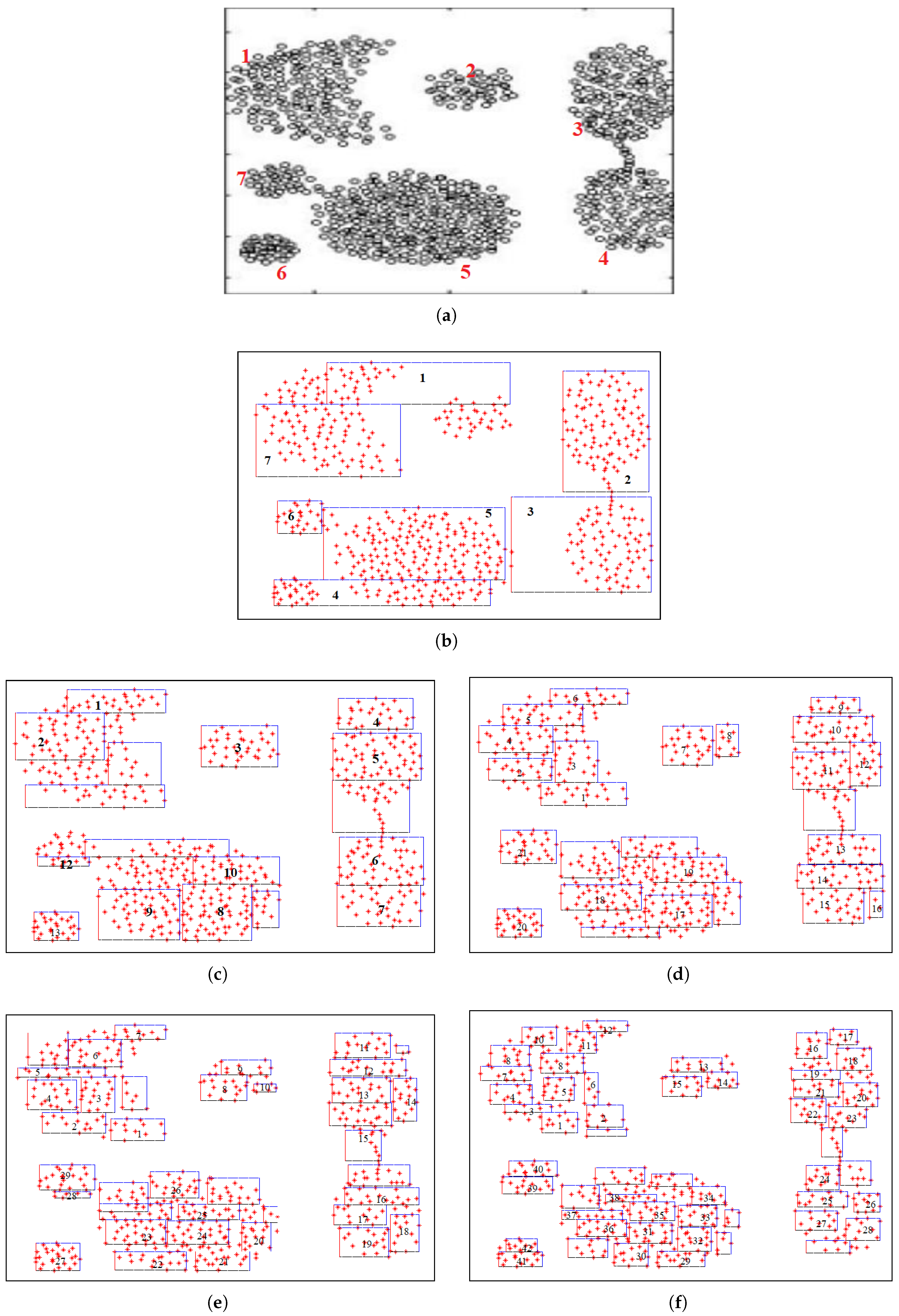
Figure 2(a) visually illustrates the limitation of the MSCFMN model during Phase 1 — the phase of defining auxiliary information (seed selection) — on the Aggregation dataset. The Aggregation dataset consists of 788 samples grouped into 7 clusters [16], as shown in
Figure 2(a).
In MSCFMN, the number of HXs generated during this initial phase is constrained to match the number of clusters. Specifically, only 7 HXs are created, as seen in
Figure 2(b). However, several of these HXs (e.g., HXs 1, 3, 4, and 5) overlap with multiple clusters, reducing the reliability of label assignment for the corresponding seed points. If seed points from such ambiguous HXs are selected and used in label propagation, the model may suffer from reduced accuracy due to mislabeling. Furthermore, this restricted partitioning approach limits the model’s ability to capture more complex or uneven data distributions within individual clusters. MSCFMN is thus highly influenced by both the number and shapes of clusters.
In contrast to MSCFMN, FA-Seed addresses these limitations by enabling a more flexible partitioning of the input space, allowing the generation of a greater number of HXs beyond the initial number of clusters. This adaptive strategy improves coverage and granularity. As illustrated in
Figure 2(c)–(d):
Figure 2(c): FA-Seed generates 17 HXs and selects 13 data points for expert query, accounting for 1.65% of the total samples.
Figure 2(d): 26 HXs with 21 queries (2.66%).
Figure 2(e): 38 HXs with 29 queries (3.68%).
Figure 2(f): 52 HXs with 40 queries (5.08%).
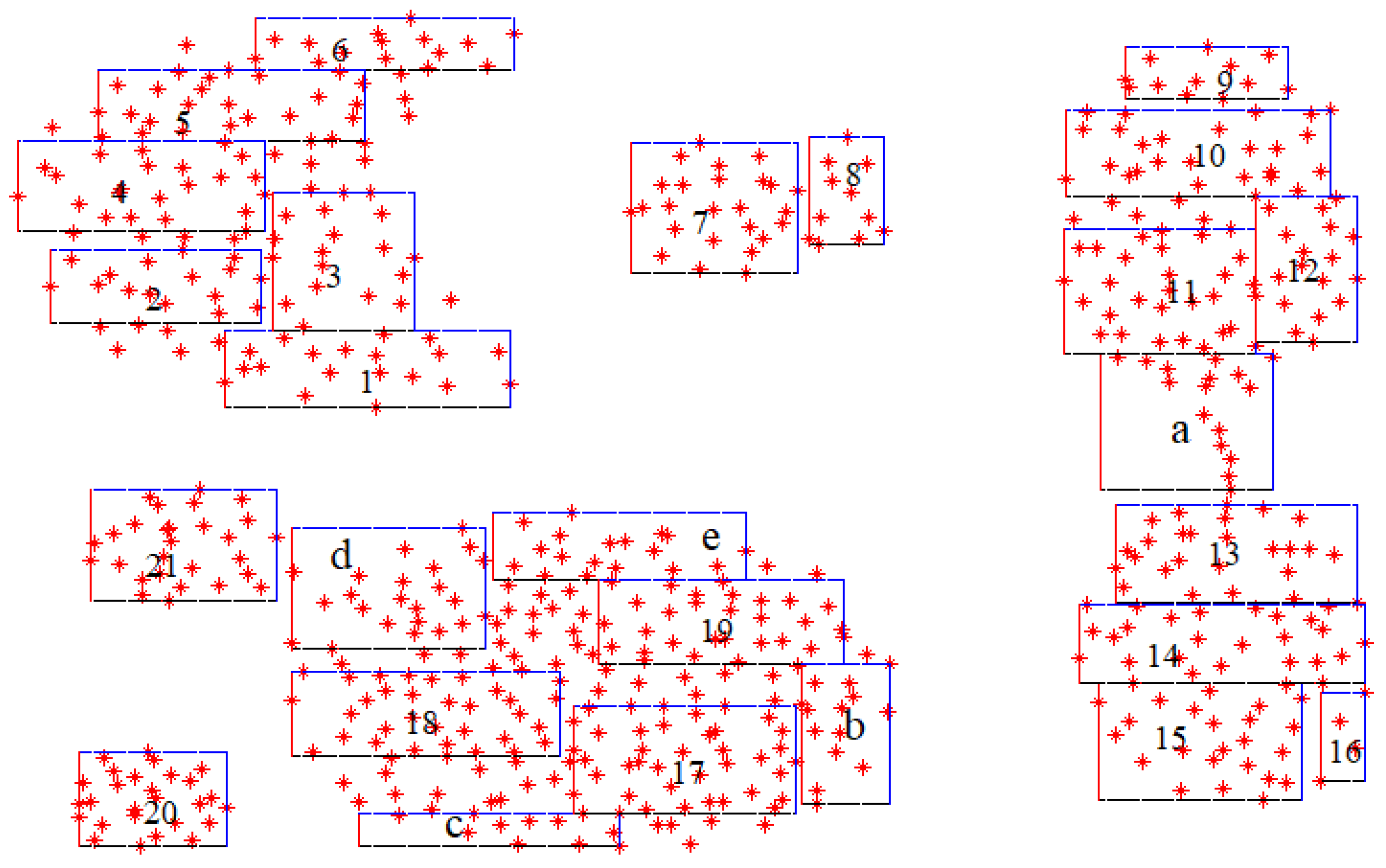
Beyond simply expanding the number of HXs, FA-Seed incorporates a quality assessment mechanism to enhance label query efficiency. Specifically, only HXs with balanced and representative data distributions are selected for expert queries. HXs with skewed distributions or very few data points are excluded to avoid wasting labeling budget and to reduce the risk of incorrect label propagation.
Figure 3 illustrates the process of calculating and selecting HXs in the proposed FA-Seed model. HXs with a high level of data balance are selected (labeled numerically from 1 to 21), whereas imbalanced HXs are excluded from selection (denoted by characters a, b, c, d, and e).
Thanks to these two improvements — adaptive HX generation and pre-query quality evaluation — FA-Seed enables more effective learning in SSC tasks involving unlabeled and non-uniform data. It also enhances clustering accuracy while reducing the model’s dependence on prior assumptions about the number or shape of clusters.
3.2. Setup and Definitions
Fuzzy hyperbox and membership. Assume we have M data instances , where each , which can be mapped into G ground-truth clusters. Let the index set be partitioned into a labeled subset and an unlabeled subset , with and , where the split is induced by partitioning the input space into HXs.
Let
denote the current set of HXs (so
). Denote the labeled and unlabeled subsets by
and
, respectively, with
A fuzzy HX
is as defined in (
2).
where
and
are the minimum and maximum vertices of the HX in the input space.
Each HX is associated with a class label , which is assigned during the learning process. For , labels are known; for , predicted labels are denoted by .
Let
x be a candidate sample. Its membership to
is
, as defined in Equation (
3). Given the current set of HXs
, the HX with the largest membership value for
x is determined by Equation(
5).
where the function
is a two-parameter function used to modulate the attenuation of the membership value
, based on the correlation between the data sample
x and the boundary of the HX
. The sensitivity parameter
directly affects the shape of this attenuation: a larger
leads to faster degradation of the membership value, thereby making the model more responsive to boundary violations. Its mathematical formulation is given in Equation (
4):
Seed set based on centers and purity. Let
be the core set used to compute the data center
as in (
6) (the
-core
). The geometric center
is defined in (
7), and the center deviation
is defined in (
8). A HX is regarded as pure if
satisfies constraint (
9) is satisfied.
Consequently,
is the index set of pure fuzzy HXs, given by (
10); the candidate query set
Q is the collection of their data centers, as in (
11); and
S denotes the set of fully contained samples, defined in (
12) and generated from
Q.
Adaptive partition and assignment with fuzzy HXs. Given the current HXs
and a candidate sample
x, first select the HX with the largest membership (as defined in Equation (
5)). Update the space of HXs in the set
B according to:
Containment rule: If such that , assign x to that (uniqueness follows from ).
-
Expansion: Form the tentative expansion of
as defined in Equation (
13). Accept it and set
iff all of the following hold:
- (1)
size constraint
(as defined in Equation (
14));
- (2)
after any necessary contraction, the overlap constraint
satisfies the constraint in Equation (
16).
If accepted, assign x to the updated .
Create a new HX: If the expansion is not accepted, initialize a new HX around
x as in Equation (
17).
where
denotes the post-expansion size of
(e.g., as in (
15)):
where
is the overlap measure.
Fuzzy HX-level labeling by inheritance. With a decreasing schedule
(
), an unlabeled HX
inherits the label of the nearest
labeled HX iff both gates hold: (i) the center-alignment gate
(with
defined in Equation (
8)); and (ii) the membership gate
where
is the data center defined in Equation (
6) and
is chosen by Equation (
19); labels are then updated according to Equation (
20).
When both gates are satisfied, update the label and the sets:
Iterate (
20) for
. Finally, submit the remaining unlabeled (outlier/atypical) HXs for expert labeling:
Edge cases: if
, skip or defer inheritance or query an expert. If no HX exists yet (
), initialize
defined as in Equation (
17); if the current sample
x has a ground-truth label, set
, otherwise leave it pending.
Training procedure:
Stage 1 (one-shot): query all seeds
(with
S defined in Equation (
11)) to obtain
.
Stage 2 (progressive): with a decreasing schedule
, for each
, let
is selected according to Equation (
19).
If the alignment gate
((
9)) and the membership gate
((
18)) both hold, then move
u to
; otherwise keep it in
. Finally, query the remainder
3.3. Model Architecture and Learning Process
3.3.1. Architecture of FA-Seed Model
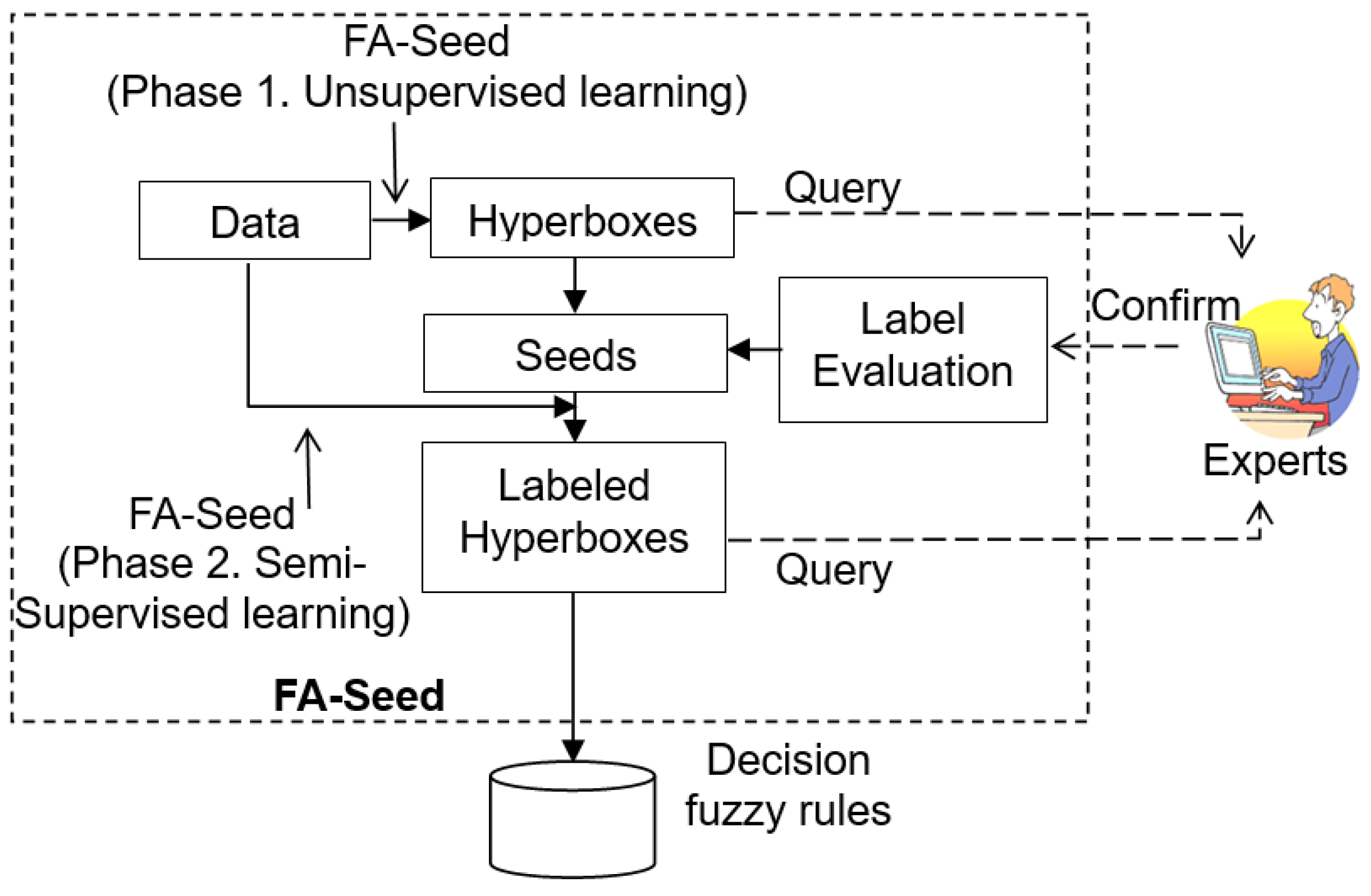
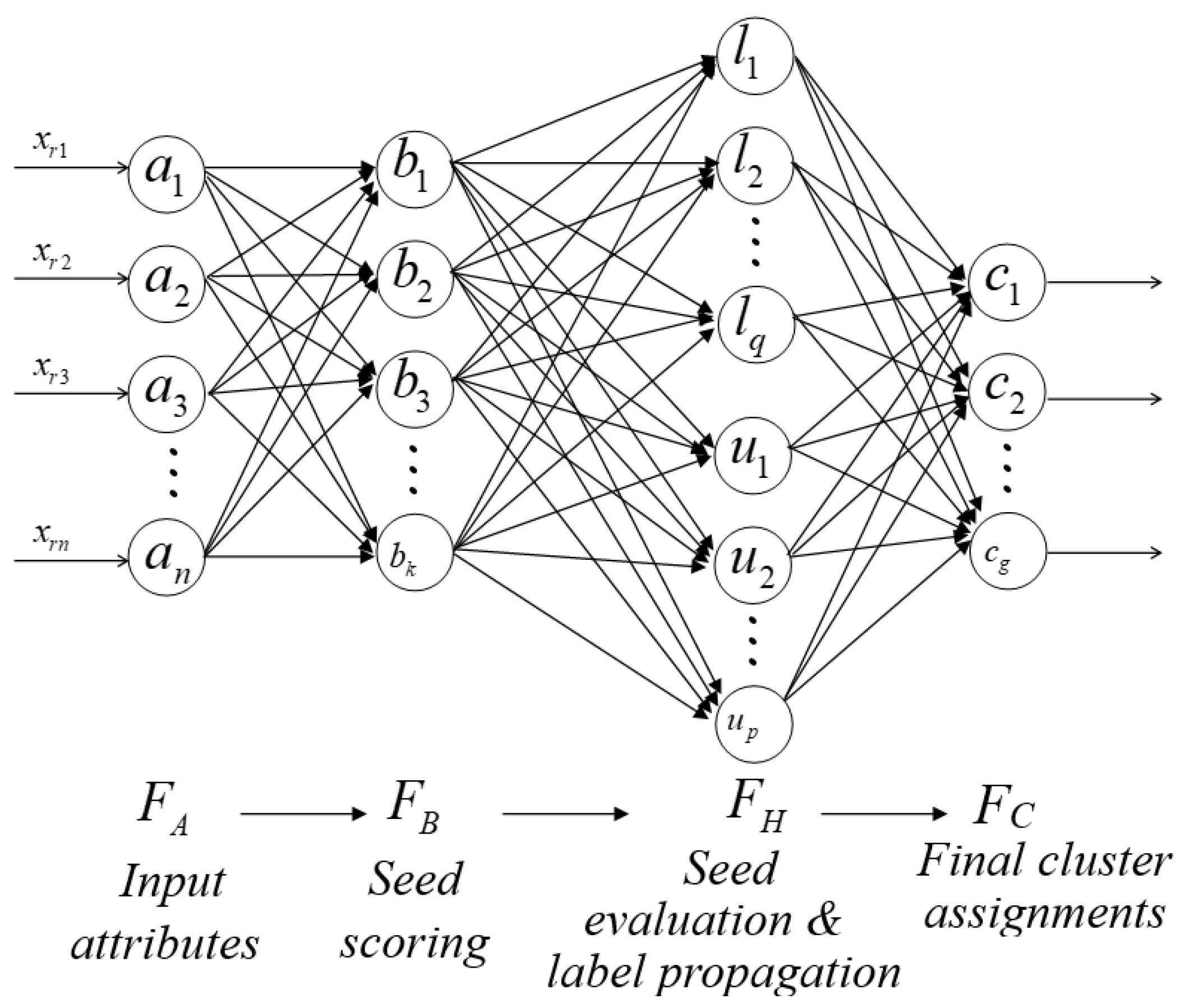
The FA-Seed model combines the architecture of the FMNN [19,20] with a semi-supervised clustering strategy, organized into two main phases for clustering on the dataset X. In the first phase, FA-Seed identifies auxiliary information (labels) for a subset of the data, denoted as S, while the remaining unlabeled samples form the subset U (). In the second phase, FA-Seed propagates labels from S to U by constructing and adapting HXs, thereby forming clusters for the entire dataset X.
As illustrated in
Figure 4, the architecture of FA-Seed consists of two main components:
Seed scoring module: Definition of auxiliary information based on the partitions the data space into HXs, evaluates candidate seeds using criteria such as data density, centroid deviation, and uncertainty, and assigns labels to reliable seeds within their respective HXs.
Controlled label propagation and extended evaluation: propagates labels from the verified seed set to the unlabeled data, ensuring consistency and accuracy across clusters. This component leverages the HX structure to minimize error propagation and actively queries uncertain samples that cannot be confidently assigned to a cluster.
3.3.2. Neural Network Architecture of FA-Seed
The FA-Seed model is structured as a four-layer neural network, as illustrated in
Figure 5. We consider four layers
, each playing a distinct role in the semi-supervised clustering process:
Input layer . This layer contains n nodes, each representing an input attribute. A sample is = and is mapped into HXs in using the min–max boundary vectors v and w.
HX scoring layer . Includes
K nodes (HXs) produced at Stage 1, each corresponding to a HX
. These HXs are constructed incrementally and capture the local data structure in the input space. The activation is the membership
Evaluation/propagation layer . HX nodes are partitioned into labeled and unlabeled sets: (size ) and (size ). This layer applies the gates and the inheritance rule to move HXs from to (at termination, so and typically ).
-
Output layer : Contains
G nodes, each representing one output cluster. The binary relationship between HX
and class
is represented by the matrix
:
The membership score of a sample to class
is computed as:
where
is the activation of HX
j. The set of HXs defining class
g is given by:
with
G denoting the index set of HXs assigned to class
g.
3.3.3. Learning Algorithm
The first stage of the learning algorithm defines auxiliary information based on the partition of the data space into fuzzy HXs. Candidate seeds are evaluated using criteria such as data density, centroid deviation, and uncertainty, and reliable seeds are then assigned labels within their respective HXs. The learning algorithm for this phase is presented in Algorithm 1.
|
Algorithm 1:Seed Set Construction Phase of FA-Seed |
|
Input: Dataset X (); ; deviation threshold
Output: Updated data set X, B
Step 1. Hyperbox generation
Perform one pass over X to construct fuzzy HX set B (, (, )) using min–max boundaries with .
Step 2. Hyperbox filtering (Identify data instances to query)
For each HX , denote by the set of samples assigned to , and by its min/max vertices.
Compute the data center defined by Equation ( 6) and the geometric center of the HX defined by Equation ( 7). Keep only those whose centroid is sufficiently close to its geometric center, and remove if condition ( 8) is violated: . Step 3. Seed set construction, partition and reordering
Build the seed set S from samples fully contained in at least one surviving HX by by Equation ( 12). Build a permutation of that places S first and D after (X is the set of unlabeled samples, i.e., points that do not fully belong to any HX), and form the reordered dataset .
return B (the surviving HXs), (the reordered dataset)
|
*
Computational Complexity of FA-Seed – Algorithm 1:
The second stage of the learning algorithm describes the propagation of labels from the verified seed set to the unlabeled data, ensuring consistency and accuracy across clusters. By leveraging the HX structure, this stage minimizes error propagation and actively queries uncertain samples that cannot be confidently assigned to a cluster. The learning algorithm for this stage is presented in Algorithm 2.
where
denotes the expansion measure when adding
x to
.
*
Computational Complexity of FA-Seed – Algorithm 2:
Computational complexity of the FA-Seed algorithm:

4. Experiments
4.1. Experimental Objectives
The goal of this section is to evaluate the effectiveness of the proposed FA-Seed model in semi-supervised clustering tasks. Specifically, we aim to investigate: (i) the ability of FA-Seed to improve clustering accuracy under limited labeled data, (ii) its efficiency in seed selection compared to baseline methods, and (iii) its applicability to benchmark datasets [16,21].
To evaluate the performance of FA-Seed, experiments were conducted to compare it against several state-of-the-art semi-supervised clustering baselines, including SSDBSCAN, SSK-Means, K-Means, SSGC [7], MSCFMN [6], and FC2 [13].
4.2. Experimental Setup
4.2.1. Datasets
We conduct experiments on benchmark datasets to evaluate the general performance and applicability of the proposed model. The benchmark datasets are drawn from standard UCI and CS collections, including Iris, Wine, Zoo, Soybean, Ecoli, Yeast, PID, Spiral, Pathbased, Thyroid, and others. These datasets span a wide range of sample sizes, dimensions, and cluster structures, thereby providing a comprehensive basis for evaluating clustering quality under diverse conditions. The detailed characteristics are summarized in
Table 1. All datasets are normalized prior to experimentation. For datasets with missing values, imputation is performed following the approach of Batista [22], where missing entries are replaced with the mean value of the corresponding attribute, as defined in Equation (
34).
4.2.2. Parameter Settings
The following settings are used unless otherwise specified: fuzziness parameter , initial threshold , threshold decay factor , and deviation threshold selected empirically based on the error correction method.
4.2.3. Evaluation Metrics
As noted earlier, clustering performance depends on both the quality and the number of selected seeds. We therefore conduct comparative experiments on benchmark datasets, comparing against state-of-the-art semi-supervised clustering baselines and evaluating performance using Rand Index (RI), accuracy, training time, number of queries (NoQ), and number of seeds (NoS) obtained via expert querying. RI is computed by (
35), and accuracy by (
36).
where:
u is the number of correct decisions when in both clustering and ground truth.
v is the number of correct decisions when in both clustering and ground truth.
n is the total number of samples.
if the prediction is correct; otherwise .
4.3. Results
As analyzed, increasing the proportion of seeds often contributes to improving clustering quality. In the first experiment, we focus on evaluating the ability to obtain seeds through expert queries.
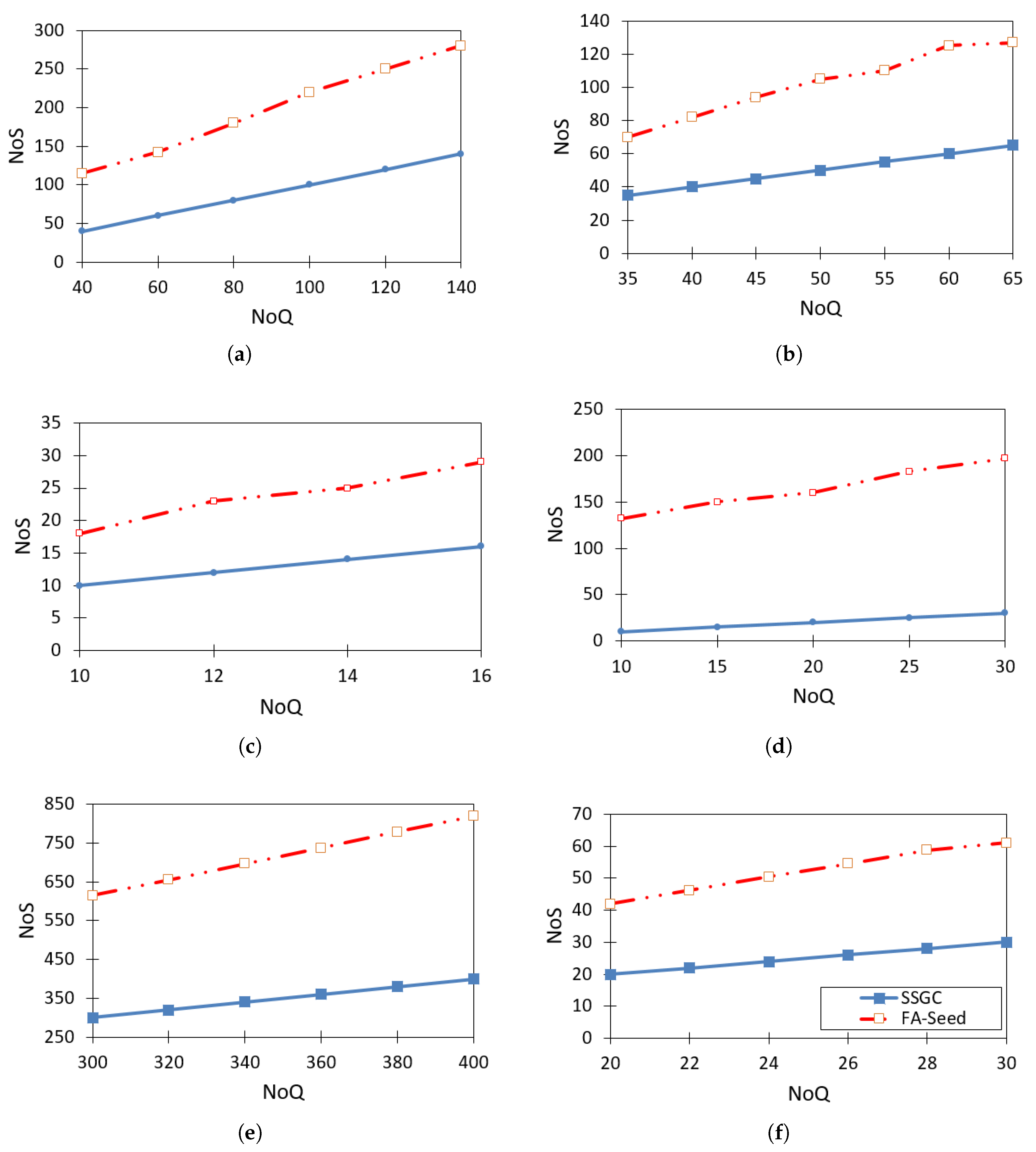
Figure 6 visually illustrates the comparison of the number of seeds obtained between the methods when using the same number of queries. The results show that, at the same NoQ level, FA-Seed consistently obtains a higher number of seeds compared to MSCFMN [6]. Notably, FA-Seed achieves a high seed ratio even when the number of queries is low. This is a significant advantage, as it greatly reduces manual labeling costs. In addition, a high seed ratio directly contributes to enhancing the model’s performance. The main reason FA-Seed achieves a larger number of seeds is its finer HX partitioning mechanism compared to MSCFMN, which allows the model to approach cluster boundaries more closely and thus more effectively exploit potential data points for labeling.
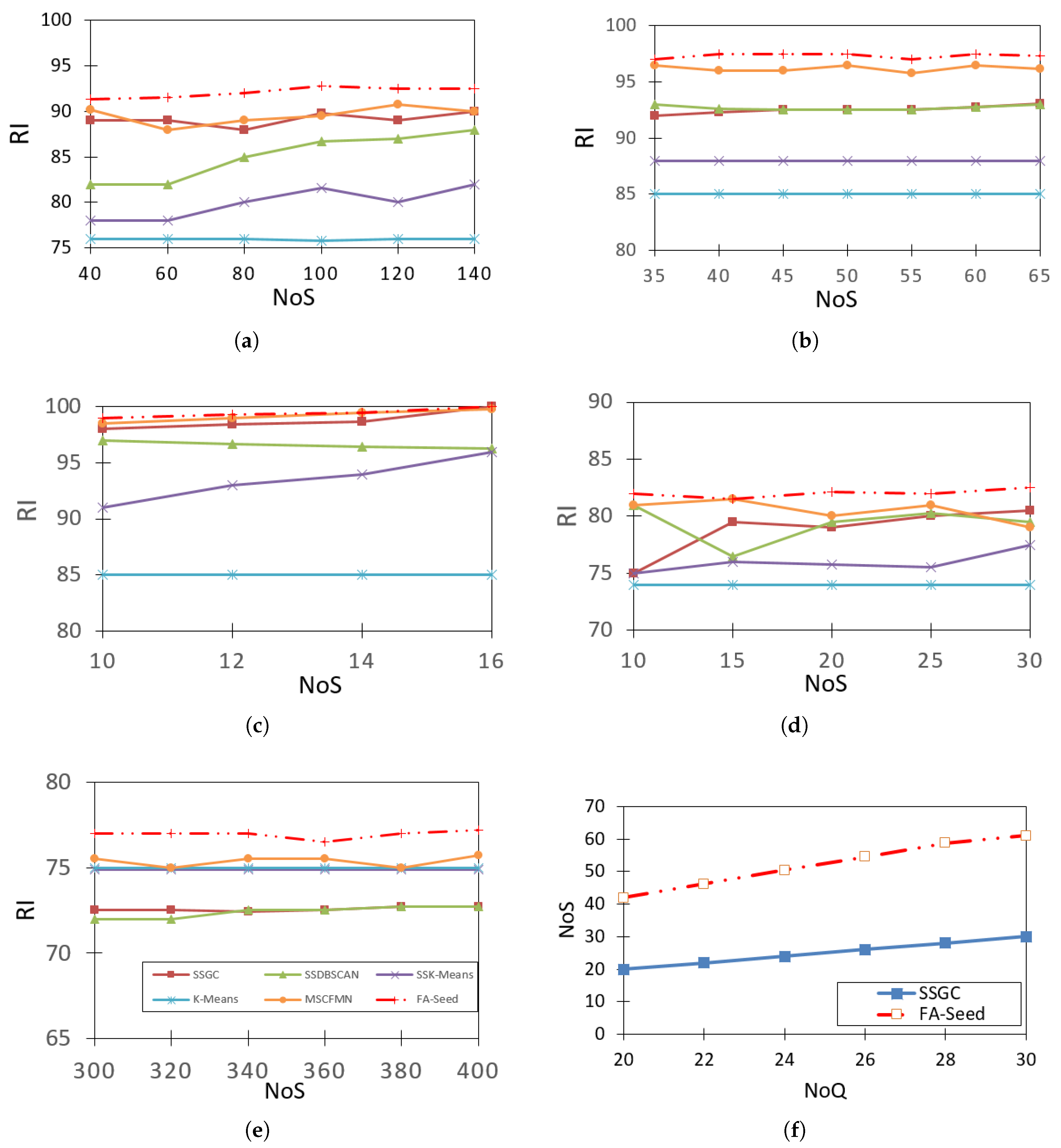
Figure 7 presents the comparison results of FA-Seed with SSGC, SSDBSCAN, SSK-Means, K-Means, MSCFMN [6,7], using the Rand Index (RI) to evaluate clustering quality. The seeds are randomly selected in each run, and the process is repeated 20 times, with the average RI value recorded for comparison. From the visual results, it can be observed that FA-Seed generally performs better or at least comparable to the other methods on most datasets. For datasets with a clear cluster structure such as Soybean, Zoo, and Iris, FA-Seed achieves near-perfect RI values and greater stability compared to other methods. This demonstrates its ability to effectively leverage seed information to assign labels accurately from the very beginning. For datasets with a large number of clusters and evenly distributed data such as Ecoli, FA-Seed still maintains high and stable RI values when the number of seeds changes. For datasets with a strong imbalance in cluster sizes such as Yeast (ranging from 5 to over 400 objects per cluster) or with high overlap levels such as Thyroid, FA-Seed achieves better results thanks to its optimal seed selection mechanism from “trusted HXs” instead of random selection, thereby reducing the negative impact of small or overlapping clusters.
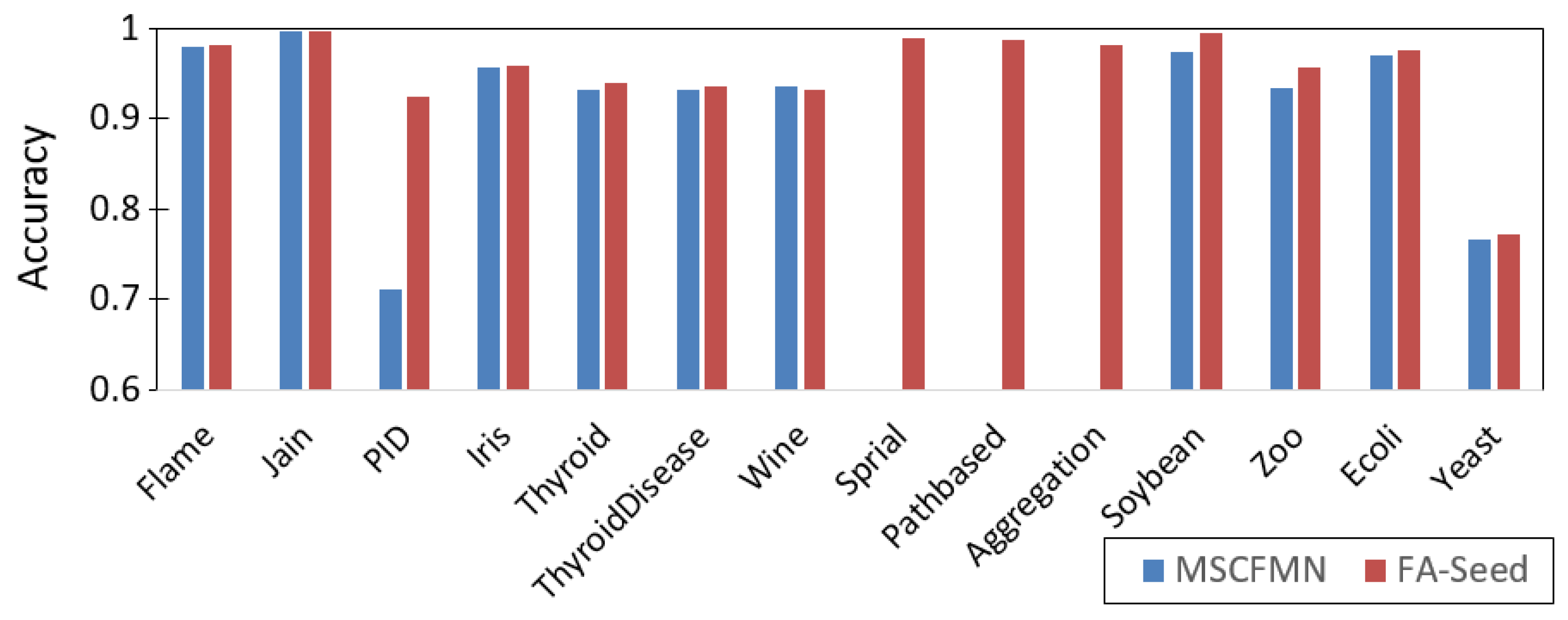
Figure 8 is a graphic comparing the Accuracy measure between FA-Seed and MSCFMN. The results indicate that FA-Seed achieves accuracy comparable to or higher than MSCFMN on most datasets, with notable improvements on datasets with imbalanced class distributions (e.g.,
Thyroid,
Pathbased,
Zoo). This demonstrates that the high-quality seed selection mechanism in FA-Seed can enhance clustering performance even under challenging data conditions.
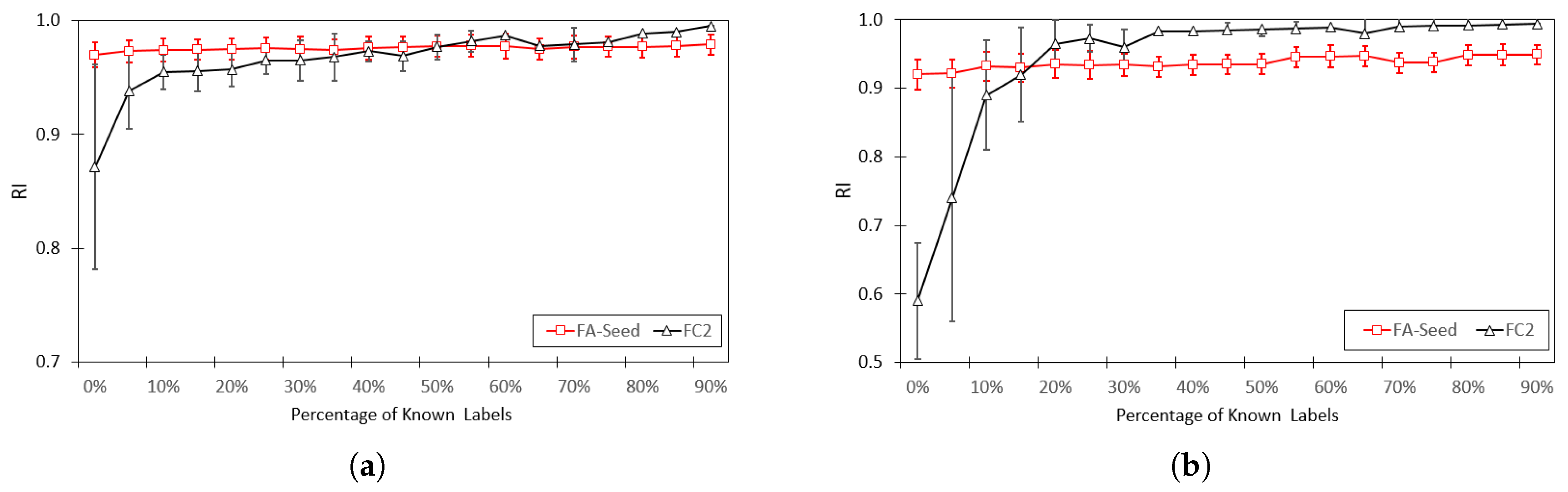
Figure 9 is a chart comparing the RI index between FA-Seed and FC2 [13]. The results show that FA-Seed consistently achieves higher RI values across all proportions of labeled data. Notably, the performance gap in RI between FA-Seed and FC2 is largest at lower labeling rates, confirming the advantage of the seed selection strategy in semi-supervised settings with limited labeled data.
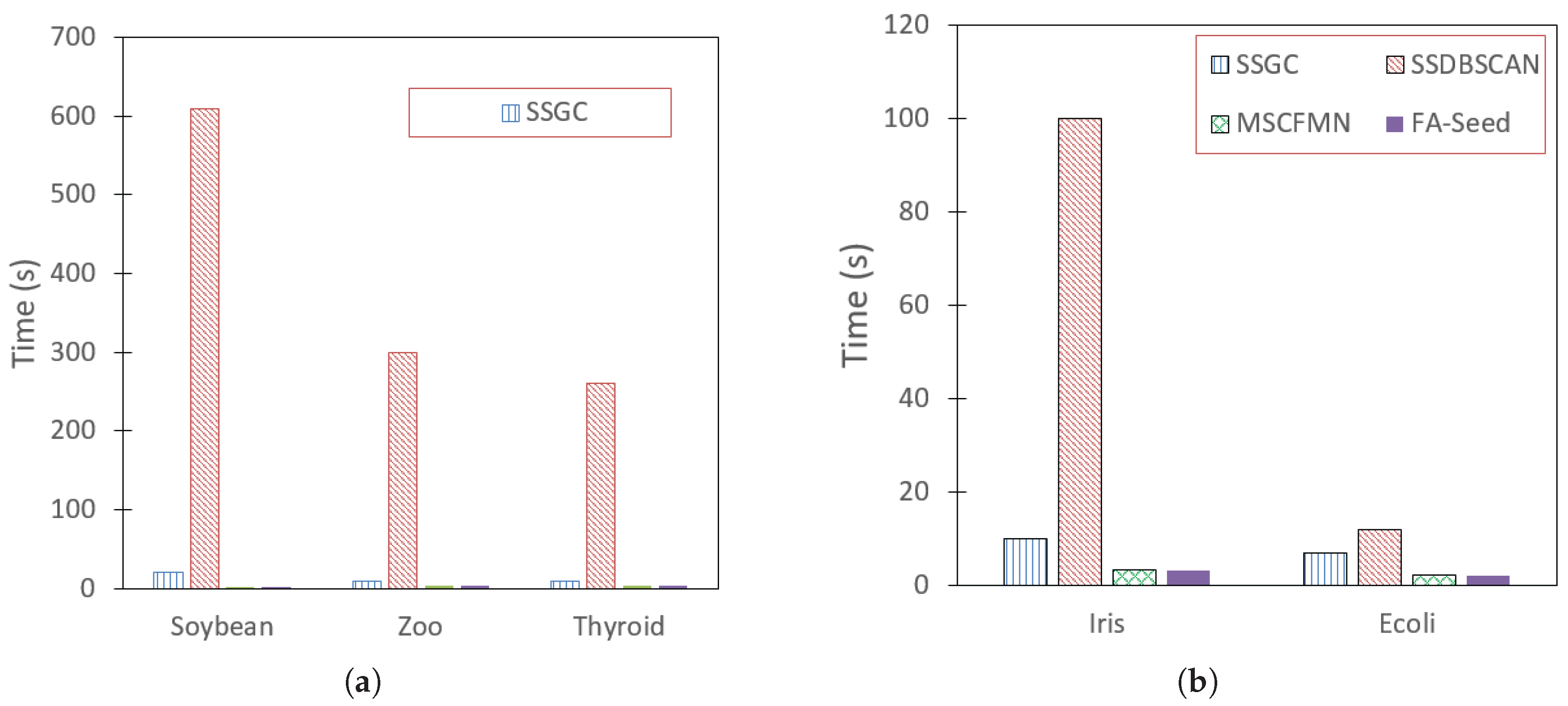
Figure 10 presents a runtime comparison between FA-Seed and the baseline algorithms on the benchmark datasets. As shown, FA-Seed is consistently the fastest (or among the fastest). In terms of time complexity, FA-Seed has a computational cost of
(Equation (
33)), where
denotes the number of data samples,
n is the number of attributes (data dimensions), and
is the number of HXs. In practice,
K is often significantly smaller than
M, allowing FA-Seed to scale efficiently even as the dataset size grows.
In contrast, SSGC—the main algorithm presented in [7]—has a complexity of due to the need to construct a k-nearest neighbor graph and perform label propagation, which becomes costly for large datasets. SSDBSCAN achieves an average complexity of when using optimal data structures, but still reaches in the worst case. For SSK-Means and semi-supervised K-Means, the complexity is , where t is the number of iterations and c is the number of clusters; the computational cost increases rapidly when either t or c is large.
Overall, FA-Seed demonstrates a clear advantage by partitioning the dataset into HXs and identifying “reliable HXs” to select high-quality seeds, while obtaining a larger number of seeds in a single independent stage. This approach allows users to proactively determine the number of labels based on the number of reliable HXs generated, thereby significantly reducing the labeling effort while maintaining, or even improving, clustering quality.
5. Conclusions
In this paper, we propose FA-Seed, a flexible and active learning-based seed selection model for semi-supervised clustering. By integrating adaptive hyperbox-based data partitioning with an active querying mechanism, FA-Seed effectively identifies high-quality seeds, reduces labeling costs, and improves clustering accuracy. Experimental results on standard benchmark datasets demonstrate that FA-Seed consistently outperforms baseline approaches such as SSGC, MSCFMN, and FC2 in terms of clustering quality, seed efficiency, and computational cost. These findings confirm the potential of FA-Seed for practical applications, particularly in medical diagnosis, where data distributions are complex and labeled data are scarce.
Despite its advantages, FA-Seed still has certain limitations. Its performance is sensitive to parameter settings (e.g., ) and remains dependent on expert feedback when clusters are highly overlapping or noisy. In the future, we plan to enhance the robustness of FA-Seed against noisy labels and uncertain expert feedback. Combining FA-Seed with deep representation learning may further improve its performance on complex data, while explainable AI techniques could enhance interpretability in sensitive domains such as healthcare. Finally, large-scale validation on multi-domain and multi-center datasets will be essential to confirm the practical applicability of FA-Seed in diagnostic systems and beyond.
Funding
Hanoi University of Industry under Grant 21-2023-RD/HD-DHCN.
Data Availability Statement
Acknowledgments
We would like to express our sincere gratitude to Hanoi University of Industry for providing the support and favorable conditions that enabled us to complete this research.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| CS-PDS |
Constraints Self-learning from Partial Discriminant Spaces |
| DPP |
Determinantal Point Processes |
| EM |
Expectation-Maximization |
| FC2 |
Finding Clusters on Constraints |
| HX |
Hyperbox |
| HXs |
Hyperboxes |
| FMNN |
Fuzzy Min–Max Neural Network |
| GFMNN |
General FMNN |
| MSCFMN |
Modified SCFMN |
| SCFMN |
Semi-supervised Clustering in FMNN |
| SSGC |
Semi-Supervised Graph-based Clustering |
| SSC |
Semi-supervised clustering |
| SSFC |
Semi-supervised Fuzzy Clustering |
References
- Li, F.; Yue, P.; Su, L. Research on the convergence of fuzzy genetic algorithm based on rough classification. In Proceedings of the International Conference on Natural Computation; Springer: [Location not specified], 2006; pp. 792–795.
- Vu, D.M.; Nguyen, V.H.; Le, B.D. Semi-supervised clustering in fuzzy min-max neural network. In Proceedings of the International Conference on Advances in Information and Communication Technology; Springer: [Location not specified], 2016; pp. 541–550.
- Pedrycz, W.; Waletzky, J. Fuzzy clustering with partial supervision. IEEE Trans. Syst. Man Cybern. Part B (Cybern.) 1997, 27, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Gabrys, B.; Bargiela, A. General fuzzy min-max neural network for clustering and classification. IEEE Trans. Neural Netw. 2000, 11, 769–783. [Google Scholar] [CrossRef] [PubMed]
- Endo, Y.; Hamasuna, Y.; Yamashiro, M.; Miyamoto, S. On semi-supervised fuzzy c-means clustering. In Proceedings of the 2009 IEEE International Conference on Fuzzy Systems; IEEE: [Location not specified], 2009; pp. 1119–1124.
- Minh, V.D.; Ngan, T.T.; Tuan, T.M.; Duong, V.T.; Cuong, N.T. An improvement in integrating clustering method and neural network to extract rules and application in diagnosis support. Iran. J. Fuzzy Syst. 2022, 19, 147–165. [Google Scholar]
- Vu, V.V. An efficient semi-supervised graph based clustering. Intell. Data Anal. 2018, 22, 297–307. [Google Scholar] [CrossRef]
- Vu, D.M.; Nguyen, T.S.; Nguyen, V.T.; Nguyen, D.L. FMN-Voting: A Semi-supervised Clustering Technique Based on Voting Methods Using FMM Neural Networks. In Proceedings of the International Conference on Advances in Information and Communication Technology; Springer: [Location not specified], 2024; pp. 137–150.
- Almazroi, A.A.; Atwa, W. Semi-Supervised Clustering Algorithms Through Active Constraints. Int. J. Adv. Comput. Sci. Appl. 2024, 15, 7. [Google Scholar] [CrossRef]
- Shen, Z.; Lai, M.J.; Li, S. Graph-based semi-supervised local clustering with few labeled nodes. arXiv preprint arXiv:2211.11114 2022.
- Tran, T.N.; Vu, D.M.; Tran, M.T.; Le, B.D. The combination of fuzzy min–max neural network and semi-supervised learning in solving liver disease diagnosis support problem. Arab. J. Sci. Eng. 2019, 44, 2933–2944. [Google Scholar] [CrossRef]
- Bajpai, N.; Paik, J.H.; Sarkar, S. Balanced seed selection for K-means clustering with determinantal point process. Pattern Recognit. 2025, 164, 111548. [Google Scholar] [CrossRef]
- Sun, X. Semi-Supervised Clustering via Constraints Self-Learning. Mathematics 2025, 13, 1535. [Google Scholar] [CrossRef]
- Yi, J.; Zhang, L.; Yang, T.; Liu, W.; Wang, J. An efficient semi-supervised clustering algorithm with sequential constraints. In Proceedings of the 21st ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; ACM: [Location not specified], 2015; pp. 1405–1414.
- Singh, Y.; Susan, S. Lung cancer subtyping from gene expression data using general and enhanced fuzzy min–max neural networks. Eng. Rep. 2023, 5, e12663. [Google Scholar] [CrossRef]
- Kämäräinen, J.K.; Kauppi, O.P.; Fränti, P. Clustering Datasets. Available online: https://cs.joensuu.fi/sipu/datasets/ (accessed on 2 August 2025).
- Khuat, T.T.; Gabrys, B. A comparative study of general fuzzy min-max neural networks for pattern classification problems. Neurocomputing 2020, 386, 110–125. [Google Scholar] [CrossRef]
- Vu, D.M.; Nguyen, T.S.; Phung, T.N.; Vu, T.H. Choosing Data Points to Label for Semi-supervised Learning Based on Neighborhood. In Proceedings of the International Conference on Advances in Information and Communication Technology; Springer: [Location not specified], 2023; pp. 134–141.
- Simpson, P.K. Fuzzy min-max neural networks-part 2: Clustering. IEEE Trans. Fuzzy Syst. 2002, 1, 32. [Google Scholar] [CrossRef]
- Simpson, P.K. Fuzzy min-max neural networks. I. Classification. IEEE Trans. Neural Netw. 1992, 3, 776–786. [Google Scholar] [CrossRef] [PubMed]
- Aha, D. UC Irvine Machine Learning Repository. Available online: https://archive.ics.uci.edu/ (accessed on 2 August 2025).
- Batista, G.E.A.P.A.; Monard, M.C. An analysis of four missing data treatment methods for supervised learning. Appl. Artif. Intell. 2003, 17, 519–533. [Google Scholar] [CrossRef]
Figure 1.
Illustration of fuzzy HXs in a two-dimensional Aggregation dataset, partitioned by the FMNN model.
Figure 1.
Illustration of fuzzy HXs in a two-dimensional Aggregation dataset, partitioned by the FMNN model.
Figure 2.
Illustration of HX partitioning on the Aggregation dataset by FMNN-based models, including: (a) Data distribution of the Aggregation dataset. (b) Data space partitioned into 7 HXs by MSCFMN (one per cluster). (c) Data space partitioned into 17 HXs by FA-Seed with 13 queried points. (d) Data space partitioned into 26 HXs bay FA-Seed with 21 queried points. (e) Data space partitioned into 38 HXs by FA-Seed with 29 queried points. (f) Data space partitioned into 52 HXs by FA-Seed with 40 queried points.
Figure 2.
Illustration of HX partitioning on the Aggregation dataset by FMNN-based models, including: (a) Data distribution of the Aggregation dataset. (b) Data space partitioned into 7 HXs by MSCFMN (one per cluster). (c) Data space partitioned into 17 HXs by FA-Seed with 13 queried points. (d) Data space partitioned into 26 HXs bay FA-Seed with 21 queried points. (e) Data space partitioned into 38 HXs by FA-Seed with 29 queried points. (f) Data space partitioned into 52 HXs by FA-Seed with 40 queried points.
Figure 3.
HX selection on the Aggregation dataset using FA-Seed. Balanced HXs (1–21) are retained, whereas imbalanced HXs (a–e) are discarded.
Figure 3.
HX selection on the Aggregation dataset using FA-Seed. Balanced HXs (1–21) are retained, whereas imbalanced HXs (a–e) are discarded.
Figure 4.
The architecture of the proposed FA-Seed model with an integrated label evaluation component.
Figure 4.
The architecture of the proposed FA-Seed model with an integrated label evaluation component.
Figure 5.
Neural Network Architecture of FA-Seed.
Figure 5.
Neural Network Architecture of FA-Seed.
Figure 6.
Comparison of NoS values between FA-Seed and baseline algorithms on benchmark datasets, including: (a) Ecoli dataset, (b) Iris dataset, (c) Soybean dataset, (d) Thyroid dataset, (e) Yeast dataset, (f) Zoo dataset.
Figure 6.
Comparison of NoS values between FA-Seed and baseline algorithms on benchmark datasets, including: (a) Ecoli dataset, (b) Iris dataset, (c) Soybean dataset, (d) Thyroid dataset, (e) Yeast dataset, (f) Zoo dataset.
Figure 7.
Comparison of RI values between FA-Seed and baseline algorithms on Benchmark datasets, including: (a) Ecoli dataset, (b) Iris dataset, (c) Soybean dataset, (d) Thyroid dataset, (e) Yeast dataset, (f) Zoo dataset.
Figure 7.
Comparison of RI values between FA-Seed and baseline algorithms on Benchmark datasets, including: (a) Ecoli dataset, (b) Iris dataset, (c) Soybean dataset, (d) Thyroid dataset, (e) Yeast dataset, (f) Zoo dataset.
Figure 8.
Visual comparison chart of Accuracy measures of FA-Seed and MSCFMN.
Figure 8.
Visual comparison chart of Accuracy measures of FA-Seed and MSCFMN.
Figure 9.
Average clustering performance RI achieved by the proposed FA-Seed and FC2, including: (a) Iris dataset, (b) Wine dataset.
Figure 9.
Average clustering performance RI achieved by the proposed FA-Seed and FC2, including: (a) Iris dataset, (b) Wine dataset.
Figure 10.
Comparison of Time (measured in Seconds) values between FA-Seed and baseline algorithms on benchmark datasets, including: (a) Soybean, Zoo, and Thyroid datasets, (b) Iris, and Ecoli datasets.
Figure 10.
Comparison of Time (measured in Seconds) values between FA-Seed and baseline algorithms on benchmark datasets, including: (a) Soybean, Zoo, and Thyroid datasets, (b) Iris, and Ecoli datasets.
Table 1.
Benchmark datasets used in the experiments.
Table 1.
Benchmark datasets used in the experiments.
| Dataset |
#Instances |
#Features |
#Clusters |
| Soybean |
47 |
35 |
4 |
| Zoo |
101 |
16 |
7 |
| Iris |
150 |
4 |
3 |
| Wine |
178 |
13 |
3 |
| Thyroid |
215 |
5 |
3 |
| Flame |
240 |
2 |
2 |
| Spiral |
312 |
2 |
3 |
| Pathbased |
317 |
2 |
3 |
| Ecoli |
336 |
8 |
8 |
| Jain |
373 |
2 |
2 |
| PID |
768 |
8 |
2 |
| Aggregation |
788 |
2 |
7 |
| Yeast |
1484 |
8 |
10 |
| ThyroidDisease |
7200 |
21 |
3 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).