Submitted:
10 September 2025
Posted:
11 September 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Related Work
3. Research Methodology
3.1. Problem Statement
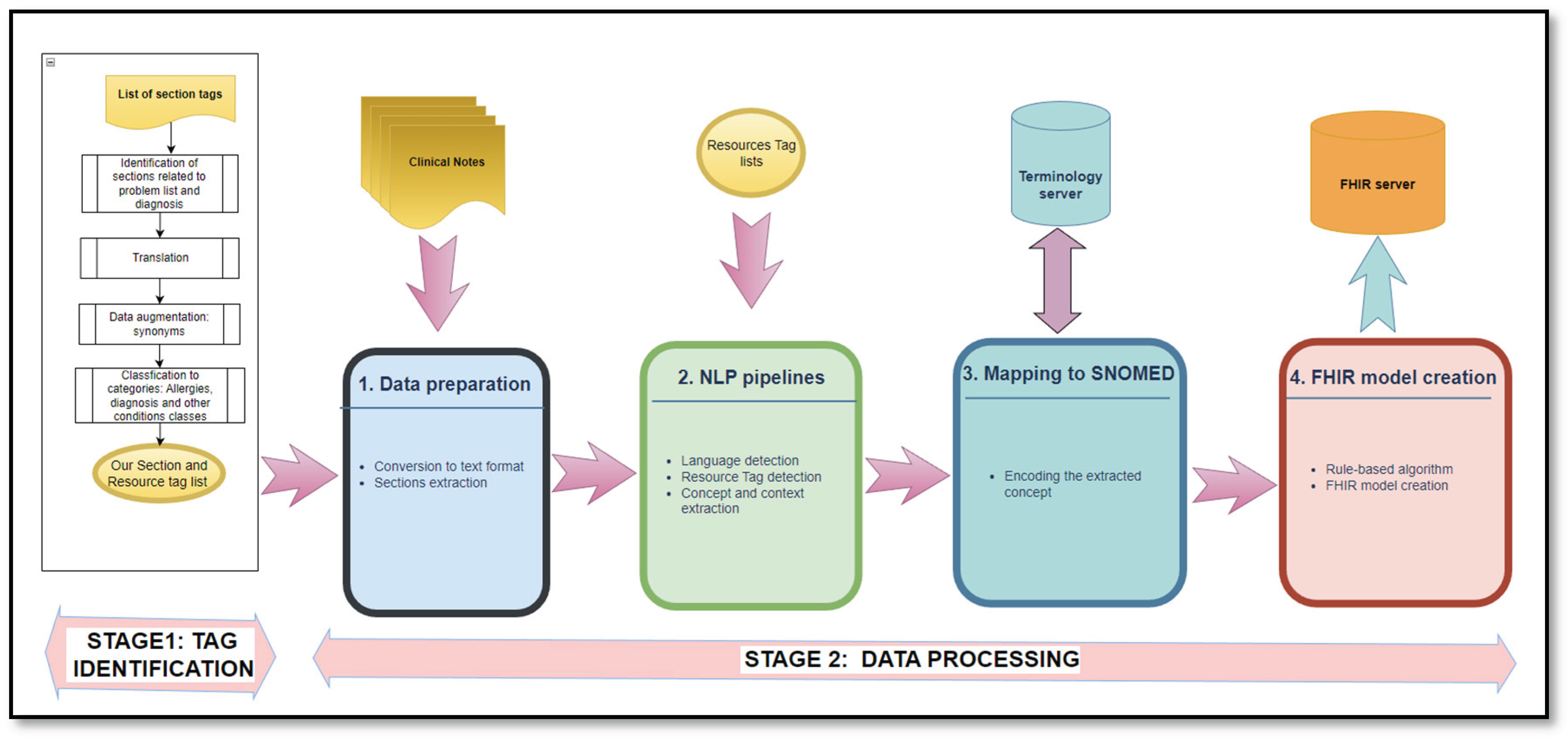
3.2. Method
- Identification of the section and resource tags – executed only once (Stage1),
- Data processing – executed for every clinical note file (Stage 2).
- Stage 1: Tag Identification
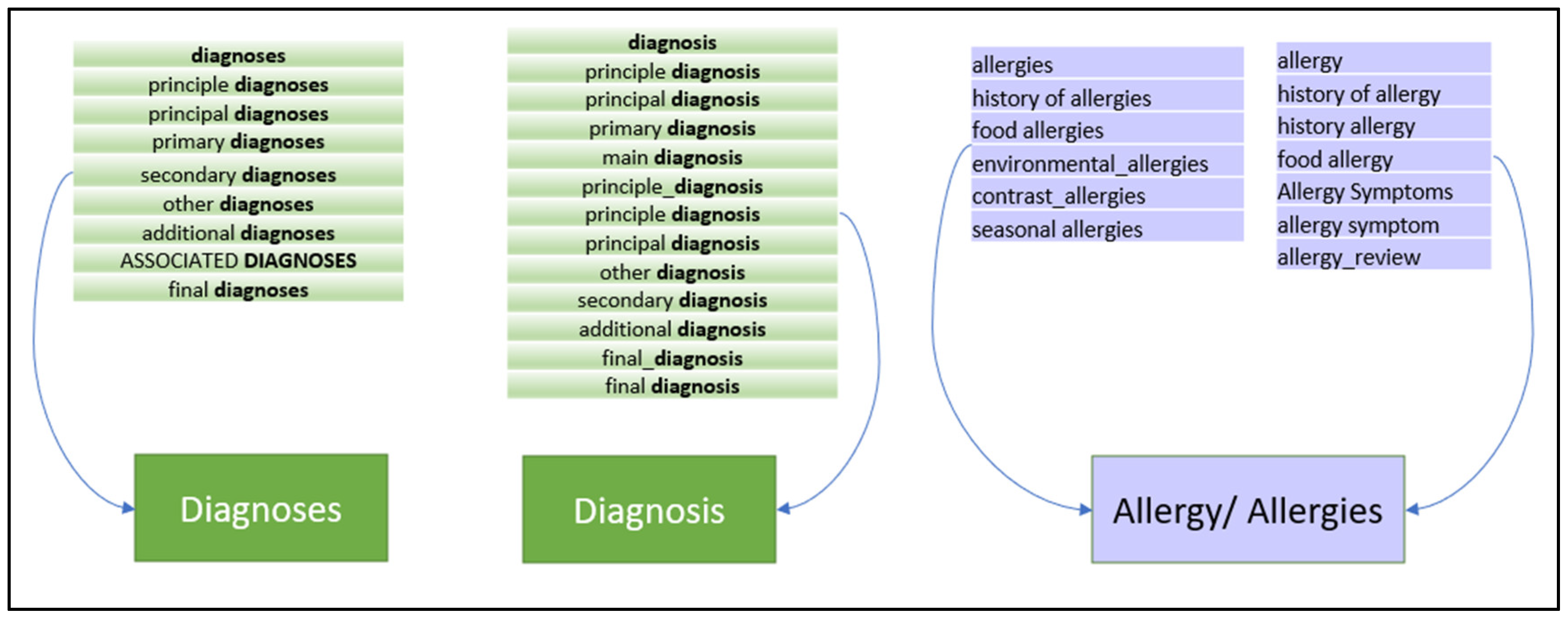
- Identification of sections related to the patient problem list and diagnosis: The list of section tags in [20] with 6773 items was used as the starting point to identify and extract section candidates that may contain information related to the patient problem list and diagnosis. The resulting list is then optimized by eliminating duplicates and semantic matching using a pivot term. For example, principal diagnosis, secondary diagnosis are replaced by diagnosis (Figure 2).
- Classification to categories: The final French tag list includes three main categories: tags related to allergies (50 elements), diagnosis (106 elements) and other conditions (156 elements)
- Stage 2: Data processing
-
Step1: Data preparationData sources of clinical notes in English or French were converted into text format when required [27]. Next, the list of section tags related to patient problem lists is used to extract the sections where possible.
- 2.
-
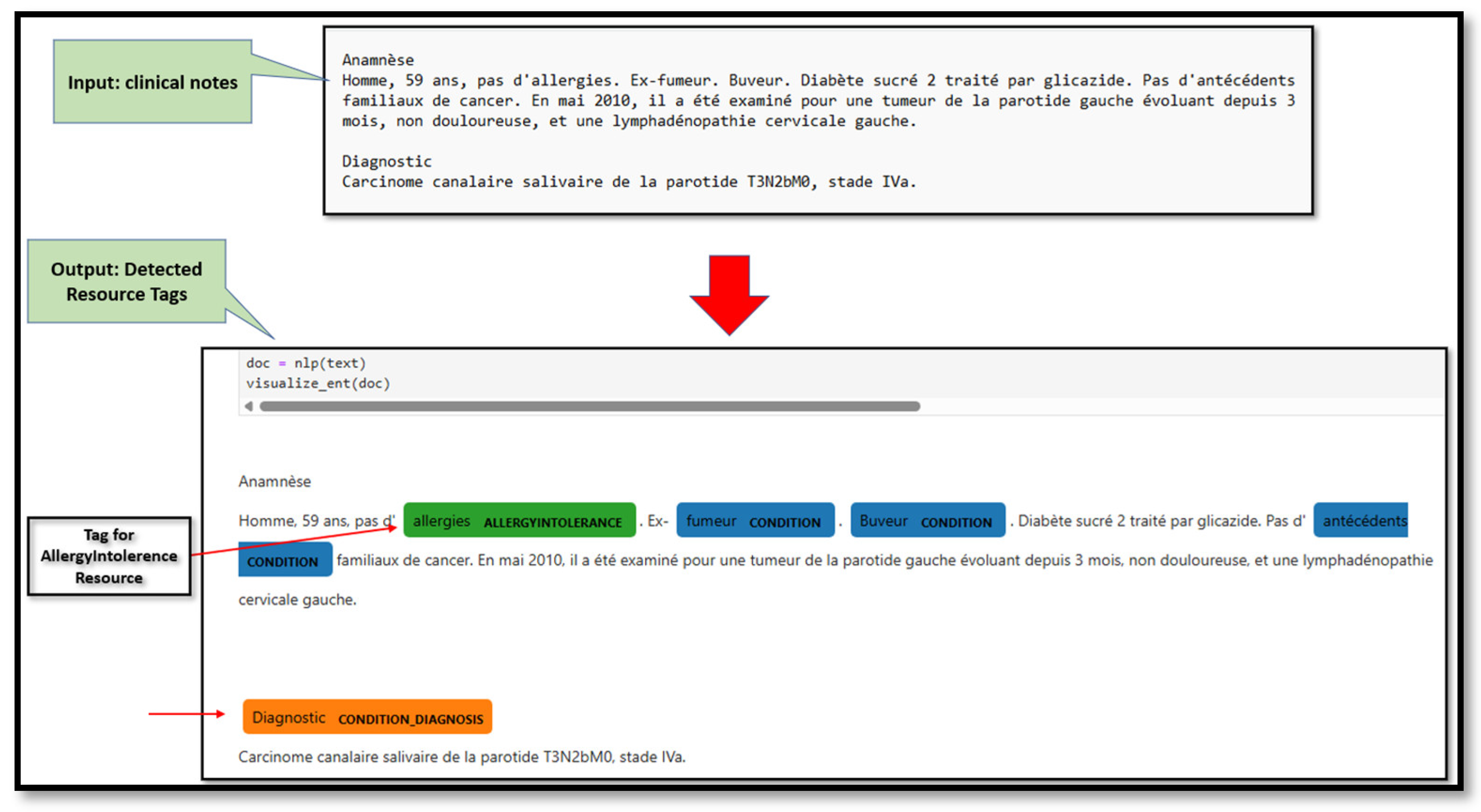
Step 2: NLP pipelinesMedSpacy is an NLP toolkit designed for processing clinical and biomedical texts [28] [29]. It is integrated within the SpaCy platform, an open-source NLP library for Python [30].Language detection: MedSpacy includes tools for language and section detection. Both tools are useful because the clinical notes are written in either English or French. This step is essential because the models are language-dependent and the study investigated French textual data.Resource Tag detection: This step is implemented with MedSpacy where the module seeks to detect more context of the concept to determine whether it is related to an Allergy or intolerance, as well as any diagnosis, based on the established resource tags list.
- 3.
-
Step 3: Mapping to SNOMED
- 4.
-
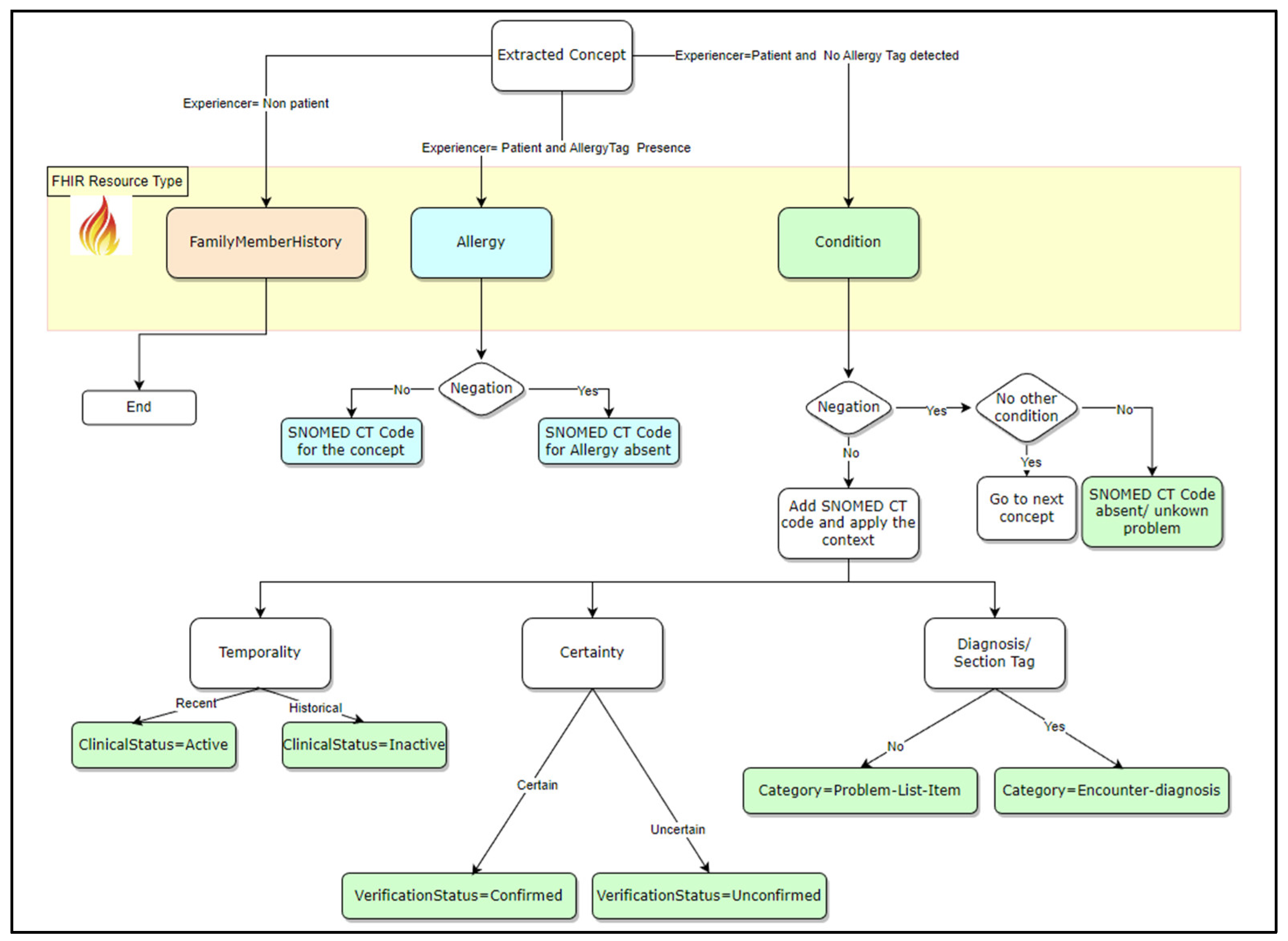
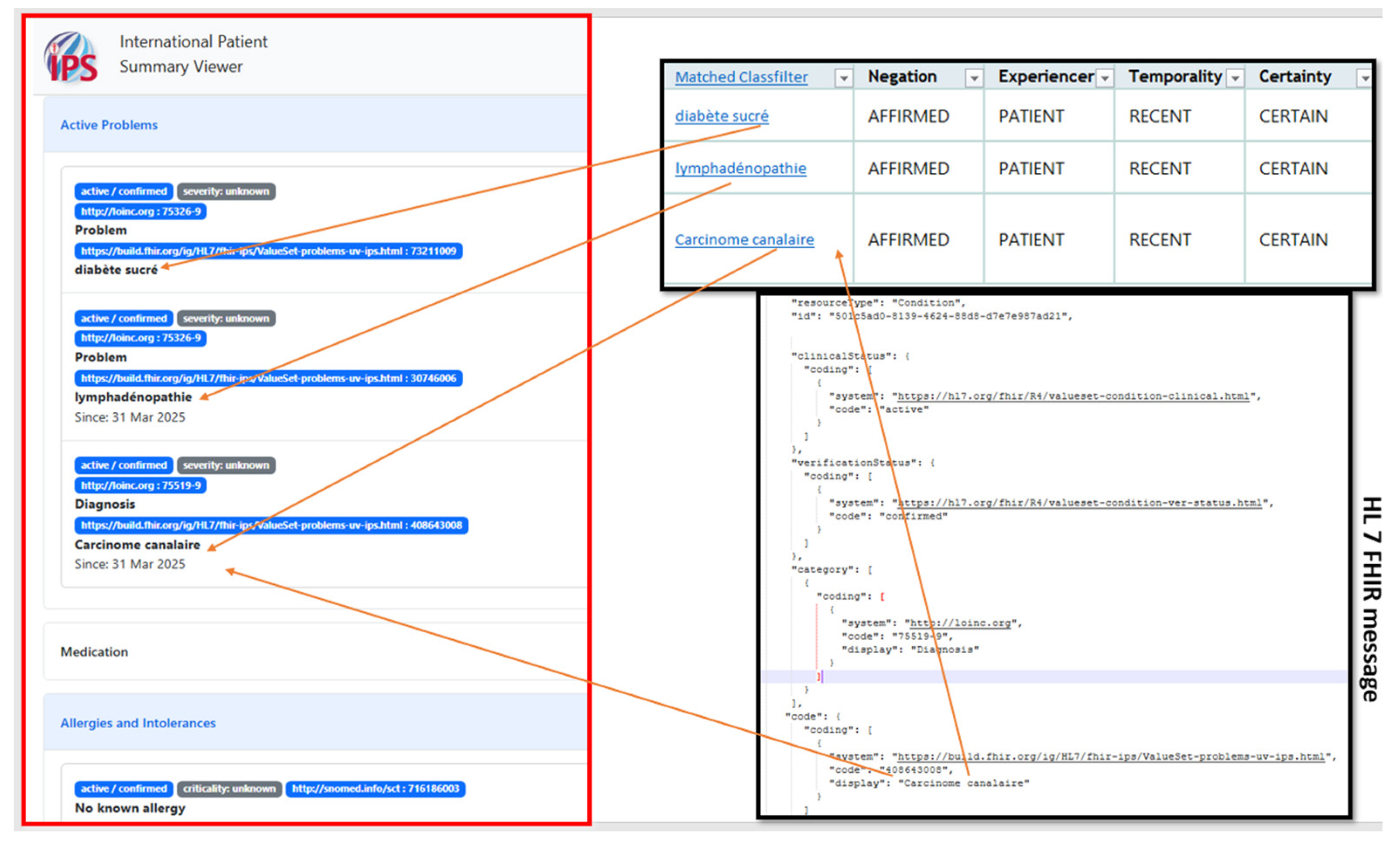
Step 4: FHIR model creationThe data output from step 3 is aggregated to construct the FHIR model. To achieve this, we used the following rule-based method that was designed to map the extracted concepts to their corresponding IPS FHIR elements (Figure 3).This rule-based method for mapping NLP output to FHIR model elements consists of:
- four rules related to context modifiers (experiencer, negation, temporality, and certainty) and
- two additional rules related to Resource tags identification (allergy, diagnosis).
- Experiencer = Non Patient: Experiencer context means that the indicated problem is for the patient or their family member. In this scenario, the FHIR resource used was FamliyMemberHistory. In this study, the focus was on the patient’s clinical problems.
- Experiencer = Patient and Allergy Tag Presence: when an Allergy tag is detected, the concept is mainly for an allergy. Therefore, the FHIR resource to be used is AllergyIntolerance. The Negation context was then used to confirm whether the patient had an allergy.
-
Experiencer = Patient and No Allergy Tag detected: The current patient problem is not related to an allergy, so the FHIR resource to be used is Condition. Next, the Negation context is used to confirm the decision rule following these alternatives:
- Negation = Yes (Concept is negated): If it is the only concept for a condition, then the patient has no known conditions and the corresponding SNOMED CT code will be added. Otherwise, the process continues with the next concept.
-
Negation = No. The next step is to add the corresponding SNOMED CT code and mapping to the corresponding FHIR Condition element following these three rules:
- ○
- Temporality confirms whether the identified problem is still active The value of the ClinicalStatus element of the Condition resource is active or inactive if temporality is Recent or Historical, respectively.
- ○
- Certainty enables us to determine whether an identified problem is confirmed or only a hypothesis. In the first case, the VerificationStatus element value is confirmed; otherwise, it is unconfirmed.
- ○
- The diagnosis section tag enables the identification of the condition category element. The possible values are Encounter-diagnosis or Problem-List-Item.
3.3. Evaluation
- -
- The rulebased approach results were manually validated by testing all possible cases because the dataset did not cover all scenarios of the context modifiers.
- -
- The result of the overall process is then viewed in IPS viewer
4. Results
- a)
- CANTEMIST-FRASIMED: The patient summary is organized into sections for medical history, physical examination, diagnosis, treatment, etc.
- b)
- DISTEMIST-FRASIMED: the summary is a text with no headers.
-
Step1: Data preparation:
- -
- Conversion to text format: the file format is text type.
- -
-
Sections extraction:
- a)
- CANTEMIST-FRASIMED Corpus: Based on the Resource and section tag list, the sections related to the patient problem list were extracted (see Table 2 for the list of section titles available in this corpus and their corresponding categories).
- b)
- DISTEMIST-FRASIMED Corpus: This step is not applicable to this corpus because there are no headers.
- 2.
-
Step 2: NLP pipelines
- -
- Language detection: The purpose of this step is to select either the French or English model to be use based as the text file language. This study focuses on French text cases.
- -
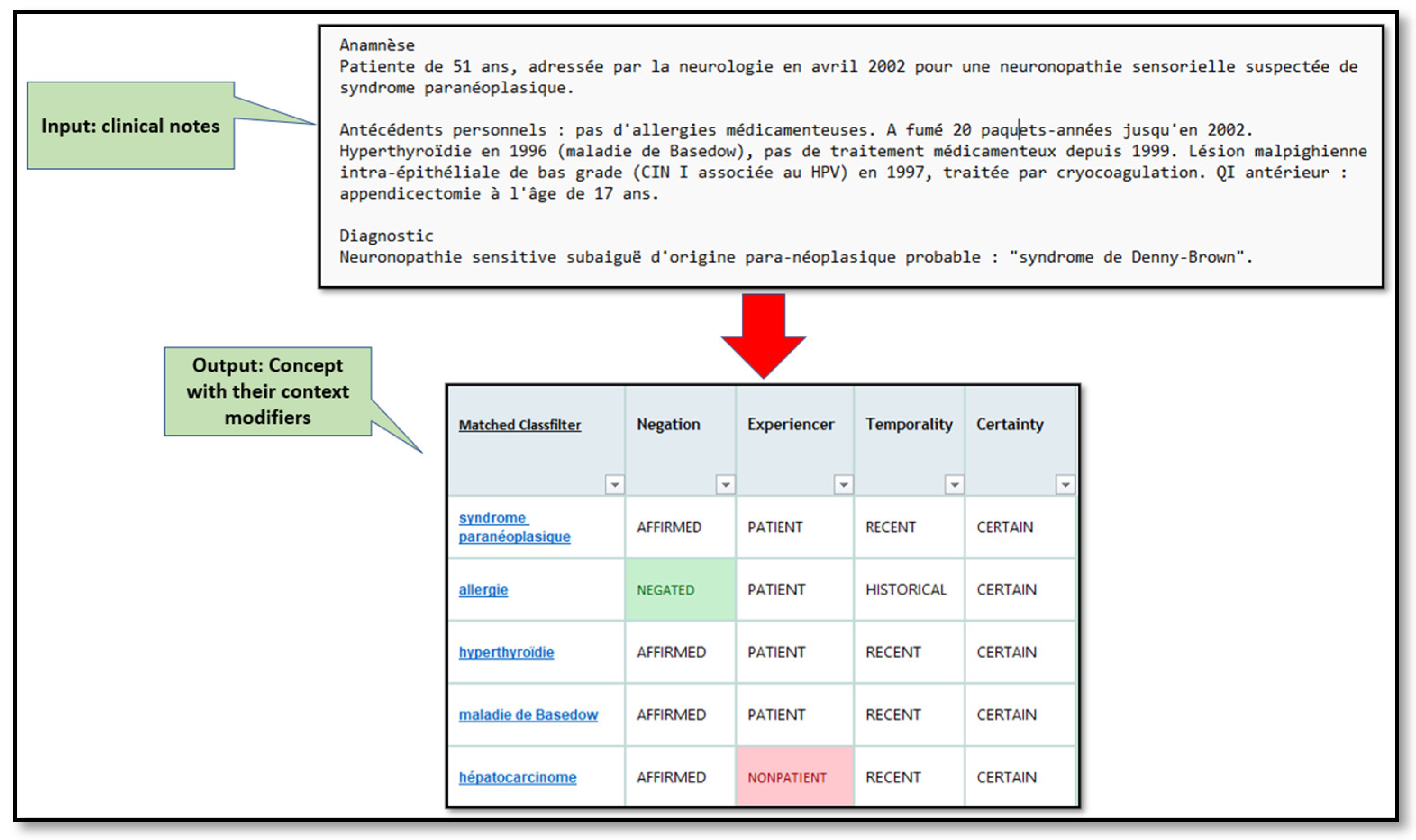
- Concepts and context extraction: Figure 4 is an example of the SIFR output for a clinical text that contains the results of context modifiers for each detected concept.
- 3.
-
Step 3: Mapping to SNOMEDThe Shrimp tool was used to find the SNOMED CT code for the extracted concepts related to the problem list.
- 4.
-
Step 4: FHIR model creationBefore delving deeper into the use cases, it is important to provide a quick overview of the FHIR specifications used.
- -
- Most of the tools used in this study were an implementation of version 4 of FHIR [36].
- -
- The HAPI FHIR server, an implementation of the FHIR specifications in JAVA, was used to test the proposed FHIR model [37].
- -
- The details of the specification profile describing the FHIR resources and their format are based on the IPS implementation guide [38].
- -
Limitations
5. Discussion and Conclusions
Conclusion
Author Contributions
Abbreviations
| API | Application Programming Interface |
| BERT | Bidirectional Encoder Representations from Transformers |
| BioBERT | Bidirectional Encoder Representations from Transformers for Biomedical Text Mining |
| CEN | European Commitee for Standardization |
| CDA | Clinical Document Architecture |
| CLAMP | Clinical Language Annotation, Modeling, and Processing |
| cTAKES | Clinical Text Analysis and Knowledge Extraction System |
| eHN | European eHealth Network |
| EHR | Electronic Health Record |
| FHIR | Fast Healthcare Interoperability Resource |
| G7 | Group of Seven Summits: Annual meeting of leaders from seven of the world’s largest advanced economies |
| GDHP | Global Digital Health Partnership |
| HL7 | Health Level Seven |
| IHE | Integrated Healthcare Exchange |
| IPS | International Patient Summary |
| ISO | International Organization for Standardization |
| PS-CA | Canadian Patient Summary |
| MedXN | Medication eXtraction and Normalization |
| MedSpacy | SpaCy-based library of core components targeting medical text |
| ML | Machine Learning |
| NLM | National Library of Medicine |
| NLP | Natural Language Processing |
| ONC | Office of the National Coordinator |
| SIFR | Ontology-based annotation web service to process biomedical text in French |
| SNOMED CT | Systematized Nomenclature of Medicine - Clinical Terms |
| UIMA | Unstructured Information Management Architecture |
| UMLS | Unified Medical language System |
References
- Amar, F.; April, A.; Abran, A. Electronic Health Record and Semantic Issues Using Fast Healthcare Interoperability Resources: Systematic Mapping Review. J Med Internet Res 2024, 26, e45209. [Google Scholar] [CrossRef] [PubMed]
- International Patient Summary. Date of access: 2025. https://international-patient-summary.net/ips-links-to-standards-and-specifications/.
- FHIR IPS Resources. Date of access: 2025. https://hl7.org/fhir/uv/ips/.
- IPS- Condition Resource. Date of access: 2025. https://build.fhir.org/ig/HL7/fhir-ips/StructureDefinition-Condition-uv-ips.html.
- Zaghir, J.; Bjelogrlic, M.; Goldman, J.P.; Aananou, S.; Gaudet-Blavignac, C.; Lovis, C. FRASIMED: a Clinical French Annotated Resource Produced through Crosslingual BERT-Based Annotation Projection. Published online 2023.
- Durango, M.C.; Torres-Silva, E.A.; Orozco-Duque, A. Named Entity Recognition in Electronic Health Records: A Methodological Review. Healthc Inform Res. 2023, 29, 286–300. [Google Scholar] [CrossRef] [PubMed]
- Gaudet-Blavignac, C.; Foufi, V.; Bjelogrlic, M.; Lovis, C. Use of the Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT) for Processing Free Text in Health Care: Systematic Scoping Review. J Med Internet Res. 2021, 23, e24594. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Yoon, W.; Kim, S.; et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Wren J, ed. Bioinformatics. 2020, 36, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Hong, N.; Wen, A.; Stone, D.J.; et al. Developing a FHIR-based EHR phenotyping framework: A case study for identification of patients with obesity and multiple comorbidities from discharge summaries. Journal of Biomedical Informatics. 2019, 99, 103310. [Google Scholar] [CrossRef] [PubMed]
- Hong, N.; Wen, A.; Shen, F.; et al. Integrating Structured and Unstructured EHR Data Using an FHIR-based Type System: A Case Study with Medication Data. Proceedings of AMIA Joint Summits on Translational Science. 2018, 2017. [Google Scholar]
- Hong, N.; Wen, A.; Shen, F.; et al. Developing a scalable FHIR-based clinical data normalization pipeline for standardizing and integrating unstructured and structured electronic health record data. JAMIA Open. 2019, 2, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Luo, Y.; Stone, D.; et al. Integration of NLP2FHIR Representation with Deep Learning Models for EHR Phenotyping: A Pilot Study on Obesity Datasets. Published online 2021.
- Soysal, E.; Wang, J.; Jiang, M.; et al. CLAMP – a toolkit for efficiently building customized clinical natural language processing pipelines. Journal of the American Medical Informatics Association. 2018, 25, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Mathews, W.C.; Pham, H.A.; Xu, H.; Zhang, Y. Opioid2FHIR: A system for extracting FHIR-compatible opioid prescriptions from clinical text. In: 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). IEEE; 2020:1748-1751. [CrossRef]
- Peterson, K.J.; Jiang, G.; Liu, H. A corpus-driven standardization framework for encoding clinical problems with HL7 FHIR. Journal of Biomedical Informatics. 2020, 110, 103541. [Google Scholar] [CrossRef] [PubMed]
- Peterson, K.J.; Liu, H. Automating the Transformation of Free-Text Clinical Problems into SNOMED CT Expressions. Published online 2020.
- Wu, H.; Toti, G.; Morley, K.I.; et al. SemEHR: A general-purpose semantic search system to surface semantic data from clinical notes for tailored care, trial recruitment, and clinical research. Journal of the American Medical Informatics Association. 2018, 25. [Google Scholar] [CrossRef] [PubMed]
- Shaitarova, A.; Zaghir, J.; Lavelli, A.; Krauthammer, M.; Rinaldi, F. Exploring the Latest Highlights in Medical Natural Language Processing across Multiple Languages: A Survey. Yearb Med Inform. 2023, 32, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Denny, J.C.; Spickard, A.; Johnson, K.B.; Peterson, N.B.; Peterson, J.F.; Miller, R.A. Evaluation of a Method to Identify and Categorize Section Headers in Clinical Documents. Journal of the American Medical Informatics Association. 2009, 16, 806–815. [Google Scholar] [CrossRef] [PubMed]
- Pomares-Quimbaya, A.; Kreuzthaler, M.; Schulz, S. Current approaches to identify sections within clinical narratives from electronic health records: a systematic review. BMC Med Res Methodol. 2019, 19, 155. [Google Scholar] [CrossRef] [PubMed]
- Deepl translator. Date of access: 2025. https://www.deepl.com/en/translator.
- French English Medical Dictionary. Date of access: 2025. https://dictionary.reverso.net/medical-french-english/.
- ChatGPT. Date of access: 2025. https://chatgpt.com/.
- Feng, S.Y.; Gangal, V.; Wei, J.; et al. A Survey of Data Augmentation Approaches for NLP. Published online December 1, 2021. [CrossRef]
- Bayer, M.; Kaufhold, M.A.; Reuter, C. A Survey on Data Augmentation for Text Classification. ACM Comput Surv. 2023, 55, 1–39. [Google Scholar] [CrossRef]
- Li, B.; Hou, Y.; Che, W. Data augmentation approaches in natural language processing: A survey. AI Open. 2022, 3, 71–90. [Google Scholar] [CrossRef]
- File Convertor PDF to Text. Date of access: 2025. https://www.freeconvert.com/pdf-to-text.
- MedSpacy githib. Date of access: 2025. https://github.com/medspacy/medspacy/blob/master/README.md.
- Eyre, H.; Chapman, A.B.; Peterson, K.S.; et al. Launching into clinical space with medspaCy: a new clinical text processing toolkit in Python.
- Spacy. Date of access: 2025. https://spacy.io/.
- Clinical French Annotator. Date of access: 2025. https://bioportal.lirmm.fr/annotator.
- Tchechmedjiev, A.; Abdaoui, A.; Emonet, V.; Zevio, S.; Jonquet, C. SIFR annotator: ontology-based semantic annotation of French biomedical text and clinical notes. BMC Bioinformatics. 2018, 19, 405. [Google Scholar] [CrossRef] [PubMed]
- Mirzapour, M.; Abdaoui, A.; Tchechmedjiev, A.; Digan, W.; Bringay, S.; Jonquet, C. French FastContext: A publicly accessible system for detecting negation, temporality and experiencer in French clinical notes. Journal of Biomedical Informatics. 2021, 117, 103733. [Google Scholar] [CrossRef] [PubMed]
- Canada Health Infoway. Canada Infoway healthcare terminology server. Date of access: 2025. https://infocentral.infoway-inforoute.ca/en/tools/standards-tools/terminology-server.
- Shrimp Tool. Date of access: 2025. https://ontoserver.csiro.au/shrimp/.
- H7 FHIR V4. Date of access: 2025. https://hl7.org/fhir/R4/resourcelist.html.
- HAPI FHIR. Date of access: 2025. https://hapi.fhir.org/.
- IPS Implementation Guide. Date of access: 2025. https://build.fhir.org/ig/HL7/fhir-ips/OperationDefinition-summary.html.
- IPS Viewer. Date of access: 2025. https://www.ipsviewer.com/classic.
| ID | Category | Challenge description |
|---|---|---|
| 1. | Data format | |
| 1.1 | Information related to the patient problem list is mainly in unstructured format. | |
| 1.2 | Most reports are in PDF file format. | |
| 2. | Language | |
| 2.1 | Clinical notes in Canada, and other French countries, are either in English or French. | |
| 2.2 | NLP models and techniques are language dependent. Selecting the appropriate NLP pipeline requires prior identification of the language used. | |
| 2.3 | Most NLP tools are for English text. There is a major need in other languages, including French which is largely used in Quebec, for the interoperability of the patient problem list. | |
| 3. | Context and modifiers | |
| 3.1 | The patient problem list may be related to an allergy/intolerance, a diagnosis or other types of related clinical conditions. It is important to distinguish between these items to ensure correct mapping to FHIR elements. | |
| 3.2 | The proposed framework needs to consider that the extracted condition may be in a negation context. | |
| 3.3 | Extracted condition may be related to the patient or their family members. | |
| 3.4 | Extracted condition may be confirmed or only an hypothesis. | |
| 3.5 | Extracted condition may be active or resolved (historical). | |
| 4. | Standard/ guidelines | |
| 4.1 | Must use a standard (e.g., SNOMED CT) to ensure semantic interoperability or common understanding and interpretability. | |
| 5. | Condition type | |
| The patient problem list may be related to an allergy or other type of health conditions. Need to distinguish allergies from the rest of condition types. |
| Section title (original list) | Selected/ Unselected | Tag category |
|---|---|---|
| Anamnèse | Selected | Other condition |
| Examen physique | Unselected | - |
| Examens complémentaires | Unselected | - |
| Tests complémentaires | Unselected | - |
| Diagnostic | Selected | Diagnosis |
| DIAGNOSTIC PRINCIPAL | Selected | Diagnosis |
| HISTOIRE DE LA FAMILLE | Selected | Other condition |
| MALADIE ACTUELLE | Selected | Diagnosis |
| CONTEXTE PERSONNEL | Selected | Other condition |
| Antécédents | Selected | Other condition |
| Antécédents oncologiques | Selected | Other condition |
| Traitement | Unselected | - |
| Évolution | Unselected | - |
| L’évolution | Unselected | - |
| Développements | Unselected | - |
| FRAMISED Dataset files | Language | Accuracy | Recall | Precision | F1 Score |
|---|---|---|---|---|---|
| CANTEMIST-FRASIMED | French | 1 | 1 | 1 | 1 |
| DISTEMIST-FRASIMED | French | 0.947 | 1 | 0.8888 | 0.9411 |
| Step | Input | Decision logic | Output |
|---|---|---|---|
| 1 | Experiencer=Patient and no associated allergy tag | The concept is related to the patient and no flag that it is an allergy | Use the Condition Resource |
| 2 | Negation | “Affirmed” means there is no negation | Three contexts to verify |
| 3 | Temporality | Apply rule for Recent | ClinicalStatus=Active |
| 4 | Certainty | Apply rule for value= Certain | VerificationStatus=Confirmed |
| 5 | Diagnostic Section Tag | Concept included in Diagnosis section | Category=Encounter-diagnosis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





