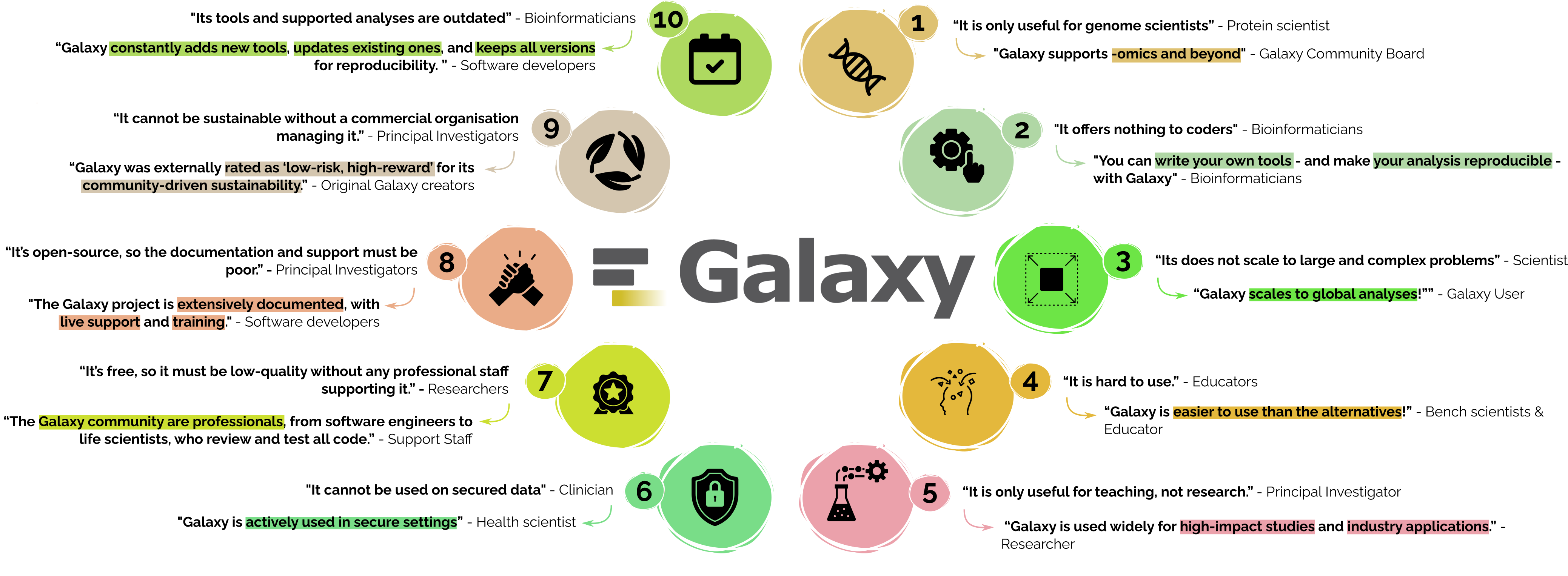
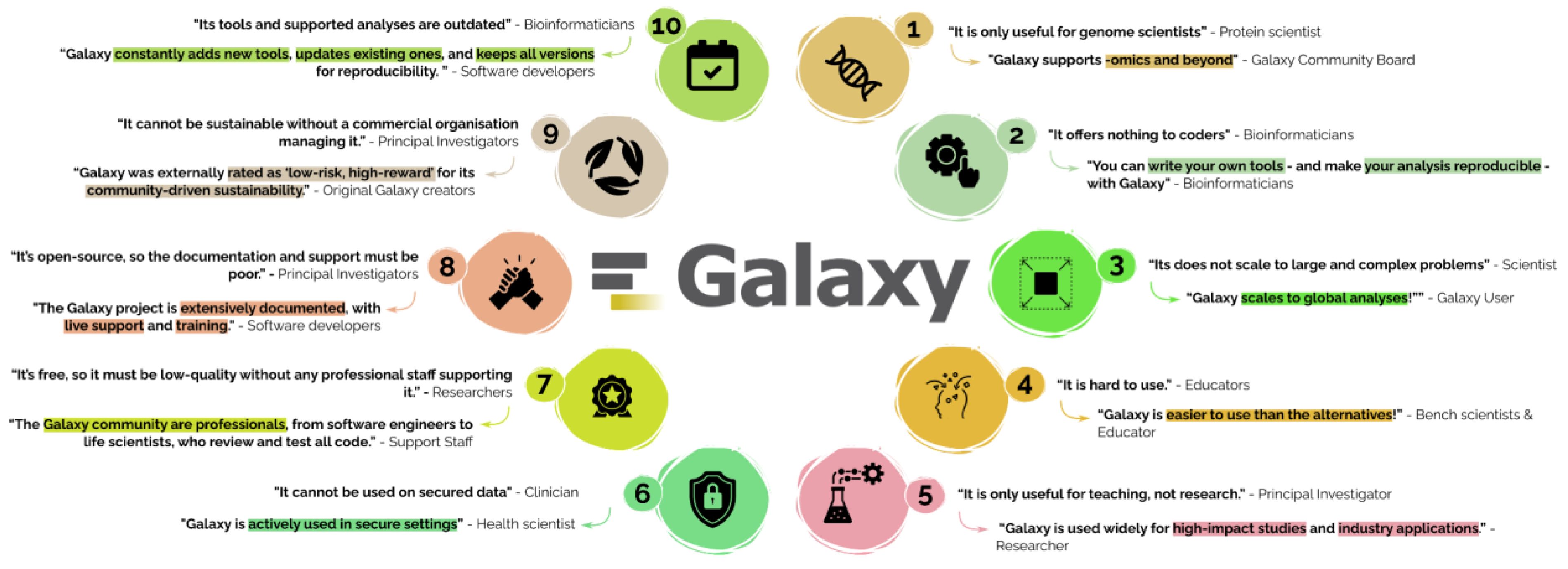
Misconceptions
We address common misconceptions and present the reality — highlighting relevant Galaxy features and proven results that directly refute these misunderstandings.
1: “It is only useful for genome scientists” - Protein scientist
“Galaxy supports -omics and beyond” - Galaxy Community Board
While Galaxy originated for genome analysis[
4] and has a long-established track record in that field[
5], its approach to visual programming and big data analysis extends beyond genomics or life sciences. Galaxy’s data-type agnostic architecture enables this broad applicability, flexibility in tool development, and curated interfaces.
Galaxy tool and workflow execution mechanisms are highly adaptable, relying on technologies such as Conda, Docker, Apptainer, and HPC schedulers like Slurm. These choices ensure compatibility with the computing environments used outside genomics. Galaxy also offers integration with interactive tools like Jupyter or RStudio, or any tool providing a user web interface (e.g., RShiny applications). Generalisable AI and machine learning resources[
6] further extend Galaxy’s capabilities.
For developers, new Galaxy tools can be created from any command-line software package. This flexibility allows Galaxy to accommodate diverse data types and analysis methods. By contributing tools to the Galaxy ToolShed (
https://toolshed.g2.bx.psu.edu/), tool developers benefit from increased visibility and discoverability of their tools within a global user community. Tools wrapped in Galaxy are not only accessible to users in the original domain but can also be reused in cross-disciplinary contexts, enabling new scientific applications and collaborations.
Due to the proliferation of domain-specific tools, Galaxy enables curation of domain-specific ‘flavours’, known as GalaxyLabs[
7] (sometimes referred to as subdomains and instances), with curated tool, workflow and resources lists for a given research field.
These features have resulted in Galaxy’s extensive datatype system, which already supports over 700 formats (
https://github.com/galaxyproject/galaxy/blob/dev/lib/galaxy/config/sample/datatypes_conf.xml.sample) and over 10,000 tools across various scientific domains. Disciplines such as proteomics[
8], single-cell[
9] and metabolomics[
10] are well established in Galaxy, while non-biological domains are rapidly growing, such as imaging, machine learning, natural language processing, ecology, climate, astronomy and physics - often supported by self-organised Special Interest Groups (
https://galaxyproject.org/community/sig/) and extensive training materials (
https://training.galaxyproject.org/). To support these communities, a large collection of GalaxyLabs (
https://galaxyproject.org/eu/subdomains/) have emerged, covering domains such as proteomics (
https://proteomics.usegalaxy.eu/), climate science (
https://climate.usegalaxy.eu/), imaging (
https://imaging.usegalaxy.eu/) and machine learning (
https://ml.usegalaxy.eu/). The European Galaxy project now maintains a list of use cases (
https://galaxyproject.eu/news?tag=UseCase) that ultimately illustrate the breadth of usage of this platform, with recent examples of astronomy (
https://galaxyproject.org/news/2025-06-11-voronoi-astronomy/), geospatial (
https://galaxyproject.org/news/2025-05-20-jupytergis/) and imaging (
https://galaxyproject.org/news/12-05-2025-galaxy-imaging-hackathon2025/) data analysis.
2: “It offers nothing to coders.” - Bioinformaticians
“You can write your own tools - and make your analysis reproducible - with Galaxy.” - Bioinformaticians
Software developers commonly prefer generating their own code rather than learning or depending on a system created by someone else, sometimes referred to as the ‘not invented here’ syndrome[
11]. As a result, research groups accumulate a plethora of often undocumented code, termed ‘in-house scripts’, which are usable to one person, who is usually on a fixed-term contract. Conversely, Galaxy brings the accumulated expertise of hundreds of different developers to drive versioned, documented, and reproducible analysis.
Galaxy analyses are easily shared with others via links. The powerful workflow manager allows metadata annotation to encourage FAIR workflows, which can be deposited (or retrieved) from WorkflowHub (
https://workflowhub.eu/). Execution is possible both through the web interface and the Galaxy API.
When an analysis has to be repeated (e.g., at a reviewer’s request), Galaxy allows for the exact environment to be reproduced, making this a straightforward task. Tools are installable with a single click. Dependency resolution is based on conda packages or containers to make deployment easier. Galaxy is cluster and cloud-agnostic, so it can utilise commonly used infrastructures that may be available, saving a scientist the need to learn all the quirks of cluster or Cloud APIs.
Finally, informaticians can write their own Galaxy tools (see Misconception #8), which then benefit from the embedded FAIR and reproducibility features.
The result is that complex pipelines can often be built much faster and with a markedly more reproducible output than a bespoke analysis could achieve - thanks to the decades of input from informaticians into the engineering of the Galaxy platform. There is a reason that data-heavy industries rely on workflow management systems[
12] - reproducibility matters.
3: “It does not scale to large and complex problems” - Scientist
“Galaxy scales to global analyses!” - Galaxy User
Galaxy computing power is commonly under-estimated - Galaxy has developed significantly over the past few years to allow for the massive scaling of datasets, analysis, and complexity.
The UseGalaxy.* instances offer ample computing power to perform data analysis, as evidenced by the availability of 9000+ CPU cores, 60+ TiB RAM, 35 GPUs, and 5+ PiB of storage on the European server alone. In addition, thanks to Pulsar (
https://pulsar.readthedocs.io/en/latest/), jobs can be sent to other places where the appropriate resources are available. On the usegalaxy.eu, .org, .org.au, and .fr instances, the platform can process more than 65,000 jobs per day.
Managing a long-term analysis with a complex experimental design can be challenging inside or outside Galaxy: naming outputs, provenance, and tracking metadata on the generated data. By design, following the FAIR principles, Galaxy traces the origin of output files back to the source data. Users can also set tags (conditions, strains, etc.) to propagate to the generated data[
13] (
Figure 1). Some tools can use tags to create contrast matrices for statistical analysis, for example[
14].
The Galaxy project supports automated data processing through its various tools and APIs. For example, the Bioblend API (bioblend.readthedocs.io/en/latest/), written in Python, enables programmatic interaction with a Galaxy instance. This capability is particularly useful for instance administration, but above all for automating processing: uploading data, launching tools and workflows, and retrieving results. This approach makes it possible to integrate Galaxy into Python scripts or offer layers on top of Galaxy, such as websites. It is also possible to run Galaxy jobs from the command line (CLI) using the planemo run tool (bioblend.readthedocs.io/en/latest/). Creating a workflow via the Galaxy user interface (tying boxes together with noodles) or importing one enables the job to be run locally or on a remote instance such as UseGalaxy.*. This means that data management, interaction with hardware, the computing environment, etc., is transparent to the user and can all be delegated to the Galaxy instance.
Galaxy’s ability to handle large datasets can be seen in its processing of COVID-19 data during the pandemic, where it has been able to analyse over 500,000 samples in near real time as the raw sequencing data was made publicly available (
infectious-diseases-toolkit.org/showcase/covid19-galaxy). Another example is the IRIDA project, built on top of Galaxy (
https://irida.ca/), or in the same principle, the ABRomics project, which aims to study and monitor antibiotic resistance in France (
abromics.fr/home/abromics-platform/). Even students are able to capitalise on the Galaxy scalability, as seen in a recent project analysing RNA in one million heart cells[
15].
4: “It’s hard to use.” - Educators
“Galaxy is easier to use than the alternatives!” - Bench scientist & educator
This misconception is relative. In comparing the complex analysis achieved with Galaxy to using simple interfaces such as the Google search engine, certainly Galaxy will be hard to use. However, when comparing Galaxy to the open-ended landscape of command line-based bioinformatics analysis, the Graphical User Interface of Galaxy assuredly lands on ‘easier to learn’. The Galaxy community achieves this usability via three mechanisms: standardised environments; a simple User Interface, and extensive training material.
First, a common barrier to any informatics analysis is inevitably the start-up: given any tutorial, workflow, or analysis framework, the challenges in re-creating an identical environment - including tool versions, packages, and even OS - prevent further analysis long before any particular non-intuitive feature of an interface can create frustration. By standardising these environments through web-browser-based access, Galaxy significantly reduces this initial barrier for users to get started.
Second, the Galaxy Community has a working group to improve the user interface and user experience (
https://galaxyproject.org/community/wg/). Community members as well as users can report errors via the Galaxy interface, post questions on the actively monitored help forums, or post issues via GitHub. All feedback is transformed into quick fixes or added to the Galaxy Roadmap. The change of the look and feel of the Galaxy webpage over the last 20 years highlights these efforts (
Figure 2).
Thirdly, Galaxy is extensively documented with a scalable training platform offering FAIR training material (see Misconception #8 for additional details).
This coordinated approach from site to support makes Galaxy relatively easy to use. Course trainers have reported that trainees learning the interface and analysis during the course successfully applied the analysis to their own datasets in the evening, demonstrating rapid up-skilling[
16]. Indeed, in one mixed-methods study evaluating the usability of Galaxy in clinical diagnostics, participants reported high satisfaction rates with the interface, in addition to its efficiency and accuracy[
17].
5: “It is only useful for teaching, not research.” - Principal Investigator
“Galaxy is used widely for high-impact studies and industry applications.” - Researcher
While it is true that many researchers’ first introduction to Galaxy is in a training environment, its capabilities extend far beyond teaching.
Galaxy scales to handle large-scale data analyses (see Misconception #3), making it a powerful tool for complex scientific research. Its emphasis on provenance and reproducibility ensures that workflows can be shared and re-run with the same input parameters, which is crucial for scientific rigour and collaboration.
The extensive toolset available in Galaxy supports various types of data analyses (see Misconception #1). These tools are continuously updated and expanded by a large and active community of users and developers, providing researchers with cutting-edge analytical capabilities (see Misconception #10).
Workflow managers and pipelines have become so complex that the ability for one research group or organisation to develop and maintain a system for the long term is long gone. By joining forces as an open source community, Galaxy has built a strong, resilient technical base, allowing data analysis to be undertaken easily at scale and reproduced (see Misconception #2).
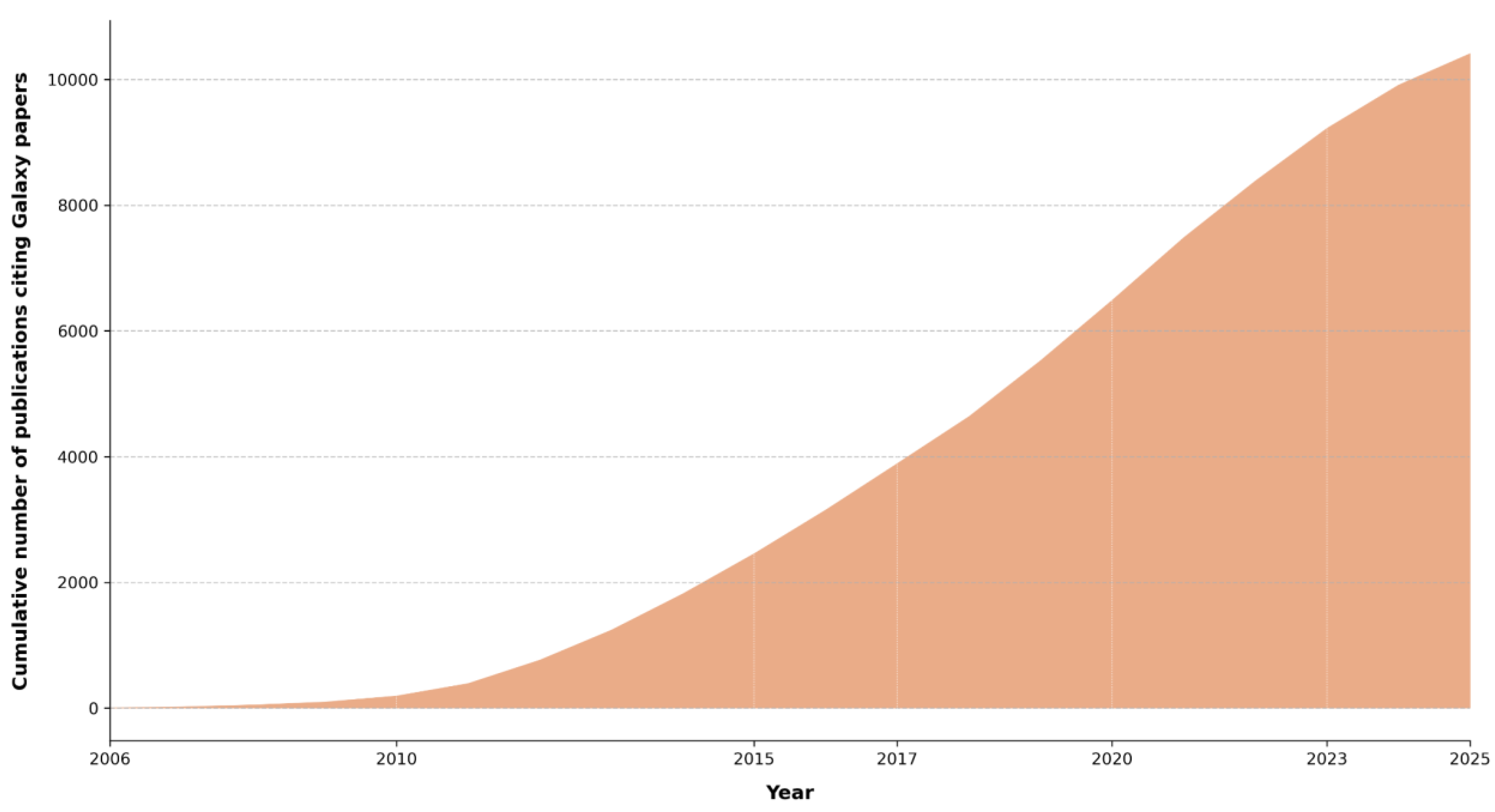
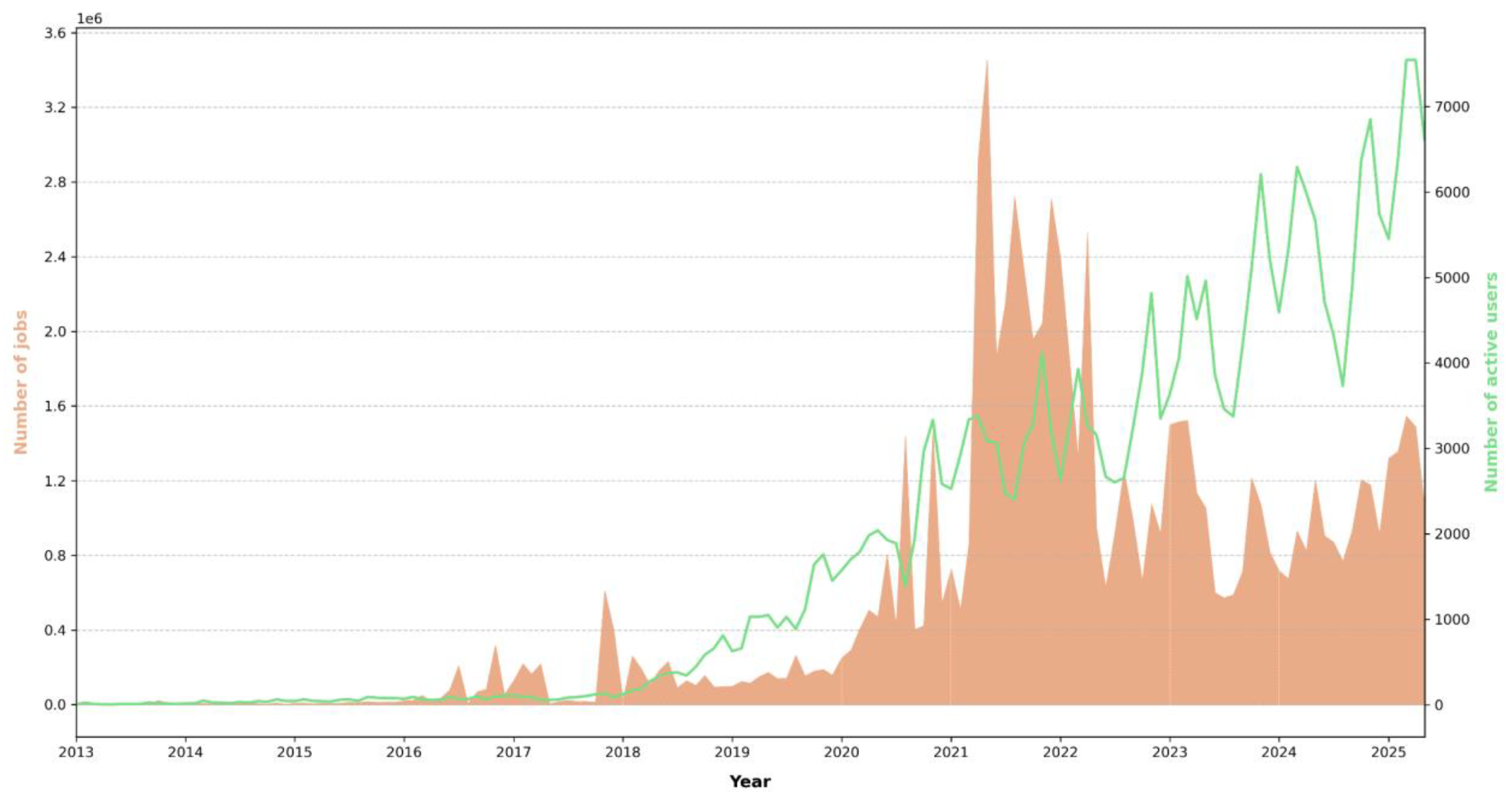
Galaxy’s utility in real-world scientific research is evident from its citation in numerous high-impact studies. Since 2016, the Galaxy Project has published biennial papers that are recommended as primary references. These publications, along with previous core works, have been cited 10,414 times (
Figure 3A), highlighting the platform’s significant impact on the scientific community. The user statistics, like the increase in the number of users and submitted jobs on the European Galaxy server alone (
Figure 3B), proves the popularity and usage of Galaxy.
Galaxy has been used in large-scale analyses, including significant projects such as COVID-19 research[
18,
19], the Vertebrate Genomes Project[
20] (VGP,
https://vertebrategenomesproject.org/), European Reference Genome Atlas (ERGA,
https://www.erga-biodiversity.eu/), the Earth Biogenome Project (
https://www.earthbiogenome.org/), BRC analytics (
https://brc-analytics.org/), the Human Cell Atlas[
21], ABRomics for antimicrobial resistance surveillance (
https://www.abromics.fr/), the Integrated Rapid Infectious Disease Analysis (IRIDA) for tracking infectious diseases in multiple countries (
https://irida.ca/), the EuroScienceGateway (ESG,
https://eosc.eu/eu-project/eurosciencegateway/) and many more. In particular, ESG nicely highlights Galaxy’s broad applicability across different scientific domains. It is also widely used by data-intensive organisations, such as the U.S. Food and Drug Administration for genomic epidemiology of foodborne pathogens with GalaxyTrackr[
22] and the Belgian national public health institute (Galaxy @Sciensano[
23]). In these cases, Galaxy servers are set up with configured solutions and toolsets for specific scientific fields. Galaxy is also the recommended platform for sharing reproducible analyses supporting scientific publications endorsed by the GigaScience journal, originating from developments within China Gene Bank (CGB) and Beijing Genome Informatics (BGI).
Galaxy’s impact extends beyond academia into the industry. Many companies recognise the value of Galaxy and actively seek professionals with Galaxy skills. Job listings that require Galaxy expertise can be found on the Galaxy Project’s careers page (
https://galaxyproject.org/careers/), demonstrating the platform’s relevance in the job market. Additionally, industry sponsors, such as Limagrain, support the annual Galaxy Community Conference, further highlighting the industry’s investment in and recognition of Galaxy’s capabilities.
6: “It cannot be used on secure data” - Clinician
“Galaxy is actively used in secure settings” - Health scientist
The increasing volume of sensitive biomedical and genomic data has necessitated secure computing environments that enable compliant, reproducible, and federated data analysis. Trusted Research Environments (TREs) and Secure Computing Environments (SCEs) ensure researchers can analyse sensitive datasets without compromising security or regulatory compliance. Galaxy has evolved into a robust framework for secure data analysis, integrating best practices in data governance, authentication, encryption, and federated computing.
Galaxy provides a flexible and scalable environment for analysing sensitive datasets, leveraging security-enhanced deployment models and access controls. Features such as Bring Your Own Compute (BYOC), Bring Your Own Storage (BYOS), Bring Your Own Data (BYOD), and deferred data enable secure data processing in Galaxy, incorporating integrations with storage services like Nextcloud, OwnCloud, Dropbox, Google Drive, and several others.
Galaxy supports federated data analysis by enabling users to integrate their compute resources, including HPC clusters, cloud environments, or on-premises servers. Storage solutions like S3 can be integrated, ensuring data remains within institutionally governed environments and reducing risks associated with external data transfers while maintaining compliance with local regulations. Security within Galaxy is enforced through role-based access control (RBAC), single sign-on (SSO), and identity federation protocols, including OpenID Connect and SAML. Additionally, Galaxy integrates with Life Science Login (LS Login), an authentication service from EOSC-Life, enabling secure authentication with institutional credentials while maintaining fine-grained control over access permissions.
Galaxy supports APIs such as the Data Repository Service (DRS), Beacon, Task Execution Service (TES), and Tool Registry Service (TRS) developed by the Global Alliance for Genomics and Health (GA4GH,
https://galaxyproject.org/ga4gh/), which promote interoperability and standardised genomic data exchange. This alignment advances medical research by enabling secure, federated analysis workflows.
These security features have led to a number of Galaxy use cases in secure settings.
The NHGRI Analysis, Visualisation, and Informatics Lab-space (AnVIL,
https://anvilproject.org/overview,
https://galaxyproject.org/use/anvil/)[
24] integrates Galaxy to provide secure access to large-scale genomic datasets under controlled data access policies, ensuring compliance while enabling analysis. Galaxy has been deployed in hospital and clinical research settings where sensitive patient genomic data is analyzed[
23,
25,
26]. By leveraging Galaxy’s authentication, encryption, and containerised execution features, these institutions can maintain full control over their datasets while enabling secure bioinformatics analysis.
Sciensano (Belgium’s national public health institute)[
23] operates a dedicated Galaxy instance for analysing whole genome sequencing (WGS) from Illumina and Oxford Nanopore data. The platform supports pathogen characterisation, outbreak detection, and diagnostics while maintaining traceability, reproducibility, and security.
For over five years, Assistance Publique–Hôpitaux de Paris (AP-HP) has used Galaxy in clinical diagnostics for more than 7,000 patient cases[
25]. Deployed on a secure server compliant with EN ISO15189:2012, the instance ensures reproducibility, traceability, secure data management, and integration of in-house tools for hereditary disease and cancer diagnostics.
Galaxy Europe is an open, domain-agnostic, publicly accessible science gateway and a federated environment supporting BYOC, BYOS, and BYOD features. It operates on an ISO 27001-certified infrastructure with encryption at rest and in transit, hosted at the University of Freiburg, Germany. The platform enables SSO and strict access control, integrating with multiple Authentication and Authorization Infrastructures (AAIs), including the German National Research Data Infrastructure (NFDI) and EGI. Galaxy Europe demonstrates Galaxy’s viability as a TRE, supporting secure, compliant, and scalable workflows across domains. Thousands of researchers actively use it to analyse datasets, from the humanities to human genomics to epidemiological surveillance, while ensuring privacy, data protection, and regulatory alignment. These examples demonstrate Galaxy’s handling of secure data.
7: “It’s free, so it must be low-quality without any professional staff supporting it.” - Researchers
“The Galaxy community are professionals, from software engineers to life scientists, who review and test all code.” - Support Staff
While it is true that much research software is untested, thrown online to support a publication, with poor - if any - documentation and no plans for support or sustainability[
27,
28], this is not the case with Galaxy. Through a combination of automation & testing support, community structure, and user engagement, the Galaxy community ensures that its product is stable and trustworthy.
Galaxy has a strong testing culture to ensure quality; the community’s dedication to testing is evident from thousands of automated tests, multiple testing frameworks and libraries, elaborate testing infrastructure, as well as formal testing policies and procedures applied to all stages of application development and system deployment. Galaxy uses numerous types of tests: unit-tests, tool-tests, framework-tests, selenium-tests, api tests, integration tests, performance tests, frontend and backend, and more. Each major Galaxy release goes through formal release testing. The testing process takes several weeks. Once any discovered bugs or issues have been addressed, the new release is deployed to usegalaxy.org, followed by extensive testing by the broader Galaxy team. Only after any issues have been resolved is the new release formally announced.
Galaxy resources, from the interface to training materials, are built by a combination of professional staff members in research, core services for institutes, or core services for national informatics infrastructures; researchers contributing significant amounts of time into Galaxy as part of their more applied research; and scientists contributing short-term support, such as for specific use cases or to deliver training. These individuals collectively are part of the Galaxy community, which is roughly divided into Working Groups and Special Interest Groups. Working Groups (
https://galaxyproject.org/community/wg/) include: Systems, Backend, User Interface/User Experience, Testing & Hardening, Workflows, and Outreach. Special Interest Groups range from scientific communities of practice (Single-cell, Microbiology, Digital Humanities, Ecology, Climate) to regional communities (India, Czech) to Service (Small Scale Admins, Support) (
https://galaxyproject.org/community/sig/). These groups work together to develop the Galaxy interface in line with user needs. While the use of Galaxy is unrestricted, it results from a distributed funding model (See Misconception #9) to support both part-time- and full-time staff, in addition to voluntary community contributions.
A particular effort is made within Galaxy to onboard and train new contributors, thereby ensuring quality in future developments and responsiveness to user needs (
https://galaxyproject.org/community/contributing/,
training-material). The Testing & Hardening Working Group even helps new contributors create tests for any contributions made.
8: “It’s open-source, so the documentation and support must be poor.” - Principal Investigators
“The Galaxy project is extensively documented, with live support and training.” - Software developers
Research software indeed is, in general, rarely well documented outside of IT fields[
29]. Such fields are full of ‘dead’ software, which is published once and unsupported[
27,
29]. In contrast, within the Galaxy community, there are real, conscientious people on the other end of bug reports, forums, Github repositories, and chat spaces who are committed to supporting Galaxy users. The Galaxy community achieves excellence in documentation and user support through a combination of: documentation standards; documentation automation; an active support team; and extensive synchronous and asynchronous training materials and delivery.
First, the Galaxy community shares a culture of promoting good documentation (
https://github.com/galaxyproject/galaxy/blob/dev/CONTRIBUTING.md#documentation), which is further reinforced during the code review process. Additionally, several aspects of Galaxy’s documentation are automatically generated from the code where feasible, and validation or linting checks are routinely performed to maintain consistency and technical correctness.
Second, Galaxy’s API is implemented using FastAPI with in-code documentation and OpenAPI specifications, enabling the automatic generation of detailed, interactive API documentation. This ensures that developers can reliably integrate and automate Galaxy through a fully documented programmatic interface. Tool developers benefit from Galaxy Tools and Language Server extensions for VSCode, providing auto-completion, validation, snippets, test generation, and embedded syntax highlighting.
Finally, to help propel researchers analysing data, developers wrapping tools, and systems administrators spinning up Galaxy servers, the Galaxy Training Network (
https://training.galaxyproject.org/) was established. Regular free, hands-on training sessions attract and support thousands of global participants, while annual global courses initiate annual updates to the milieu of tutorials and training resources documented across the Galaxy Training Network.
These practices result in excellent documentation of the Galaxy project across use levels, from administration to platform development to tool wrapping (
docs.galaxyproject.org). The Galaxy ecosystem offers Planemo, an SDK designed to streamline the wrapping of tools and integrating them with Galaxy. It is also comprehensively documented and encourages test-driven development best practices. BioBlend, the Python library for interacting with Galaxy’s API, offers both standard and object-oriented interfaces, and is well documented and structured to match Galaxy services. Frequent, detailed release notes (
https://docs.galaxyproject.org/en/master/releases/index.htm) document rapid development, while thousands of GitHub issues and pull requests provide in-depth design discussions.
The evolution of Galaxy’s code base, as well as Galaxy’s numerous satellite projects (such as Planemo[
30], Pulsar[
31], Bioblend[
32], etc.) that comprise the Galaxy ecosystem, is thoroughly documented in thousands of in-depth discussions in issues and pull requests by hundreds of contributors in public repositories on GitHub. The Galaxy help forum receives an average of 3,000-5,000 page views per month from individuals with an account, and an additional 1k per day of anonymous page views.
Finally, the Galaxy Training Network[
33] (
https://training.galaxyproject.org/) provides high-quality training material to empower scientists to analyse data[
33]. It contains over 400 tutorials, 200 videos, 450 contributors, 29 scientific topics, 2.4 million visitors since 2021 (
https://training.galaxyproject.org/training-material/stats/), a 100% FAIRness score, an excellent Accessibility compliance (
https://training.galaxyproject.org/training-material/about.html), and over 20,000 scientists trained since 2018 on the free Training Infrastructure as a Service (
https://usegalaxy.eu/tiaas/stats/). The Galaxy Training Network additionally provides extensive training on administrating Galaxy servers, contributing features, wrapping tools, and creating training materials, thereby ensuring a full circle of Galaxy documentation from user to contributor.
9: “It cannot be sustainable without a commercial organisation managing it.” - Principal Investigators
“Galaxy was externally rated as ‘low-risk, high-reward’ for its community-driven sustainability.” - Original Galaxy creators
Sustainability does not require corporate ownership: many successful open-source projects thrive through community-driven governance, diverse funding, and widespread adoption. For example, Apache (
https://www.apache.org/foundation/governance/) and Jupyter (
https://jupyter.org/governance/overview.html) are governed by non-profit organisations. NumPy, SciPy, and Pandas are maintained by a diverse group of contributors under the umbrella of the NumFOCUS non-profit, which supports community governance and fundraising. These examples demonstrate that open, transparent, and inclusive governance can foster both longevity and innovation. In fact, commercial software can be riskier, as companies may discontinue products if they are no longer profitable, leaving users without support or access.
Galaxy ensures sustainability through its forward-thinking and adaptive governance and administrative structures (
https://galaxyproject.org/community/governance/). This governance framework includes dedicated working groups (
https://galaxyproject.org/community/wg/) focused on technical development, training, and outreach, as well as a community board (
https://galaxyproject.org/community/governance/gcb/) representing diverse scientific domains. Additionally, Galaxy maintains published technical development and strategic roadmaps (
https://galaxyproject.org/roadmap/), ensuring transparency and a clear vision for the future. Financially, Galaxy benefits from a diverse and distributed funding model across continents, including research grants, institutional support, and industry collaborations. Unlike commercial software, which can be abruptly discontinued if unprofitable, Galaxy’s open-source nature guarantees continuous development, long-term accessibility, and community-driven support.
This collaborative community model has made Galaxy one of the most active open-source projects, placing it in the top 2% on Open Hub (
https://openhub.net/p/galaxybx/factoids#FactoidTeamSizeVeryLarge). What started as an academic tool has evolved into a mature, global collaboration with a solid engineering foundation, robust governance, and mechanisms to manage personnel and technological evolution. Committed to sustainability, the Galaxy community requested an external evaluation by the EGI, where Galaxy was ultimately described as “low-risk, high-reward investment in the future of data-driven research”[
34].
10: “Its tools and supported analyses are outdated” - Bioinformaticians
“Galaxy constantly adds new tools, updates existing ones, and keeps all versions for reproducibility. “ - Software developers
Bioinformatics tool quality is notoriously poor - with scientists often creating bespoke code on short-term contracts, then abandoning it for other roles[
27,
28]. Indeed, scientific tools and workflows are often difficult to keep up to date, as they are typically maintained by researchers rather than professional developers and may not follow standard software development practices. This issue is further compounded by the slow release cycles of repositories like Bioconductor. Galaxy ensures tool maintenance by open-source structure, semi-automation, integrations, and community responsibility.
Galaxy tools are developed and maintained by the global Galaxy community and groups such as the Intergalactic Utilities Commission (IUC) (
https://galaxyproject.org/iuc/), with their source code hosted in open-source, publicly available GitHub repositories.
Galaxy’s semi-automatic update infrastructure helps keep tools as current as possible while balancing stability and reproducibility. Rather than relying solely on automated updates, which can introduce breaking changes, Galaxy combines automation with manual oversight to ensure controlled and reliable updates. This process is powered by GitHub Continuous Integration (CI) pipelines and Planemo[
30], the Galaxy Software Development Kit.
A critical component of this system is its integration with Bioconda and BioContainers, which manage dependencies and allow tools to be installed in isolated environments. The benefits of this community engagement extend beyond Galaxy, as workflow management systems like Nextflow and Snakemake also rely on Bioconda and BioContainers for tool packaging.
When these dependencies are updated, tools maintained by IUC or other community tool GitHub repositories undergo a semi-automated updating process. Once approved by a community member, the updated tool becomes publicly available on ToolShed and is automatically updated on the major Galaxy servers where they are installed. This process ensures that users have access to the latest versions of tools while retaining legacy versions to support reproducibility and maintain standardised workflows, enabling users to repeat analyses even as tools evolve. However, some tools cannot be updated automatically when new features alter their logic or outputs. In these cases, Galaxy tool updates depend on community contributions and require expert reviews to ensure quality and correctness.
Similar to tools, workflows offered by the Intergalactic Workflow Commission (IWC) (
https://github.com/galaxyproject/iwc) are stored in the IWC GitHub repository and supported by a semi-automated updating system. When a Galaxy tool used in the workflow is updated, a continuous integration pipeline tests the workflow with the updated tool version. Once a community member approves, the updated workflow becomes publicly available on WorkflowHub, Dockstore, and the major Galaxy instances.
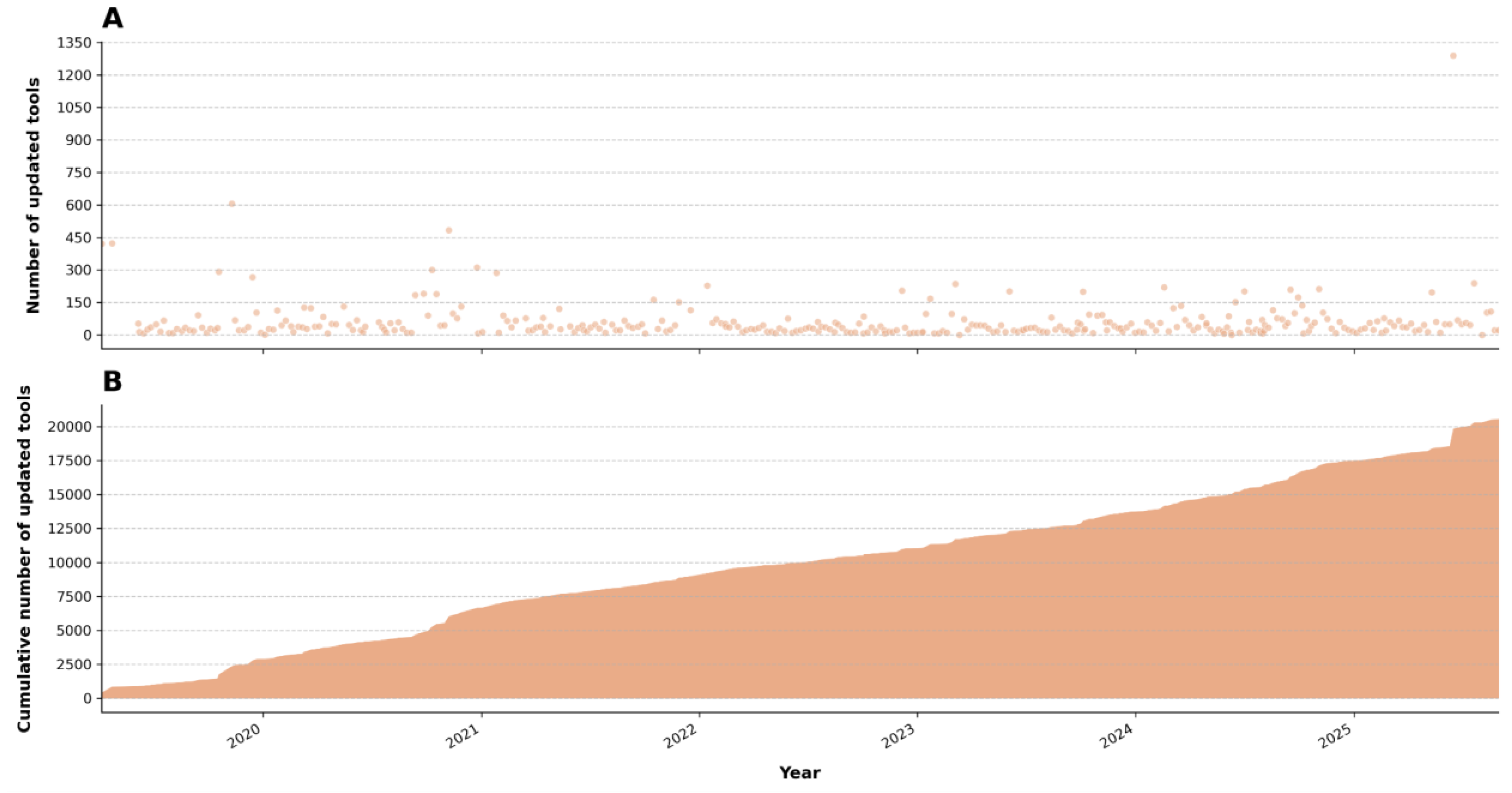
The collective efforts of these community-driven initiatives—with 277 contributors for tools and 50 for workflows—are instrumental in maintaining the functionality of tools (
Figure 4) and workflows within the Galaxy ecosystem. However, these efforts are not without their limitations, as they are constrained by the availability of human resources. Outdated tools are a ubiquitous issue in computational pipelines, regardless of whether they are built with a workflow editor or custom command-line solutions. Therefore, sustaining an active and engaged community that continually adapts to advancements in the field is essential for addressing this ongoing challenge.