Submitted:
30 August 2025
Posted:
01 September 2025
You are already at the latest version
Abstract
Background/Objectives: Autism Spectrum Disorder (ASD) is a complex neurodevelopmental condition that remains challenging to diagnose using traditional clinical methods. Recent advances in artificial intelligence, particularly transformer-based deep learning models, have shown considerable potential for improving diagnostic accuracy in neuroimaging applications. This study aims to develop and evaluate a transformer-based framework for automated ASD detection using structural and functional brain MRI data. Methods: We developed a deep learning framework using neuroimaging data comprising structural MRI (sMRI) and functional MRI (fMRI) from the ABIDE repository. Each modality was analyzed independently through transfer learning with transformer-based pretrained architectures, including Vision Transformer (ViT), MaxViT, and Transformer-in-Transformer (TNT), fine-tuned for binary classification between ASD and typically developing controls. Data augmentation techniques were applied to address the limited sample size. Model interpretability was achieved using SHAP to identify the influential brain regions that contribute to classification decisions. Results: Our approach significantly outperformed traditional CNN-based methods and state-of-the-art baseline approaches across all evaluation metrics. The MaxViT model achieved the highest performance on sMRI (98.51% accuracy and F1-score), while both TNT and MaxViT reached 98.42% accuracy and F1-score on fMRI. SHAP analysis provided clinically relevant insights into brain regions most associated with ASD classification. Conclusions: These results demonstrate that transformer-based models, coupled with explainable AI, can deliver accurate and interpretable ASD detection from neuroimaging data. These findings highlight the potential of explainable DL frameworks to assist clinicians in diagnosing ASD and provide valuable insights into associated brain abnormalities.

Keywords:
1. Introduction
- End-to-End AI Pipeline: Proposing a unified framework that integrates preprocessing, data augmentation, feature extraction, classification, and explainability into a single streamlined workflow for ASD detection.
- Comparative Modality Evaluation: Conducting a systemic comparison of sMRI and fMRI modalities, demonstrating that fMRI alone yields superior diagnostic performance compared to sMRI.
- Leveraging Transformer-based Models: Exploring the performance of advanced transformer-based pretrained architectures to capture both complex spatial patterns and long-range dependencies in neuroimaging data.
- Model Interpretability: Implementing SHAP-based explainable AI techniques to provide transparent, clinically relevant interpretability insights into model predictions, and ensuring clinical trust.
2. Related Work
2.1. Overview of AI Paradigms

2.2. MRI-Based Approaches
2.2.1. fMRI-Based Approaches
2.2.2. sMRI-Based Approaches
2.2.3. Multimodal MRI Approaches (sMRI + fMRI)
2.3. Literature Gaps
3. Materials
- ABIDE I includes data from 539 individuals with ASD and 573 TC participants, aged 7-64 years, collected across 17 international sites.
- ABIDE II extends the first dataset with an additional 521 ASD and 593 TC participants, aged 5-64 years, collected from 19 sites worldwide.
- It contains one of the largest subject pools with 79 ASD and 105 TC participants,
- The imaging data undergoes reliable preprocessing steps and exhibits consistency in imaging protocols, making them suitable for model training and evaluation. Using data from a single, high-quality site reduces variability caused by site-dependent confounding factors, which is particularly important for training DL models and ensuring evaluation reliability.
4. Methods
4.1. Proposed Method
- Data Acquisition: sMRI and fMRI scans were obtained from the preprocessed ABIDE repository.
- Data Preprocessing: Multiple preprocessing operations were performed separately on the sMRI and fMRI, including slicing, normalization, resizing, and channel conversion. For fMRI, 3D volumes were converted into 2D slices to match the input requirements of pretrained 2D models.
- Data Augmentation: Classical augmentation techniques were applied to increase the diversity and volume of the training data and mitigate overfitting due to limited dataset size.
- Feature Extraction with Pretrained Models: Instead of adapting 3D models, we extracted features by applying state-of-the-art pretrained models directly to 2D slices of fMRI and sMRI data. Separate experiments were conducted for each modality to evaluate their individual effectiveness.
- Classification: The extracted features were passed through fully connected layers to classify subjects as either ASD or TC, with performance evaluated for each modality.
- Explainable AI (XAI): XAI techniques were employed to interpret the model’s predictions by highlighting the most influential image regions, thereby supporting model transparency and clinical relevance.
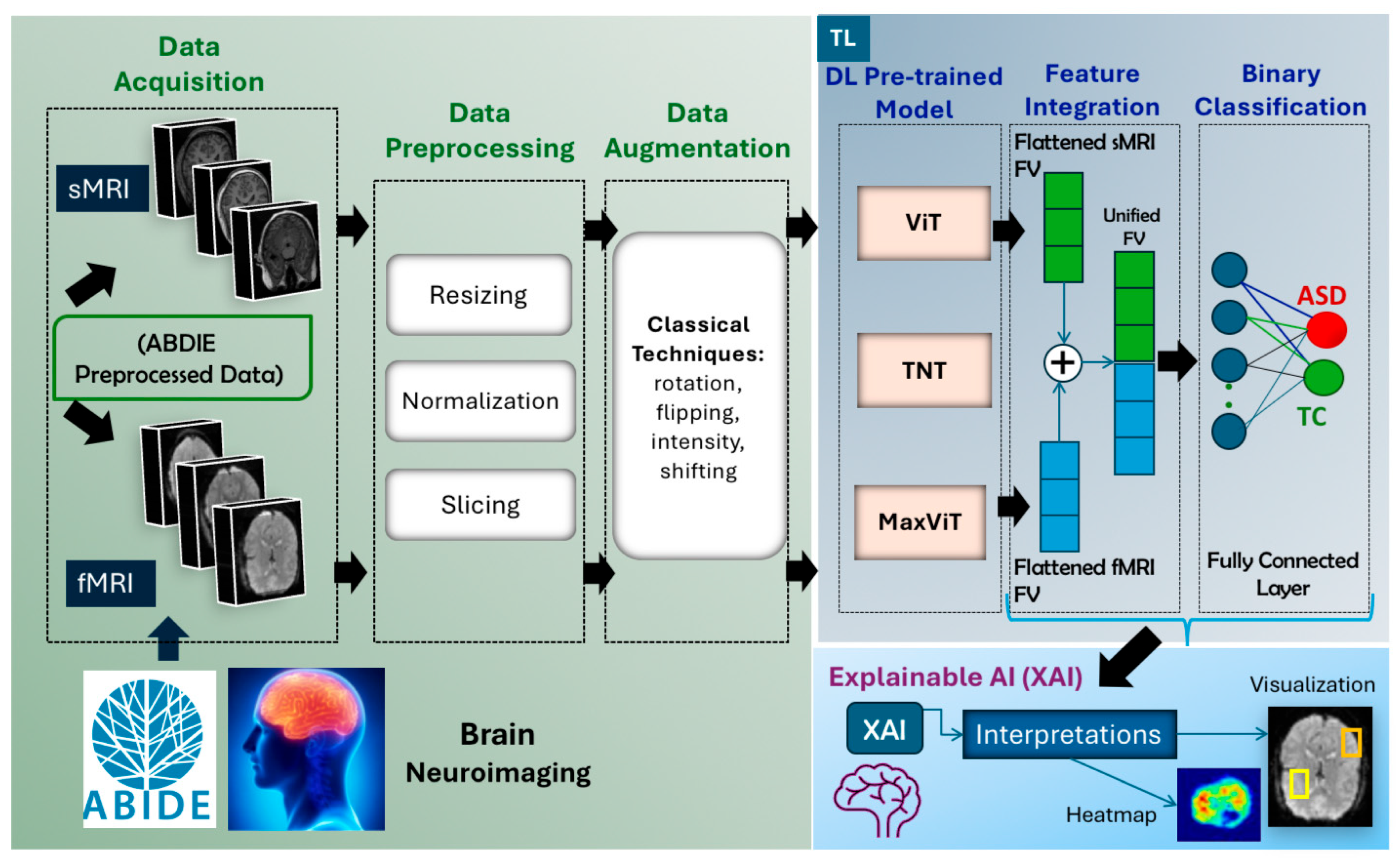
4.2. Data Preprocessing
4.2.1. fMRI Data Preprocessing
- 4D to 3D Conversion: To enable compatibility with 2D pretrained models while maintaining spatial consistency, the 4D fMRI data were converted into 3D static volumes through temporal mean calculation. In particular, the voxel-wise mean intensity was computed across all 176 time points, resulting in a 3D static volume of size (61 × 73 × 61) representing average voxel-wise activation.
- Fuzzy C-Means (FCM) Volume-level Normalization: The 3D volumes were normalized using FCM segmentation, which separates tissues into distinct regions, such as gray matter, white matter, and cerebrospinal fluid. This approach enhances tissue-specific representation, improving downstream feature extraction during model training for slice-based modeling using 2D pretrained architectures
- Slice Extraction: The normalized 3D volumes (61 × 73 × 61) were decomposed into 2D slices to analyze certain regions of interest in the MRI image while reducing computational complexity. To ensure consistent representation of brain structures, slices were extracted from the central positions of each axis: axial plane → mid_z, sagittal plane → mid_x, and coronal plane → mid_y. This strategy captures the most representative cross-sectional views of the brain.
- Slice-level Normalization: Min-Max Normalization was applied to each 2D slice, scaling pixel intensities to the range [0,255] using the formula:where: I(x, y) is the original pixel intensity at position (x, y), and and are the minimum and maximum intensity values in the image, respectively. This step standardizes the image’s intensity scale, preparing it for digital display and subsequent processing while retaining the relative distribution of the original intensities.
- Resizing: All fMRI slices were resized from their original dimensions to (pixels to meet the input requirements of the selected pretrained models. The step standardizes image resolution across the dataset.
- Channel Conversion: Since the pretrained models require three-channel RGB input, the grayscale slices were transformed into three-channel images by duplicating the single-channel intensity values across all three channels. The resulting input shape for each slice became (3 × 224 × 224).
4.2.2. sMRI Data Preprocessing
- Slicing: Unlike fMRI, sMRI volumes are already in 3D format. Therefore, 2D slices were directly extracted from the volumes along the axial, coronal, and sagittal planes, following the same central slice selection strategy as fMRI (mid_z, mid_x, mid_y) to obtain consistent representations of brain structures.
- Slice-Level Normalization: As with fMRI slices, Min-Max normalization was applied to scale pixel intensities into a uniform range of [0,255] using the same formula provided above.
- Resizing: Each sMRI slice was resized from its original resolutions to a standard size of pixels to meet the input size requirements of the selected pretrained transformer models.
- Channel Conversion: The grayscale sMRI slices were transformed into three-channel RGB images following the same step applied to fMRI, resulting in an input shape of (3 224 224).
4.3. Pretrained Models and Feature Extraction
4.3.1. Vision Transformers (ViT)
4.3.2. Transformer-in-Transformer (TNT) Applied to fMRI and sMRI
- The inner block models local-level features within each image patch, such as the connectivity inside each brain region (i.e., intra-regional connectivity).
- The outer block captures global relationships between patches, such as connectivity between different brain regions (i.e., inter-regional connectivity across the brain).
- In this study, we applied TNT in two experimental settings:
- 2D fMRI slices: TNT’s ability to model both spatial and temporal dependencies (i.e., dual-level feature learning) makes it effective for analyzing and understanding functional connectivity between brain regions is essential for detecting ASD-related abnormalities.
- 2D sMRI slices: TNT extracts fine-grained structural features from high-resolution anatomical scans, capturing both local tissue details and global anatomical relationships

4.3.3. Maximizing Vision Transformer (MaxViT)- Applied to sMRI and fMRI
- Stem Stage (S0): Two convolutional layers with kernel size of 3×3 to downsample the input.
-
Core Stages S1–S4: Each stage contains a MaxViT block composed of three key modules:
-
Mobile inverted residual bottleneck convolution (MBConv) Module:MBConv expands, processes, and compresses features using depth wise convolutions and Squeeze-and-Excitation mechanisms by computing global statistics through average pooling and channel-wise scaling using fully connected layers.
-
Block Attention Module:The feature map is partitioned into non-overlapping windows, where self-attention mechanisms capture localized temporal and spatial interactions within each window. A feedforward network is then applied to introduce non-linear transformations to further refine the localized representations.
-
Grid Attention Module:Distributes attention across a global grid structure, enabling the model to capture long-range interdependencies.
-
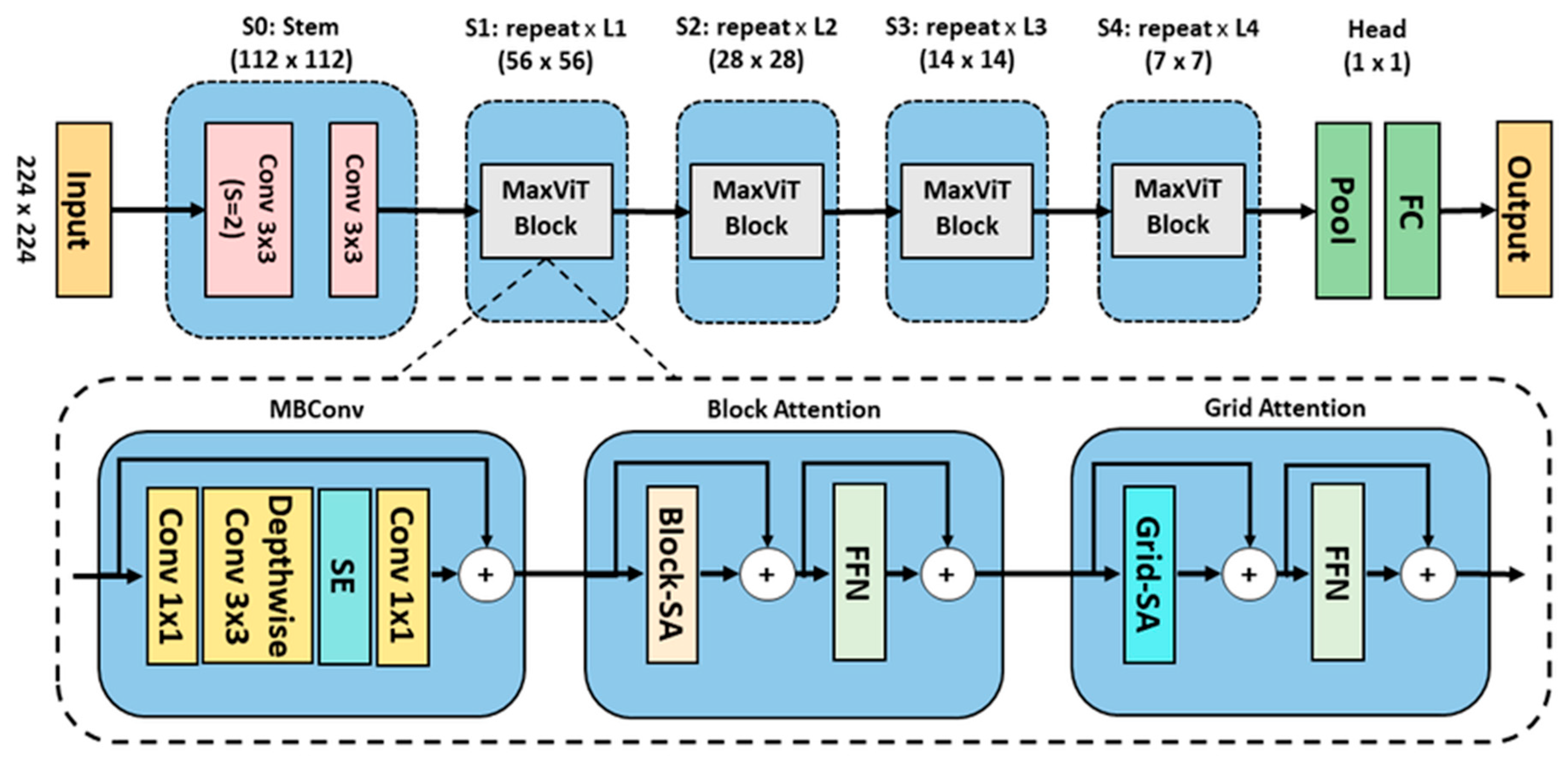
4.4. Transfer Learning
4.4.1. Fine Tuning
- Adapting the Input Layer: We prepared the MRI slices to match the input requirements of the selected models. The first convolutional layer was adjusted accordingly to accept the input format.
- Freezing Base Layers: Initially, the convolutional layers were frozen to preserve low-level features such as edges, textures, and patterns that transfer across domains. The freezing step prevents unnecessary parameter updates during backpropagation.
- Customizing the Fully Connected (FC) layers: We replaced the original FC layers with new layers tailored to MRI slice classification. We also add dense layers with dropout regularization to prevent overfitting.
4.4.2. Classification Model
4.5. Explainable AI (XAI) Technique
5. Experiments and Evaluation
5.1. Training and Testing Strategy
5.2. Implementation Details
- Scikit-learn for stratified data splitting and performance evaluation.
- Hugging Face Transformers and Timm (PyTorch Image Models) for loading and fine tuning pretrained transformer models using timm.create_model.
- Nibabel and Nilearn for reading and processing neuroimaging data in NIfTI format
- PIL and NumPy for image manipulation and preprocessing.
- Intensity-normalization (FCM-based) for standardizing voxel intensities across MR images.
- SHAP for model interpretability and explainability.
5.3. Evaluation Measures
5.4. Experimental Protocol
6. Results and Discussion
6.1. Prediction Performance Using 2D fMRI Slices
- ViT (baseline): Although it achieved the lowest results among the three models, its performance remains remarkably strong, obtaining 98.03% accuracy and 98.02% F1-score. Its attention-based mechanism effectively captured spatial relationships across image patches, making it a solid benchmark for comparison.
- TNT achieved the best and most powerful performance, leveraging inner and outer attention mechanisms within and between image patches to capture fine-grained local features.
- MaxViT, our focus model, matched TNT’s performance, reaching 98.42% accuracy and an F1-score. This outstanding performance is due to its hierarchical integration of multi-scale and multi-axis attention.
6.2. Prediction Performance Using 2D sMRI Slices
- ViT (baseline): uses global attention across image patches, which helps capture overall spatial patterns. However, this approach may overlook finer local details that are important for analyzing brain structure. Consequently, it achieved moderate performance with 81.34% accuracy and 80.81% F1-score.
- TNT: incorporates attention mechanism within and between patches, allowing it to capture more detailed structural features. It performed second best, improving over ViT by around 10% in accuracy and 11% in F1-score, reaching 91.62% accuracy and 91.65% F1-score.
- MaxViT: once again, achieved the best performance by combining local and global attention in a hierarchical structure. This design enabled the model to deliver superior accuracy (98.51%), high F1-score (98.51%), and a very low loss (0.0409).
6.3. Comparison with the State-of-the-Art Methods
6.4. Explainable AI Results and Findings
- 1. SHAP Value Colors Interpretation:
- Red regions indicate positive SHAP values. These features contribute strongly to the predicted class (ASD or TC).
- Blue regions represent negative SHAP values. These features reduce the model’s confidence in the prediction.
- White or neutral regions have minimal influence.
- 2. Anatomical View Insights:
- Sagittal view (side): In ASD cases, the activity was stronger on one side of the brain, especially in areas related to memory and social understanding. In contrast, in the TC cases the activity appeared more balanced and spread across both sides.
- Axial view (top-down): In ASD cases, strong activations appear in the frontal and lateral regions, linked to executive function and cognitive control. In TC cases, activations are more evenly distributed across hemispheres, reflecting balanced neural activity.
- Coronal view (front-facing): In ASD cases, there was stronger activity in the middle and lower parts of the brain, which may affect movement and certain cognitive functions. In TC cases, the model highlighted contributions on both sides, showing patterns linked to typical memory and motor skills.
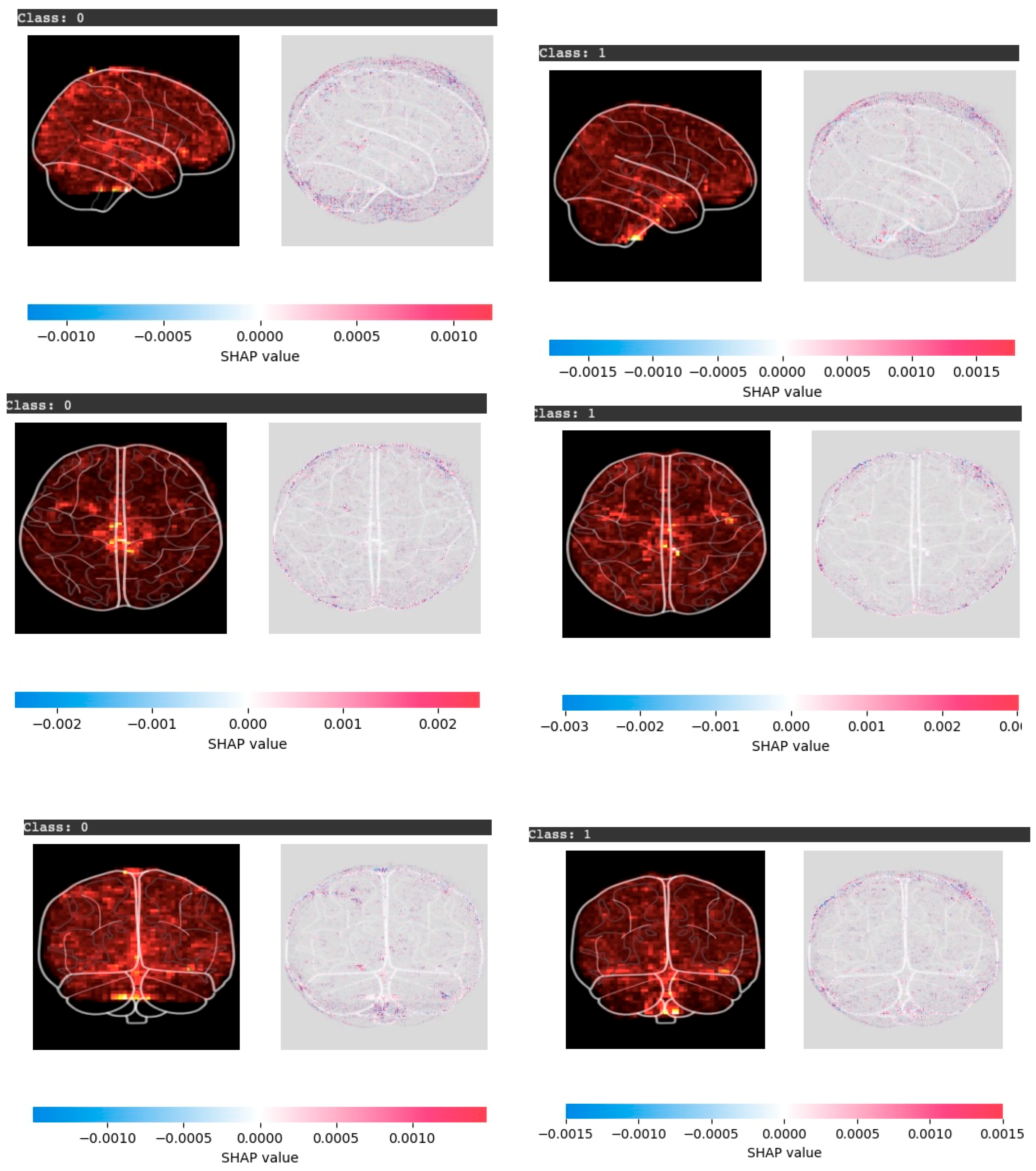
6.5. Limitations
7. Conclusion
- Apply data-level fusion by combining MRI modalities with phenotypic data to further boost prediction accuracy.
- Utilize GAN-based augmentation for generating synthetic samples and to explore pretrained models trained on large neuroimaging datasets to improve generalizability.
- Investigate more efficient architectures with less computational complexity, such as DeiT (Data-Efficient Image Transformers).
- Finally, incorporate attention-based XAI techniques, such as attention maps, to further improve model transparency.
Author Contributions
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ASD | Autism Spectrum Disorder |
| fMRI | Functional magnetic resonance imaging |
| sMRI | Structural magnetic resonance imaging |
| WHO | World Health Organization |
| ADHD | Attention deficit hyperactivity disorder |
| EEG | Electroencephalography |
| BOLD | Blood-oxygen-level-dependent |
| TC | Typically controls |
| DMN | Default mode network |
| ROI | Regions of Interest |
| ML | Machine learning |
| DL | Deep learning |
| CNNs | Convolutional neural networks |
| XAI | Explainable artificial intelligence |
| TL | Transfer learning |
| SVMs | Support vector machines |
| ANNs | Artificial neural networks |
| REF | Recursive Feature Elimination |
| CPAC | Configurable pipeline for the analysis of connectomes |
| ABIDE | Autism brain imaging data exchange |
| CCS | Connectome computation system |
| MLPs | Multilayer perceptrons |
| SLP | Single layer perceptron |
| PCC | Pearson’s correlation coefficient |
| DCNN | Deep convolutional neural networks |
| ResNet | Residual network |
| Grad-CAM | Gradient-weighted class activation mapping |
| NIfTI | Neuroimaging informatics technology initiative |
| FCM | Fuzzy C-means |
| ViT | Vision transformer |
| TNT | Transformer in transformer |
| MaxViT | Maximizing vision transformer |
| SHAP | Shapley additive explanations |
| ACC | Accuracy |
| BCE | Binary cross entropy |
| DeiT | Data efficient image transformers |
References
- Hirota, T.; King, B. Autism Spectrum Disorder: A Review. JAMA 2023, 329, 157–168.
- World Health Organization Available online: https://www.who.int/news-room/fact-sheets/detail/autism-spectrum-disorders.
- Rogers, S.J.; Vismara, L.A.; Dawson, G. Coaching Parents of Young Children with Autism: Promoting Connection, Communication, and Learning; Guilford Publications, 2021; ISBN 9781462545728.
- Bougeard, C.; Picarel-Blanchot, F.; Schmid, R.; Campbell, R.; Buitelaar, J. Prevalence of Autism Spectrum Disorder and Co-Morbidities in Children and Adolescents: A Systematic Literature Review. Focus (Am Psychiatr Publ) 2024, 22, 212–228. [CrossRef]
- Hodges, H.; Fealko, C.; Soares, N. Autism Spectrum Disorder: Definition, Epidemiology, Causes, and Clinical Evaluation. Transl. Pediatr. 2020, 9, S55–S65. [CrossRef]
- Gelbar, N.W. Adolescents with Autism Spectrum Disorder: A Clinical Handbook; Oxford University Press, 2018; ISBN 9780190624828.
- Flickr, F. us on What Are the Treatments for Autism? Available online: https://www.nichd.nih.gov/health/topics/autism/conditioninfo/treatments (accessed on 20 October 2024).
- Ahmed, Z.A.T.; Albalawi, E.; Aldhyani, T.H.H.; Jadhav, M.E.; Janrao, P.; Obeidat, M.R.M. Applying Eye Tracking with Deep Learning Techniques for Early-Stage Detection of Autism Spectrum Disorders. Data (Basel) 2023, 8, 168. [CrossRef]
- Taha Ahmed, Z.A.; Jadhav, M.E. A Review of Early Detection of Autism Based on Eye-Tracking and Sensing Technology. In Proceedings of the 2020 International Conference on Inventive Computation Technologies (ICICT); IEEE, February 2020; pp. 160–166.
- Cilia, F.; Carette, R.; Elbattah, M.; Dequen, G.; Guérin, J.-L.; Bosche, J.; Vandromme, L.; Le Driant, B. Computer-Aided Screening of Autism Spectrum Disorder: Eye-Tracking Study Using Data Visualization and Deep Learning. JMIR Hum. Factors 2021, 8, e27706. [CrossRef]
- Awatramani, J.; Hasteer, N. Facial Expression Recognition Using Deep Learning for Children with Autism Spectrum Disorder. In Proceedings of the 2020 IEEE 5th International Conference on Computing Communication and Automation (ICCCA); IEEE, October 30 2020; pp. 35–39.
- Derbali, M.; Jarrah, M.; Randhawa, P. Autism Spectrum Disorder Detection: Video Games Based Facial Expression Diagnosis Using Deep Learning. Int. J. Adv. Comput. Sci. Appl. 2023, 14, . [CrossRef]
- Thapaliya, S.; Jayarathna, S.; Jaime, M. Evaluating the EEG and Eye Movements for Autism Spectrum Disorder. In Proceedings of the 2018 IEEE International Conference on Big Data (Big Data); IEEE, December 2018; pp. 2328–2336.
- Ibrahim, S.; Djemal, R.; Alsuwailem, A. Electroencephalography (EEG) Signal Processing for Epilepsy and Autism Spectrum Disorder Diagnosis. Biocybern. Biomed. Eng. 2018, 38, 16–26. [CrossRef]
- Bosl, W.J.; Tager-Flusberg, H.; Nelson, C.A. EEG Analytics for Early Detection of Autism Spectrum Disorder: A Data-Driven Approach. Sci. Rep. 2018, 8, 6828. [CrossRef]
- Sinha, T.; Munot, M.V.; Sreemathy, R. An Efficient Approach for Detection of Autism Spectrum Disorder Using Electroencephalography Signal. IETE J. Res. 2022, 68, 824–832. [CrossRef]
- Hashemian, M.; Pourghassem, H. Decision-Level Fusion-Based Structure of Autism Diagnosis Using Interpretation of EEG Signals Related to Facial Expression Modes. Neurophysiology 2017, 49, 59–71. [CrossRef]
- Hassouneh, A.; Mutawa, A.M.; Murugappan, M. Development of a Real-Time Emotion Recognition System Using Facial Expressions and EEG Based on Machine Learning and Deep Neural Network Methods. Inform. Med. Unlocked 2020, 20, 100372. [CrossRef]
- Mehdizadehfar, V.; Ghassemi, F.; Fallah, A.; Pouretemad, H. EEG Study of Facial Emotion Recognition in the Fathers of Autistic Children. Biomed. Signal Process. Control 2020, 56, 101721. [CrossRef]
- Feng, M.; Xu, J. Detection of ASD Children through Deep-Learning Application of FMRI. Children 2023, 10, . [CrossRef]
- Suri, J.; El-Baz, A.S. Neural Engineering Techniques for Autism Spectrum Disorder, Volume 2: Diagnosis and Clinical Analysis; Academic Press, 2022; ISBN 9780128244227.
- Rakić, M.; Cabezas, M.; Kushibar, K.; Oliver, A.; Lladó, X. Improving the Detection of Autism Spectrum Disorder by Combining Structural and Functional MRI Information. Neuroimage Clin 2020, 25, 102181. [CrossRef]
- Koc, E.; Kalkan, H.; Bilgen, S. Autism Spectrum Disorder Detection by Hybrid Convolutional Recurrent Neural Networks from Structural and Resting State Functional MRI Images. Autism Res. Treat. 2023, 2023, 4136087. [CrossRef]
- Yang, X.; Paul; Zhang, N. A Deep Neural Network Study of the ABIDE Repository on Autism Spectrum Classification. Int. J. Adv. Comput. Sci. Appl. 2020, 11, . [CrossRef]
- Rane, P.; Cochran, D.; Hodge, S.M.; Haselgrove, C.; Kennedy, D.N.; Frazier, J.A. Connectivity in Autism: A Review of MRI Connectivity Studies. Harv. Rev. Psychiatry 2015, 23, 223–244.
- Riva, D.; Bulgheroni, S.; Zappella, M. Neurobiology, Diagnosis and Treatment in Autism: An Update; John Libbey Eurotext, 2013; ISBN 9782742008360.
- Blackmon, K.; Ben-Avi, E.; Wang, X.; Pardoe, H.R.; Di Martino, A.; Halgren, E.; Devinsky, O.; Thesen, T.; Kuzniecky, R. Periventricular White Matter Abnormalities and Restricted Repetitive Behavior in Autism Spectrum Disorder. NeuroImage Clin. 2016, 10, 36–45. [CrossRef]
- Alamro, H.; Thafar, M.A.; Albaradei, S.; Gojobori, T.; Essack, M.; Gao, X. Exploiting Machine Learning Models to Identify Novel Alzheimer’s Disease Biomarkers and Potential Targets. 2023, 13, 4979. [CrossRef]
- Swarnkar, S.K.; Guru, A.; Chhabra, G.S.; Devarajan, H.R. Artificial Intelligence Revolutionizing Cancer Care: Precision Diagnosis and Patient-Centric Healthcare; CRC Press, 2025; ISBN 9781040271230.
- Zhang, H.-Q.; Arif, M.; Thafar, M.A.; Albaradei, S.; Cai, P.; Zhang, Y.; Tang, H.; Lin, H. PMPred-AE: A Computational Model for the Detection and Interpretation of Pathological Myopia Based on Artificial Intelligence. Front. Med. (Lausanne) 2025, 12, 1529335. [CrossRef]
- Ehsan, K.; Sultan, K.; Fatima, A.; Sheraz, M.; Chuah, T.C. Early Detection of Autism Spectrum Disorder Through Automated Machine Learning. Diagn. (Basel) 2025, 15, . [CrossRef]
- Alharthi, A.G.; Alzahrani, S.M. Do It the Transformer Way: A Comprehensive Review of Brain and Vision Transformers for Autism Spectrum Disorder Diagnosis and Classification. Comput Biol Med 2023, 167, 107667. [CrossRef]
- Ahmed, M.; Hussain, S.; Ali, F.; Gárate-Escamilla, A.K.; Amaya, I.; Ochoa-Ruiz, G.; Ortiz-Bayliss, J.C. Summarizing Recent Developments on Autism Spectrum Disorder Detection and Classification through Machine Learning and Deep Learning Techniques. Appl. Sci. (Basel) 2025, 15, 8056. [CrossRef]
- Chaddad, A.; Li, J.; Lu, Q.; Li, Y.; Okuwobi, I.P.; Tanougast, C.; Desrosiers, C.; Niazi, T. Can Autism Be Diagnosed with Artificial Intelligence? A Narrative Review. Diagn. (Basel) 2021, 11, . [CrossRef]
- Ali, M.T.; Elnakieb, Y.A.; Shalaby, A.; Mahmoud, A.; Switala, A.; Ghazal, M.; Khelifi, A.; Fraiwan, L.; Barnes, G.; El-Baz, A. Autism Classification Using SMRI: A Recursive Features Selection Based on Sampling from Multi-Level High Dimensional Spaces. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI); IEEE, April 13 2021.
- Shao, L.; Fu, C.; You, Y.; Fu, D. Classification of ASD Based on FMRI Data with Deep Learning. Cogn. Neurodyn. 2021, 15, 961–974. [CrossRef]
- Almuqhim, F.; Saeed, F. ASD-SAENet: A Sparse Autoencoder, and Deep-Neural Network Model for Detecting Autism Spectrum Disorder (ASD) Using FMRI Data. Front. Comput. Neurosci. 2021, 15, 654315. [CrossRef]
- Alharthi, A.G.; Alzahrani, S.M. Multi-Slice Generation SMRI and FMRI for Autism Spectrum Disorder Diagnosis Using 3D-CNN and Vision Transformers. Brain Sci. 2023, 13, 1578. [CrossRef]
- Tang, M.; Kumar, P.; Chen, H.; Shrivastava, A. Deep Multimodal Learning for the Diagnosis of Autism Spectrum Disorder. J. Imaging Sci. Technol. 2020, 6, . [CrossRef]
- Huang, Z.-A.; Zhu, Z.; Yau, C.H.; Tan, K.C. Identifying Autism Spectrum Disorder From Resting-State FMRI Using Deep Belief Network. IEEE Trans Neural Netw Learn Syst 2021, 32, 2847–2861. [CrossRef]
- Yousefian, A.; Shayegh, F.; Maleki, Z. Detection of Autism Spectrum Disorder Using Graph Representation Learning Algorithms and Deep Neural Network, Based on FMRI Signals. Front. Syst. Neurosci. 2022, 16, 904770. [CrossRef]
- Zhang, J.; Feng, F.; Han, T.; Gong, X.; Duan, F. Detection of Autism Spectrum Disorder Using FMRI Functional Connectivity with Feature Selection and Deep Learning. Cognit. Comput. 2023, 15, 1106–1117. [CrossRef]
- Subah, F.Z.; Deb, K.; Dhar, P.K.; Koshiba, T. A Deep Learning Approach to Predict Autism Spectrum Disorder Using Multisite Resting-State FMRI. Appl. Sci. 2021, 11, 3636. [CrossRef]
- Wang, C.; Xiao, Z.; Xu, Y.; Zhang, Q.; Chen, J. A Novel Approach for ASD Recognition Based on Graph Attention Networks. Front Comput Neurosci 2024, 18, 1388083. [CrossRef]
- Eslami, T.; Raiker, J.S.; Saeed, F. Explainable and Scalable Machine Learning Algorithms for Detection of Autism Spectrum Disorder Using FMRI Data. In Neural Engineering Techniques for Autism Spectrum Disorder; Elsevier, 2021; pp. 39–54 ISBN 9780128228227.
- Eslami, T.; Mirjalili, V.; Fong, A.; Laird, A.R.; Saeed, F. ASD-DiagNet: A Hybrid Learning Approach for Detection of Autism Spectrum Disorder Using FMRI Data. Front. Neuroinform. 2019, 13, 70. [CrossRef]
- Heinsfeld, A.S.; Franco, A.R.; Craddock, R.C.; Buchweitz, A.; Meneguzzi, F. Identification of Autism Spectrum Disorder Using Deep Learning and the ABIDE Dataset. Neuroimage Clin 2018, 17, 16–23. [CrossRef]
- Duan, Y.; Zhao, W.; Luo, C.; Liu, X.; Jiang, H.; Tang, Y.; Liu, C.; Yao, D. Identifying and Predicting Autism Spectrum Disorder Based on Multi-Site Structural MRI With Machine Learning. Front. Hum. Neurosci. 2021, 15, 765517. [CrossRef]
- Mishra, M.; Pati, U.C. A Classification Framework for Autism Spectrum Disorder Detection Using SMRI: Optimizer Based Ensemble of Deep Convolution Neural Network with on-the-Fly Data Augmentation. Biomed. Signal Process. Control 2023, 84, 104686. [CrossRef]
- Sharif, H.; Khan, R.A. A Novel Machine Learning Based Framework for Detection of Autism Spectrum Disorder (ASD). Appl. Artif. Intell. 2022, 36, 1–33. [CrossRef]
- Gao, J.; Chen, M.; Li, Y.; Gao, Y.; Li, Y.; Cai, S.; Wang, J. Multisite Autism Spectrum Disorder Classification Using Convolutional Neural Network Classifier and Individual Morphological Brain Networks. Front Neurosci 2020, 14, 629630. [CrossRef]
- Nogay, H.S.; Adeli, H. Multiple Classification of Brain MRI Autism Spectrum Disorder by Age and Gender Using Deep Learning. J Med Syst 2024, 48, 15. [CrossRef]
- Ali, M.T.; ElNakieb, Y.; Elnakib, A.; Shalaby, A.; Mahmoud, A.; Ghazal, M.; Yousaf, J.; Abu Khalifeh, H.; Casanova, M.; Barnes, G.; et al. The Role of Structure MRI in Diagnosing Autism. Diagn. (Basel) 2022, 12, . [CrossRef]
- Mostafa, S.; Wu, F.-X. Diagnosis of Autism Spectrum Disorder with Convolutional Autoencoder and Structural MRI Images. In Neural Engineering Techniques for Autism Spectrum Disorder; Elsevier, 2021; pp. 23–38 ISBN 9780128228227.
- Yakolli, N.; Anusha, V.; Khan, A.A.; Shubhashree, A.; Chatterjee, S. Enhancing the Diagnosis of Autism Spectrum Disorder Using Phenotypic, Structural, and Functional MRI Data. Innov. Syst. Softw. Eng. 2023, . [CrossRef]
- Manikantan, K.; Jaganathan, S. A Model for Diagnosing Autism Patients Using Spatial and Statistical Measures Using Rs-FMRI and SMRI by Adopting Graphical Neural Networks. Diagn. (Basel) 2023, 13, . [CrossRef]
- Dekhil, O.; Ali, M.; Haweel, R.; Elnakib, Y.; Ghazal, M.; Hajjdiab, H.; Fraiwan, L.; Shalaby, A.; Soliman, A.; Mahmoud, A.; et al. A Comprehensive Framework for Differentiating Autism Spectrum Disorder From Neurotypicals by Fusing Structural MRI and Resting State Functional MRI. Semin Pediatr Neurol 2020, 34, 100805.
- Jain, S.; Tripathy, H.K.; Mallik, S.; Qin, H.; Shaalan, Y.; Shaalan, K. Autism Detection of MRI Brain Images Using Hybrid Deep CNN with DM-Resnet Classifier. IEEE Access 2023, 11, 117741–117751. [CrossRef]
- Itani, S.; Thanou, D. Combining Anatomical and Functional Networks for Neuropathology Identification: A Case Study on Autism Spectrum Disorder. Med Image Anal 2021, 69, 101986. [CrossRef]
- ABIDE Available online: https://fcon_1000.projects.nitrc.org/indi/abide/ (accessed on 17 December 2024).
- Cameron, C.; Yassine, B.; Carlton, C.; Francois, C.; Alan, E.; András, J.; Budhachandra, K.; John, L.; Qingyang, L.; Michael, M.; et al. The Neuro Bureau Preprocessing Initiative: Open Sharing of Preprocessed Neuroimaging Data and Derivatives. Front. Neuroinform. 2013, 7, . [CrossRef]
- Ahammed, M.S.; Niu, S.; Ahmed, M.R.; Dong, J.; Gao, X.; Chen, Y. DarkASDNet: Classification of ASD on Functional MRI Using Deep Neural Network. Front Neuroinform 2021, 15, 635657. [CrossRef]
- Yang, M.; Cao, M.; Chen, Y.; Chen, Y.; Fan, G.; Li, C.; Wang, J.; Liu, T. Large-Scale Brain Functional Network Integration for Discrimination of Autism Using a 3-D Deep Learning Model. Front Hum Neurosci 2021, 15, 687288. [CrossRef]
- Wang, Y.; Sheng, H.; Wang, X. Recognition and Diagnosis of Alzheimer’s Disease Using T1-Weighted Magnetic Resonance Imaging via Integrating CNN and Swin Vision Transformer. Clin. (Sao Paulo) 2025, 80, 100673. [CrossRef]
- Asiri, A.A.; Shaf, A.; Ali, T.; Shakeel, U.; Irfan, M.; Mehdar, K.; Halawani, H.; Alghamdi, A.H.; Alshamrani, A.F.A.; Alqhtani, S.M. Exploring the Power of Deep Learning: Fine-Tuned Vision Transformer for Accurate and Efficient Brain Tumor Detection in MRI Scans. Diagn. (Basel) 2023, 13, . [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image Is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv [cs.CV] 2020.
- Han, K.; Xiao, A.; Wu, E.; Guo, J.; Xu, C.; Wang, Y. Transformer in Transformer. Neural Inf Process Syst 2021, 34, 15908–15919.
- Tu, Z.; Talebi, H.; Zhang, H.; Yang, F.; Milanfar, P.; Bovik, A.; Li, Y. MaxViT: Multi-Axis Vision Transformer. arXiv [cs.CV] 2022.
- Ong, K.L.; Lee, C.P.; Lim, H.S.; Lim, K.M.; Alqahtani, A. MaxMViT-MLP: Multiaxis and Multiscale Vision Transformers Fusion Network for Speech Emotion Recognition. IEEE Access 2024, 12, 18237–18250. [CrossRef]
- Lundberg, S.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. arXiv [cs.AI] 2017.
- Redmon, J.; Farhadi, A. YOLO9000: Better, Faster, Stronger. arXiv [cs.CV] 2016.
- Liu, M.; Wang, Z.; Chen, C.; Hu, S. Functional and Structural Brain Network Construction, Representation and Application; Frontiers Media SA, 2023; ISBN 9782832520017. [CrossRef]
- Nielsen, J.A.; Zielinski, B.A.; Fletcher, P.T.; Alexander, A.L.; Lange, N.; Bigler, E.D.; Lainhart, J.E.; Anderson, J.S. Multisite Functional Connectivity MRI Classification of Autism: ABIDE Results. Front. Hum. Neurosci. 2013, 7, 599. [CrossRef]
| Feature Name | Definition & Link to Autism | Associated Brain Regions |
| Functional connectivity | Measures correlation of activation time series between brain regions. ASD shows altered connectivity, especially in social-communication networks [24,25]. | default mode network (DMN) and salience network. |
| Regions of Interest (ROIs) | Selected regions with known cognitive roles. Abnormal ROI activity is linked to social and communication deficits. | prefrontal cortex, amygdala, and superior temporal sulcus. |
| Time Series Analysis | Evaluates BOLD signal fluctuations over time. Irregular patterns may indicate dysfunctional connectivity. | Multiple cortical regions |
| Temporal Resolution | Captures brain dynamics at finer time intervals. Higher resolution reveals subtle neural differences in ASD. | Default mode network, Visual cortex |
| Feature Name | Definition & Link to Autism | Associated Brain Regions |
| Frontal & Temporal Lobes Volume | Abnormal volumes correlate with social and cognitive impairments. | Frontal cortex, Temporal cortex |
| Cortical Thickness | Thickness of the cerebral cortex, related to higher-order cognitive functions. | Prefrontal cortex, Temporal lobe |
| Cerebrospinal Fluid Volume | Volume of the fluid surrounding the brain, involved in protection and waste removal. Increased volume may indicate early brain development abnormalities predictive of autism. | Subarachnoid space |
| Cortical Surface Area | Altered surface area affects connectivity and cognition. | Prefrontal cortex, Parietal lobe |
| Gray Matter Volume and Density | Abnormalities relate to deficits in social-emotional functions | Amygdala, Hippocampus, Prefrontal cortex [26]. |
| White Matter Integrity | Reduced integrity weakens inter-regional communication | Corpus callosum, Superior longitudinal fasciculus [27]. |
| Modality | ASD | TC |
| fMRI | 74 | 98 |
| sMRI | 79 | 105 |
| Hyperparameters | Values |
| Learning rate | 1e-3, 1e-4, 1e-5 |
| Batch size | 8, 16, 32 |
| Dropout rate | 0.2, 0.3, 0.4 |
| Optimizer | Adam, AdamW |
| Model | Accuracy % | F1-Score % | Loss |
| ViT (baseline) | 98.03 | 98.02 | 0.0788 |
| TNT | 98.42 | 98.42 | 0.1242 |
| MaxViT | 98.42 | 98.42 | 0.13 |
| Model | Accuracy % | F1-Score % | Loss |
| ViT (baseline) | 81.34 | 80.81 | 0.4456 |
| TNT | 91.62 | 91.65 | 0.2928 |
| MaxViT | 98.51 | 98.51 | 0.0409 |
| Study | Model | Best Accuracy | F1-score |
| DarkASDNet, Ahammed et al. 2021. [78] | CNN | 94.7% | 95% |
|
Alharthi and Alzahrani, 2023. [38] |
3D-CNN |
87% |
82% |
|
Our proposed method in this study |
ViT | 98.03% | 98.02% |
| TNT | 98.42% | 98.42% | |
| MaxViT | 98.42% | 98.42% |
| Study | Model | Best Accuracy | F1-score |
| Alharthi and Alzahrani, 2023. [38] | ConvNeXt | 77% | 76% |
| Our proposed method in this study | ViT | 81.34% | 80.81% |
| TNT | 91.62% | 91.65% | |
| MaxViT | 98.51% | 98.51% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




