Submitted:
19 August 2025
Posted:
20 August 2025
You are already at the latest version
Abstract
Inactivation of the dnd gene, which is essential for the development of primordial germ cells (PGCs), leads to the loss of gametes and halts further reproductive development in vertebrates. Studies on the gonads of sterile fish are useful for identifying GC-related genes, as well as the structures and processes dependent on this lineage. We identified genes affected in GC-ablated testes of Atlantic salmon following temporal silencing of dnd and tracked their expression during normal gonadal development. In sterile testes, transcripts of several GC markers were detected at low levels, suggesting the presence of cells with a GC-related expression profile that failed to initiate spermatogenesis. We found 260 genes silenced in the gonads of sterile males and females, and 61.5% of these were also inactivated during first maturation of fertile testes. This group was enriched with genes highly expressed in the brain, including those involved in endocrine and paracrine regulation, synaptic transmission, and numerous genes critical for brain development, among them, 45 genes encoding homeobox proteins. Another group of 229 genes showed increased expression in developing testes and included genes involved in neurosecretion and brain development regulation. Genes upregulated in GC-ablated testes included reproductive regulators such as amh and sdy, as well as numerous genes related to the innate and adaptive immune systems, suggesting a reprogramming of germ cell-depleted testes. Temporal silencing of dnd revealed the presence of a complex neural structure in the Atlantic salmon testis that becomes inactive at first maturation. Potential roles for this structure may include PGC homing, the creation of a specific environment required for spermatogenesis, or facilitating communication between the gonads and brain to signal readiness for reproduction, among other possibilities.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Fish Material
2.2. Gene Expression
3. Results
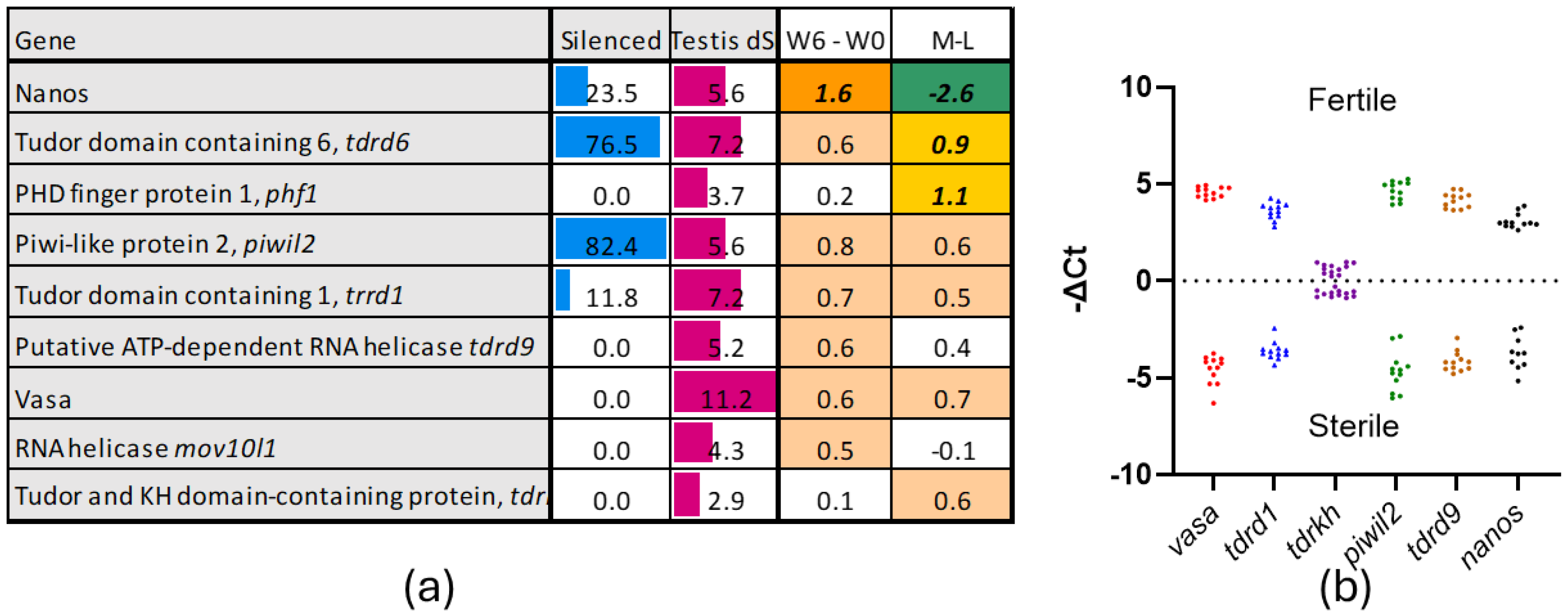
3.1. Gene Markers of Germ Cells
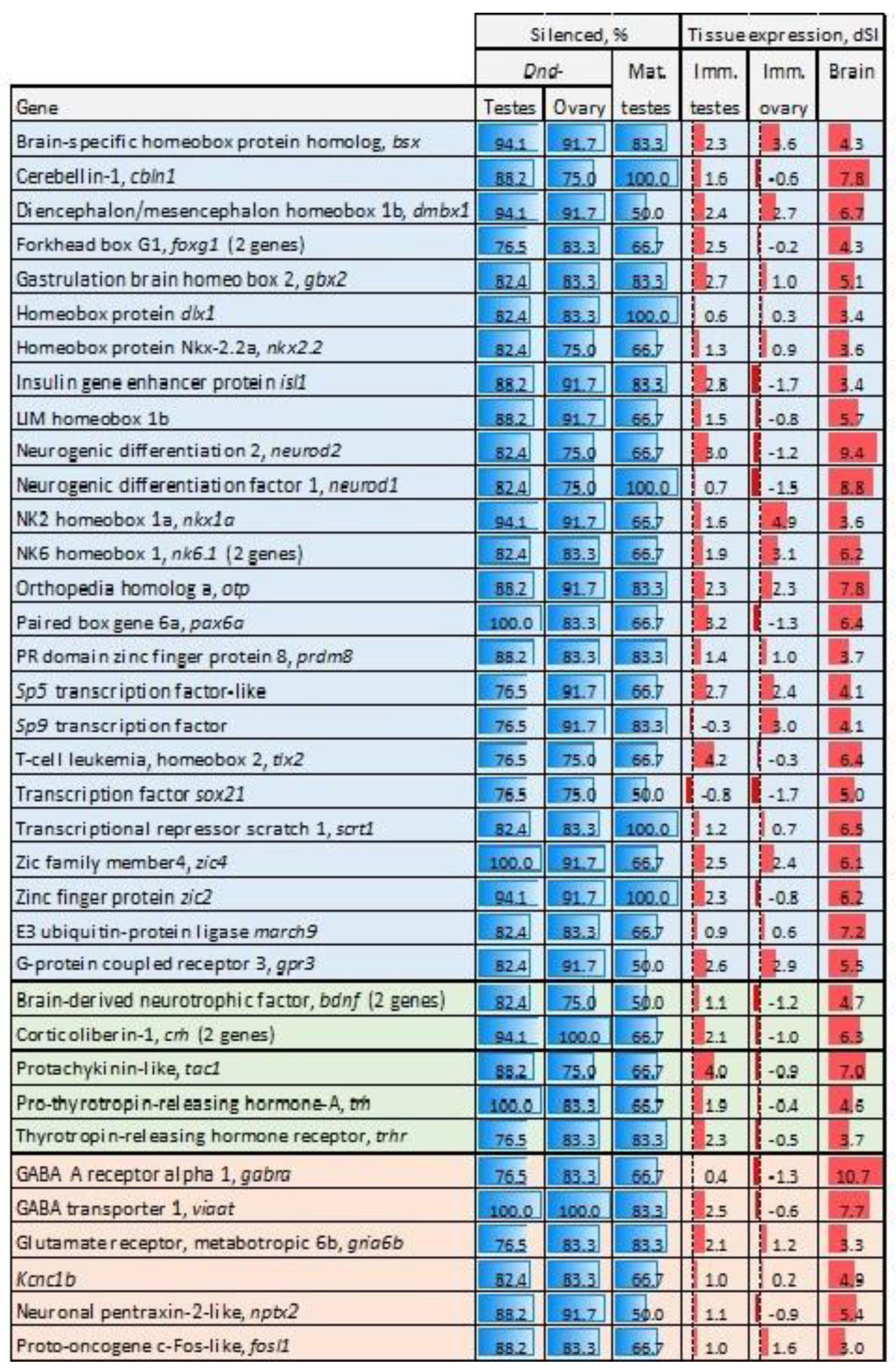
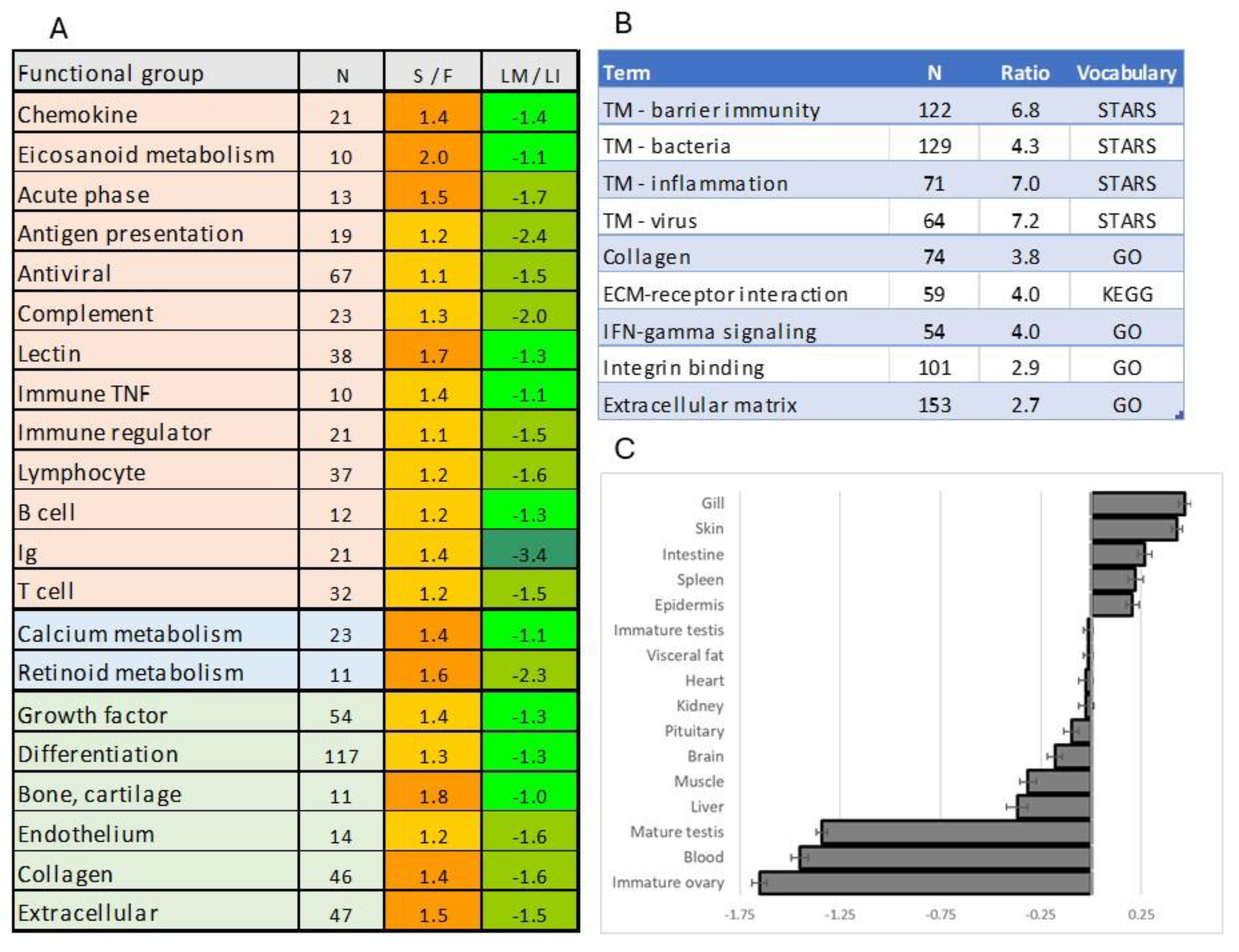
3.2. Genes Silenced or Downregulated in Sterile Testes
3.2.1. Silenced Genes
3.2.2. Genes Downregulated in sterile GC-Depleted Testes
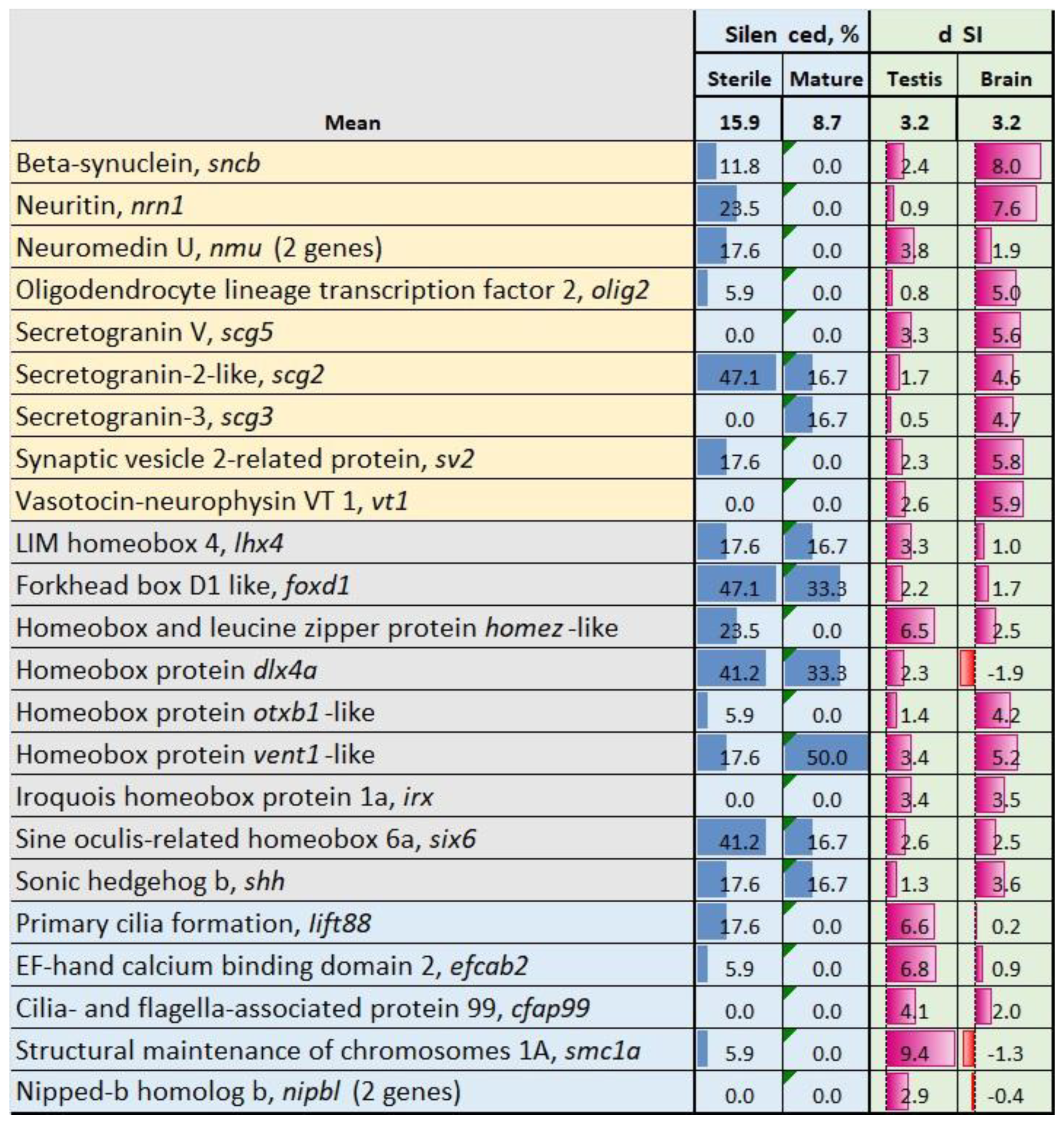
3.3. Genes with Stable Expression During Gonadal Growth and Maturation
3.4. Genes with Expression Changes at Maturation
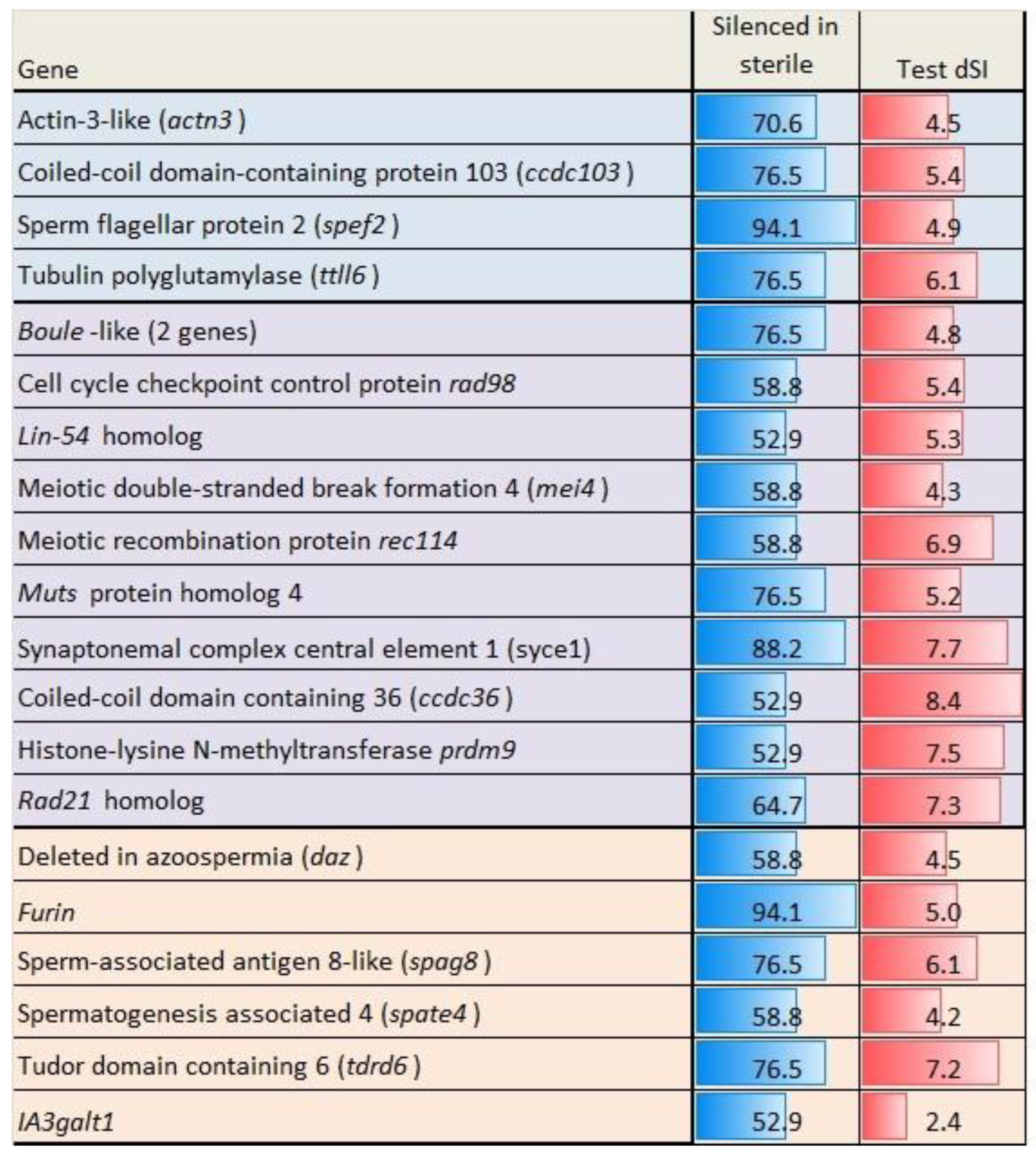
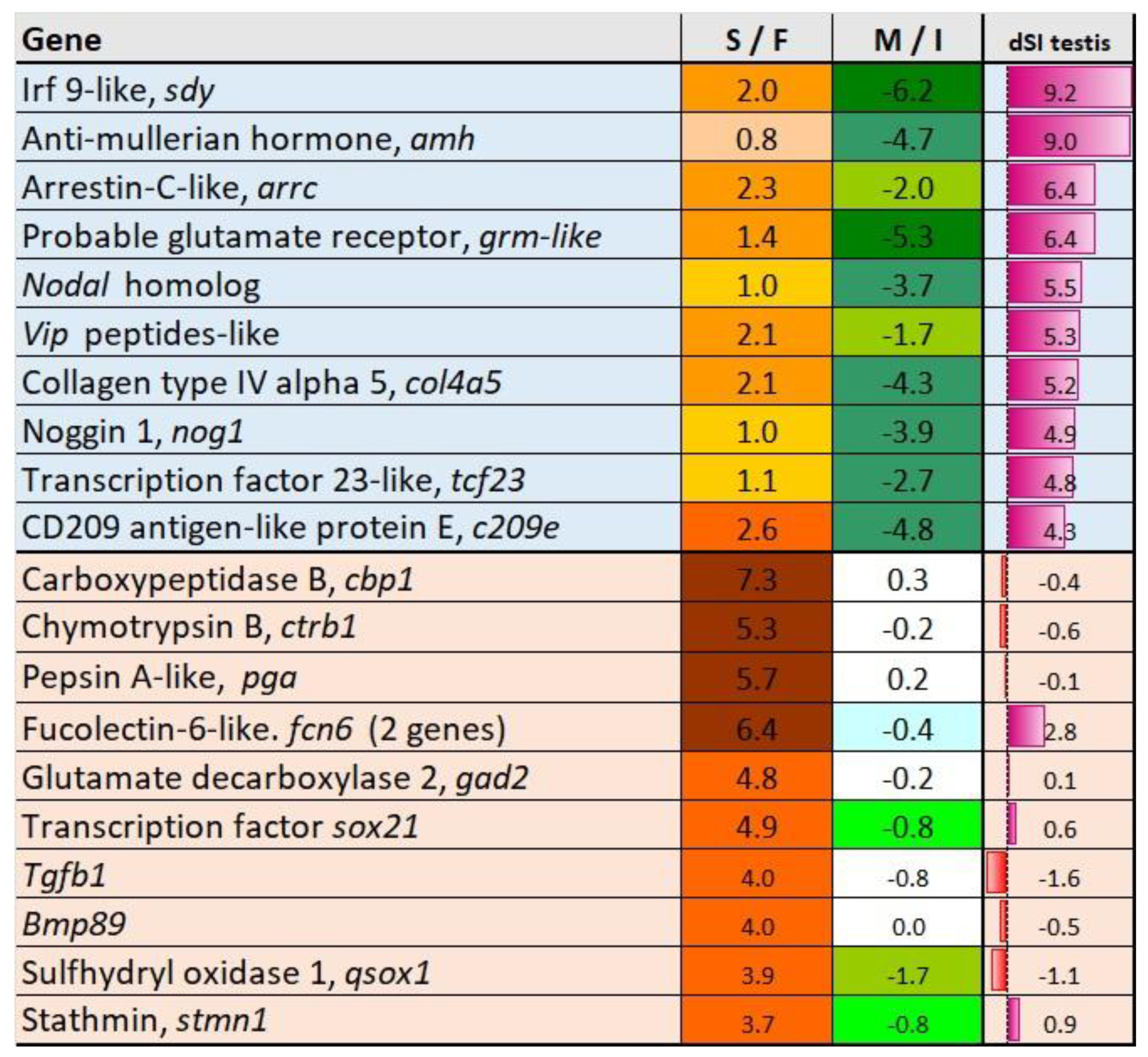
3.5. Genes Upregulated in Sterile Testes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Abbreviations
| DEG | differentially expressed genes |
| ER | expression ratio |
| GC | germ cells |
| HPA | hypothalamic-pituitary-adrenal (HPA) axis |
| PGC | primordial germ cells |
References
- Kedde, M.; Strasser, M.J.; Boldajipour, B.; Vrielink, J.A.F.O.; Slanchev, K.; le Sage, C.; Nagel, R.; Voorhoeve, P.M.; van Duijse, J.; Ørom, U.A.; et al. RNA-Binding Protein Dnd1 Inhibits MicroRNA Access to Target mRNA. Cell 2007, 131, 1273–1286. [Google Scholar] [CrossRef]
- Gross-Thebing, T.; Yigit, S.; Pfeiffer, J.; Reichman-Fried, M.; Bandemer, J.; Ruckert, C.; Rathmer, C.; Goudarzi, M.; Stehling, M.; Tarbashevich, K.; et al. The Vertebrate Protein Dead End Maintains Primordial Germ Cell Fate by Inhibiting Somatic Differentiation. Dev. Cell 2017, 43, 704–715.e5. [Google Scholar] [CrossRef]
- Xu, C.; Li, Y.; Wen, Z.; Jawad, M.; Gui, L.; Li, M. Spinyhead Croaker Germ Cells Gene dnd Visualizes Primordial Germ Cells in Medaka. Life 2022, 12, 1226. [Google Scholar] [CrossRef]
- Liu, W.; Collodi, P. Zebrafish dead end possesses ATPase activity that is required for primordial germ cell development. FASEB J. 2010, 24, 2641–2650. [Google Scholar] [CrossRef]
- Yang, X.; Yue, H.; Ye, H.; Li, C.; Wei, Q. Identification of a germ cell marker gene, the dead end homologue, in Chinese sturgeon Acipenser sinensis. Gene 2015, 558, 118–125. [Google Scholar] [CrossRef]
- Zhu, T.; Gui, L.; Zhu, Y.; Li, Y.; Li, M. Dnd is required for primordial germ cell specification in Oryzias celebensis. Gene 2018, 679, 36–43. [Google Scholar] [CrossRef]
- Kawamura, W.; Tani, R.; Yahagi, H.; Kamio, S.; Morita, T.; Takeuchi, Y.; Yazawa, R.; Yoshizaki, G. Suitability of hybrid mackerel (Scomber australasicus × S. japonicus) with germ cell-less sterile gonads as a recipient for transplantation of bluefin tuna germ cells. Gen. Comp. Endocrinol. 2020, 295, 113525. [Google Scholar] [CrossRef]
- Kurokawa, H.; Saito, D.; Nakamura, S.; Katoh-Fukui, Y.; Ohta, K.; Baba, T.; Morohashi, K.-I.; Tanaka, M. Germ cells are essential for sexual dimorphism in the medaka gonad. Proc. Natl. Acad. Sci. 2007, 104, 16958–16963. [Google Scholar] [CrossRef]
- Siegfried, K.R.; Nüsslein-Volhard, C. Germ line control of female sex determination in zebrafish. Dev. Biol. 2008, 324, 277–287. [Google Scholar] [CrossRef]
- Slanchev, K.; Stebler, J.; de la Cueva-Méndez, G.; Raz, E. Development without germ cells: The role of the germ line in zebrafish sex differentiation. Proc. Natl. Acad. Sci. 2005, 102, 4074–4079. [Google Scholar] [CrossRef]
- Weltzien, F.-A.; Andersson, E.; Andersen, Ø.; Shalchian-Tabrizi, K.; Norberg, B. The brain–pituitary–gonad axis in male teleosts, with special emphasis on flatfish (Pleuronectiformes). Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 2004, 137, 447–477. [Google Scholar] [CrossRef]
- Fujihara, R.; Katayama, N.; Sadaie, S.; Miwa, M.; Matias, G.A.S.; Ichida, K.; Fujii, W.; Naito, K.; Hayashi, M.; Yoshizaki, G. Production of Germ Cell-Less Rainbow Trout by dead end Gene Knockout and their Use as Recipients for Germ Cell Transplantation. Mar. Biotechnol. 2022, 24, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, T.; Nishimura, T.; Goto-Kazeto, R.; Kawakami, Y.; Yamaha, E.; Arai, K. Sexual dimorphism of gonadal structure and gene expression in germ cell-deficient loach, a teleost fish. Proc. Natl. Acad. Sci. 2010, 107, 17211–17216. [Google Scholar] [CrossRef] [PubMed]
- Goto, R.; Saito, T.; Takeda, T.; Fujimoto, T.; Takagi, M.; Arai, K.; Yamaha, E. Germ cells are not the primary factor for sexual fate determination in goldfish. Dev. Biol. 2012, 370, 98–109. [Google Scholar] [CrossRef]
- Wargelius, A.; Leininger, S.; Skaftnesmo, K.O.; Kleppe, L.; Andersson, E.; Taranger, G.L.; Schulz, R.W.; Edvardsen, R.B. Dnd knockout ablates germ cells and demonstrates germ cell independent sex differentiation in Atlantic salmon. Sci. Rep. 2016, 6, 21284. [Google Scholar] [CrossRef]
- Yoshikawa, H.; Ino, Y.; Kishimoto, K.; Koyakumaru, H.; Saito, T.; Kinoshita, M.; Yoshiura, Y. Induction of germ cell-deficiency in grass puffer by dead end 1 gene knockdown for use as a recipient in surrogate production of tiger puffer. Aquaculture 2020, 526. [Google Scholar] [CrossRef]
- Yoshizaki G, Takashiba K, Shimamori S, Fujinuma K, Shikina S, Okutsu T, et al. Production of germ cell-deficient salmonids by dead end gene knockdown, and their use as recipients for germ cell transplantation. Mol Reprod Dev. 2016;83(4):298-311.
- Braat, A.K.; E Speksnijder, J.; Zivkovic, D. Germ line development in fishes. . 1999, 43, 745–60. [Google Scholar]
- Hansen, C.L.; Pelegri, F. Primordial Germ Cell Specification in Vertebrate Embryos: Phylogenetic Distribution and Conserved Molecular Features of Preformation and Induction. Front. Cell Dev. Biol. 2021, 9. [Google Scholar] [CrossRef]
- Xu, L.; Zhao, M.; Ryu, J.H.; Hayman, E.S.; Fairgrieve, W.T.; Zohar, Y.; Luckenbach, J.A.; Wong, T. Reproductive sterility in aquaculture: A review of induction methods and an emerging approach with application to Pacific Northwest finfish species. Rev. Aquac. 2022, 15, 220–241. [Google Scholar] [CrossRef]
- Booncherd, K.; Sreebun, S.; Pasomboon, P.; Boonanuntanasarn, S. Effects of CRISPR/Cas9-mediated dnd1 knockout impairs gonadal development in striped catfish. Animal 2023, 18, 101039. [Google Scholar] [CrossRef]
- Škugor, A.; Tveiten, H.; Krasnov, A.; Andersen, Ø. Knockdown of the germ cell factor Dead end induces multiple transcriptional changes in Atlantic cod (Gadus morhua) hatchlings. Anim. Reprod. Sci. 2014, 144, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Tveiten H, Karlsen K, Thesslund T, Johansson GS, Thiyagarajan DB, Andersen Ø. Impact of germ cell ablation on the activation of the brain-pituitary-gonadal axis in precocious Atlantic salmon (Salmo salar L.) males. Mol Reprod Dev. 2022;89(10):471-84.
- Skjold, V.; Afanasyev, S.; Burgerhout, E.; Sveen, L.; Rørvik, K.-A.; Mota, V.F.C.N.; Dessen, J.-E.; Krasnov, A. Endocrine and Transcriptome Changes Associated with Testicular Growth and Differentiation in Atlantic Salmon (Salmo salar L.). Curr. Issues Mol. Biol. 2024, 46, 5337–5351. [Google Scholar] [CrossRef] [PubMed]
- Pauli, A.; Montague, T.G.; Lennox, K.A.; Behlke, M.A.; Schier, A.F.; Neuhauss, S.C. Antisense Oligonucleotide-Mediated Transcript Knockdown in Zebrafish. PLOS ONE 2015, 10, e0139504. [Google Scholar] [CrossRef] [PubMed]
- Krasnov, A.; Timmerhaus, G.; Afanasyev, S.; Jørgensen, S.M. Development and assessment of oligonucleotide microarrays for Atlantic salmon (Salmo salar L.). Comp. Biochem. Physiol. Part D: Genom. Proteom. 2011, 6, 31–38. [Google Scholar] [CrossRef]
- Kortner, T.M.; Afanasyev, S.; Koppang, E.O.; Bjørgen, H.; Krogdahl, Å.; Krasnov, A. A comprehensive transcriptional body map of Atlantic salmon unveils the vital role of the intestine in the immune system and highlights functional specialization within its compartments. Fish Shellfish. Immunol. 2024, 146, 109422. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Krasnov, A.; Afanasyev, S.; Nylund, S.; Rebl, A. Multigene Expression Assay for Assessment of the Immune Status of Atlantic Salmon. Genes 2020, 11, 1236. [Google Scholar] [CrossRef]
- Vasileva, A.; Tiedau, D.; Firooznia, A.; Müller-Reichert, T.; Jessberger, R. Tdrd6 Is Required for Spermiogenesis, Chromatoid Body Architecture, and Regulation of miRNA Expression. Curr. Biol. 2009, 19, 630–639. [Google Scholar] [CrossRef]
- Lee, J.H.; Engel, W.; Nayernia, K. Stem cell protein Piwil2 modulates expression of murine spermatogonial stem cell expressed genes. Mol. Reprod. Dev. 2005, 73, 173–179. [Google Scholar] [CrossRef]
- Qu, Y.; Wang, Y.; Qiao, J. PHF1 is required for chromosome alignment and asymmetric division during mouse meiotic oocyte maturation. Cell Cycle 2018, 17, 2447–2459. [Google Scholar] [CrossRef]
- Nonogaki, K.; Kaji, T. Role of homeobox genes in the hypothalamic development and energy balance. Front. Biosci. 2013, 18, 740–7. [Google Scholar] [CrossRef]
- Merlo, G.R.; Zerega, B.; Paleari, L.; Trombino, S.; Mantero, S.; Levi, G. Multiple functions of Dlx genes. . 2000, 44, 619–26. [Google Scholar]
- Urbach R, Technau GM. Dorsoventral patterning of the brain: a comparative approach. Adv Exp Med Biol. 2008;628:42-56.
- Holland, P.W.; Takahashi, T. The evolution of homeobox genes: Implications for the study of brain development. Brain Res. Bull. 2005, 66, 484–490. [Google Scholar] [CrossRef]
- Aruga, J. The role of Zic genes in neural development. Mol. Cell. Neurosci. 2004, 26, 205–221. [Google Scholar] [CrossRef]
- Broccoli, V.; Colombo, E.; Cossu, G. Dmbx1 is a paired-box containing gene specifically expressed in the caudal most brain structures. Mech. Dev. 2002, 114, 219–223. [Google Scholar] [CrossRef] [PubMed]
- Borghini, S.; Bachetti, T.; Fava, M.; Di Duca, M.; Cargnin, F.; Fornasari, D.; Ravazzolo, R.; Ceccherini, I. The TLX2 homeobox gene is a transcriptional target of PHOX2B in neural-crest-derived cells. Biochem. J. 2006, 395, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Kumamoto, T.; Hanashima, C. Evolutionary conservation and conversion of Foxg1 function in brain development. Dev. Growth Differ. 2017, 59, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Gogoi, R.N.; Schubert, F.R.; Martinez-Barbera, J.-P.; Acampora, D.; Simeone, A.; Lumsden, A. The paired-type homeobox gene Dmbx1 marks the midbrain and pretectum. Mech. Dev. 2002, 114, 213–217. [Google Scholar] [CrossRef]
- Wang, Z.; Nakayama, Y.; Tsuda, S.; Yamasu, K. The role of gastrulation brain homeobox 2 (gbx2) in the development of the ventral telencephalon in zebrafish embryos. Differentiation 2018, 99, 28–40. [Google Scholar] [CrossRef]
- Ashery-Padan, R.; Gruss, P. Pax6 lights-up the way for eye development. Curr. Opin. Cell Biol. 2001, 13, 706–714. [Google Scholar] [CrossRef]
- Whittington, N.; Cunningham, D.; Le, T.-K.; De Maria, D.; Silva, E.M. Sox21 regulates the progression of neuronal differentiation in a dose-dependent manner. Dev. Biol. 2015, 397, 237–247. [Google Scholar] [CrossRef]
- Liang, X.; Song, M.-R.; Xu, Z.; Lanuza, G.M.; Liu, Y.; Zhuang, T.; Chen, Y.; Pfaff, S.L.; Evans, S.M.; Sun, Y. Isl1 Is required for multiple aspects of motor neuron development. Mol. Cell. Neurosci. 2011, 47, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Huang, H.; Chen, Z.; Zhang, Z.; Lu, W.; Qiu, M. The transcription factor NKX2-2 regulates oligodendrocyte differentiation through domain-specific interactions with transcriptional corepressors. J. Biol. Chem. 2020, 295, 1879–1888. [Google Scholar] [CrossRef] [PubMed]
- Leal, G.; Comprido, D.; Duarte, C.B. BDNF-induced local protein synthesis and synaptic plasticity. Neuropharmacology 2014, 76 Pt C, 639–656. [Google Scholar] [CrossRef] [PubMed]
- Aubrey, K.R. Presynaptic control of inhibitory neurotransmitter content in VIAAT containing synaptic vesicles. Neurochem. Int. 2016, 98, 94–102. [Google Scholar] [CrossRef]
- Conde-Sieira M, Ceinos RM, Velasco C, Comesaña S, López-Patiño MA, Míguez JM, et al. Response of rainbow trout's (Oncorhynchus mykiss) hypothalamus to glucose and oleate assessed through transcription factors BSX, ChREBP, CREB, and FoxO1. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2018;204(11):893-904.
- Herrera DG, Robertson HA. Activation of c-fos in the brain. Prog Neurobiol. 1996;50(2-3):83-107.
- Tsigos C, Chrousos GP. Hypothalamic-pituitary-adrenal axis, neuroendocrine factors and stress. J Psychosom Res. 2002;53(4):865-71.
- Jean, D.; Bernier, G.; Gruss, P. Six6 (Optx2) is a novel murine Six3-related homeobox gene that demarcates the presumptive pituitary/hypothalamic axis and the ventral optic stalk. Mech. Dev. 1999, 84, 31–40. [Google Scholar] [CrossRef]
- Roisman-Geller, N.; Pisanty, O.; Weinberger, A.; Gajbhiye, D.S.; Golan, M.; Gothilf, Y. Combined Pituitary Hormone Deficiency in lhx4-Knockout Zebrafish. Int. J. Mol. Sci. 2024, 25, 7332. [Google Scholar] [CrossRef]
- Rodríguez-Seguel, E.; Alarcón, P.; Gómez-Skarmeta, J.L. The Xenopus Irx genes are essential for neural patterning and define the border between prethalamus and thalamus through mutual antagonism with the anterior repressors Fezf and Arx. Dev. Biol. 2009, 329, 258–268. [Google Scholar] [CrossRef]
- Guo, R.; Ge, K.; Wang, Y.; Lu, M.; Li, F.; Tian, L.; Gan, L.; Sheng, D. LIM Homeobox 4 (lhx4) regulates retinal neural differentiation and visual function in zebrafish. Sci. Rep. 2021, 11, 1–10. [Google Scholar] [CrossRef]
- Stühmer, T.; Puelles, L.; Ekker, M.; Rubenstein, J.L. Expression from a Dlx Gene Enhancer Marks Adult Mouse Cortical GABAergic Neurons. Cereb. Cortex 2002, 12, 75–85. [Google Scholar] [CrossRef]
- Goodson, J.L.; Bass, A.H. Social behavior functions and related anatomical characteristics of vasotocin/vasopressin systems in vertebrates. Brain Res. Rev. 2001, 35, 246–265. [Google Scholar] [CrossRef]
- Lu, Q.R.; Sun, T.; Zhu, Z.; Ma, N.; Garcia, M.; Stiles, C.D.; Rowitch, D.H. Common Developmental Requirement for Olig Function Indicates a Motor Neuron/Oligodendrocyte Connection. Cell 2002, 109, 75–86. [Google Scholar] [CrossRef]
- Howard, A.D.; Wang, R.; Pong, S.-S.; Mellin, T.N.; Strack, A.; Guan, X.-M.; Zeng, Z.; Williams, D.L.; Feighner, S.D.; Nunes, C.N.; et al. Identification of receptors for neuromedin U and its role in feeding. Nature 2000, 406, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Naeve, G.S.; Ramakrishnan, M.; Kramer, R.; Hevroni, D.; Citri, Y.; Theill, L.E. Neuritin: A gene induced by neural activity and neurotrophins that promotes neuritogenesis. Proc. Natl. Acad. Sci. 1997, 94, 2648–2653. [Google Scholar] [CrossRef]
- Mahata SK, Mahapatra NR, Mahata M, O'Connor DT. Neuroendocrine cell type-specific and inducible expression of chromogranin/secretogranin genes: crucial promoter motifs. Ann N Y Acad Sci. 2002;971:27-38.
- Reijo, R.; Lee, T.-Y.; Salo, P.; Alagappan, R.; Brown, L.G.; Rosenberg, M.; Rozen, S.; Jaffe, T.; Straus, D.; Hovatta, O.; et al. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA–binding protein gene. Nat. Genet. 1995, 10, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Xu, E.Y.; Moore, F.L.; Pera, R.A.R. A gene family required for human germ cell development evolved from an ancient meiotic gene conserved in metazoans. Proc. Natl. Acad. Sci. 2001, 98, 7414–7419. [Google Scholar] [CrossRef] [PubMed]
- Grey, C.; Baudat, F.; de Massy, B.; Cohen, P.E. PRDM9, a driver of the genetic map. PLOS Genet. 2018, 14, e1007479. [Google Scholar] [CrossRef]
- Morales, C.; Losada, A. Establishing and dissolving cohesion during the vertebrate cell cycle. Curr. Opin. Cell Biol. 2018, 52, 51–57. [Google Scholar] [CrossRef]
- Yano, A.; Guyomard, R.; Nicol, B.; Jouanno, E.; Quillet, E.; Klopp, C.; Cabau, C.; Bouchez, O.; Fostier, A.; Guiguen, Y. An Immune-Related Gene Evolved into the Master Sex-Determining Gene in Rainbow Trout, Oncorhynchus mykiss. Curr. Biol. 2012, 22, 1423–1428. [Google Scholar] [CrossRef]
- Krasnov, A.; Johansen, L.-H.; Karlsen, C.; Sveen, L.; Ytteborg, E.; Timmerhaus, G.; Lazado, C.C.; Afanasyev, S. Transcriptome Responses of Atlantic Salmon (Salmo salar L.) to Viral and Bacterial Pathogens, Inflammation, and Stress. Front. Immunol. 2021, 12. [Google Scholar] [CrossRef]
- Matos B, Publicover SJ, Castro LFC, Esteves PJ, Fardilha M. Brain and testis: more alike than previously thought? Open Biol. 2021;11(6):200322.
- Doitsidou, M.; Reichman-Fried, M.; Stebler, J.; Köprunner, M.; Dörries, J.; Meyer, D.; Esguerra, C.V.; Leung, T.; Raz, E. Guidance of Primordial Germ Cell Migration by the Chemokine SDF-1. Cell 2002, 111, 647–659. [Google Scholar] [CrossRef]
- Ara, T.; Nakamura, Y.; Egawa, T.; Sugiyama, T.; Abe, K.; Kishimoto, T.; Matsui, Y.; Nagasawa, T. Impaired colonization of the gonads by primordial germ cells in mice lacking a chemokine, stromal cell-derived factor-1 (SDF-1). Proc. Natl. Acad. Sci. 2003, 100, 5319–5323. [Google Scholar] [CrossRef]
- Di Carlo A, De Felici M. A role for E-cadherin in mouse primordial germ cell development. Dev Biol. 2000;226(2):209-19.
- Blaser, H.; Reichman-Fried, M.; Castanon, I.; Dumstrei, K.; Marlow, F.L.; Kawakami, K.; Solnica-Krezel, L.; Heisenberg, C.-P.; Raz, E. Migration of Zebrafish Primordial Germ Cells: A Role for Myosin Contraction and Cytoplasmic Flow. Dev. Cell 2006, 11, 613–627. [Google Scholar] [CrossRef] [PubMed]
- Nelson JO, Chen C, Yamashita YM. Germline stem cell homeostasis. Curr Top Dev Biol. 2019;135:203-44.
- de Michele F, Vermeulen M, Wyns C. Fertility restoration with spermatogonial stem cells. Curr Opin Endocrinol Diabetes Obes. 2017;24(6):424-31.
- Köse S, Yersal N, Önen S, Korkusuz P. Comparison of Hematopoietic and Spermatogonial Stem Cell Niches from the Regenerative Medicine Aspect. Adv Exp Med Biol. 2018;1107:15-40.
- Oatley, J.M.; Brinster, R.L. The Germline Stem Cell Niche Unit in Mammalian Testes. Physiol. Rev. 2012, 92, 577–595. [Google Scholar] [CrossRef] [PubMed]
- Sakib, S.; Goldsmith, T.; Voigt, A.; Dobrinski, I. Testicular organoids to study cell–cell interactions in the mammalian testis. Andrology 2019, 8, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.D.; Alberio, R. Primordial germ cells: the first cell lineage or the last cells standing? Development 2015, 142, 2730–2739. [Google Scholar] [CrossRef]
- Barson, N.J.; Aykanat, T.; Hindar, K.; Baranski, M.; Bolstad, G.H.; Fiske, P.; Jacq, C.; Jensen, A.J.; Johnston, S.E.; Karlsson, S.; et al. Sex-dependent dominance at a single locus maintains variation in age at maturity in salmon. Nature 2015, 528, 405–408. [Google Scholar] [CrossRef]
| Genbank | Gene | Forward primer | Reverse primer |
|---|---|---|---|
| RT qPCR | |||
| XM_014143380 | Vasa | CCCAGTACAGAAGCATGGCA | CACTGGACGCACACAAGTTC |
| XM_014157064 | Tdrd1 | AGCTCCCTTTCCAGATTGCC | AGCTGTGGCGATGATGGG |
| XM_014158129 | Tdrkh | AGGCCAAGGAGCTCATCCTA | CCCCTTCCTCCATCCAGAGA |
| XM_014171837 | Piwil1 | ATGACATTGCGTGGGACCAT | AGCACGCATCTTGTCAGTCA |
| XM_014199425 | Dnd1 | TTTGCCTACGCCAAGTACGA | GGAGGCATAGACCACCACTG |
| XM_014189524 | Tdrd9 | GAACAGGACCATCTGCCACA | CCTCTGAGAAAGGTGCCAGG |
| RT-PCR | |||
| NM_001165390 | Viaat | TCGACGTCGCCATATTCGTT | TGGTATGTCGACAGAACCTGC |
| XM_014139471 | Hr6 | TGGAGAAGAGAAGACGTGCG | AATCCCGCTCTGTACTTGCC |
| XM_014206751 | Soho | CATCTCCCAGTTCACCCACC | CCTCTTATGAGTGTGGCCGT |
| XM_014140645 | Gbh | TGTCCGATCAACTTGACAACAT | TCTCACAAACAGCTGGCGAA |
| XM_014148127 | Dbx1 | AGCAAGCACTCCGACTTCTC | CACAACCAATATGGCCCCTAC |
| NM_001141590 | Crf1 | ACCTGACGTTCCATCTGCTG | GAAAGAACGAAGAAAGTTAACCA |
| CA049789 | Rpol2 | TAACGCCTGCCTCTTCAGTTGA | ATGAGGGACCTTGTAGCCAGCAA |
| DW542195 | Eif-3 | CAGGATGTTGTTGCTGGATGGG | ACCCAACTGGGCAGGTCAAGA |
| Functional Group | DEG | Vocabulary |
|---|---|---|
| DNA replication Cell cycle Gene silencing by RNA DNA repair Double-strand break repair Base-excision repair G1/S transition RNA polymerase RNA degradation Chromosome Histone binding Pyrimidine metabolism Cyclin binding Basal transcription factors Mitochondrion |
30 16 10 36 11 6 22 10 14 23 17 17 8 7 97 |
GO KEGG GO GO GO GO GO KEGG KEGG GO GO KEGG GO KEGG GO |
| Functional Group | DEG | Vocabulary |
|---|---|---|
| Upregulated (790 genes) DNA replication Mitosis Meiosis Cell division Chromosome Chromosome segregation DNA repair Nucleotide excision repair Mismatch repair Cilia Downregulated (689 genes) DNA replication Homologous recombination Mismatch repair Peptidase inhibitor activity Carbohydrate binding Metabolism Calcium Growth factor Midbrain development Notochord development Differentiation homeobox |
45 21 39 94 41 21 49 12 13 48 9 7 7 25 22 19 20 10 12 23 |
GO GO STARS GO GO GO GO KEGG GO STARS KEGG KEGG KEGG GO GO STARS STARS GO GO STARS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





