Submitted:
18 August 2025
Posted:
19 August 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
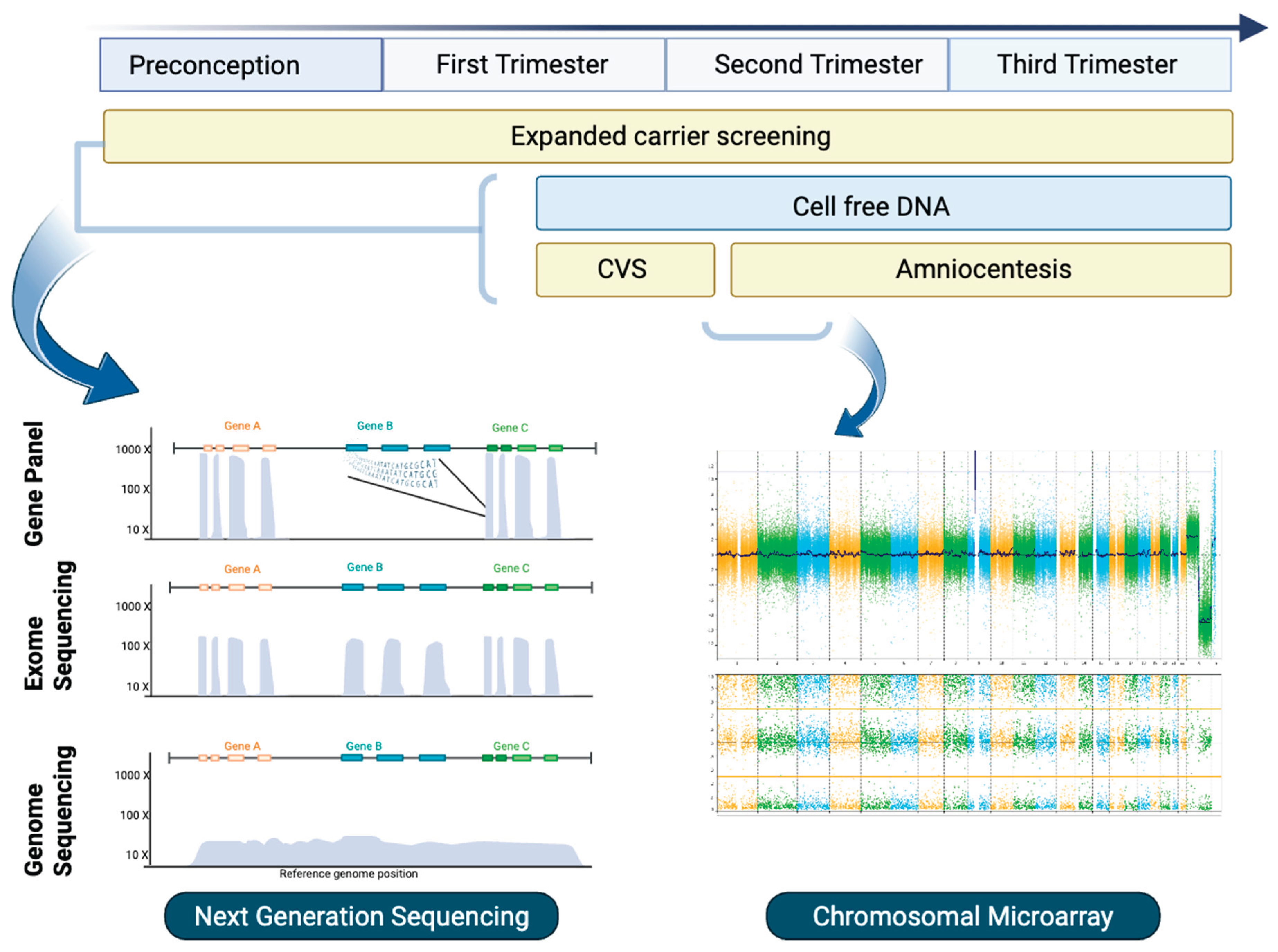
2. Expanded Carrier Screening
2.1. Overview of Carrier Screening
2.2. Technological Evolution, Challenges and Limitations
2.3. Guidelines and Ethical Considerations
2.4. Carrier Screening by Genome Sequencing
3. Cell-Free DNA
3.1. Overview of cfDNA Screening for Aneuploidies and Copy Number Variants
3.2. Maternal cfDNA Testing and Screening for Fetal Single Gene Disorders
3.3. Developments in cfDNA Analysis
4. Chromosomal Microarray and Copy Number Variant Detection
5. Diagnostic Sequencing
5.1. Targeted Gene Panels
5.2. Exome Sequencing
5.3. Genome Sequencing
5.4. Considerations for Data Interpretation in Prenatal Sequencing
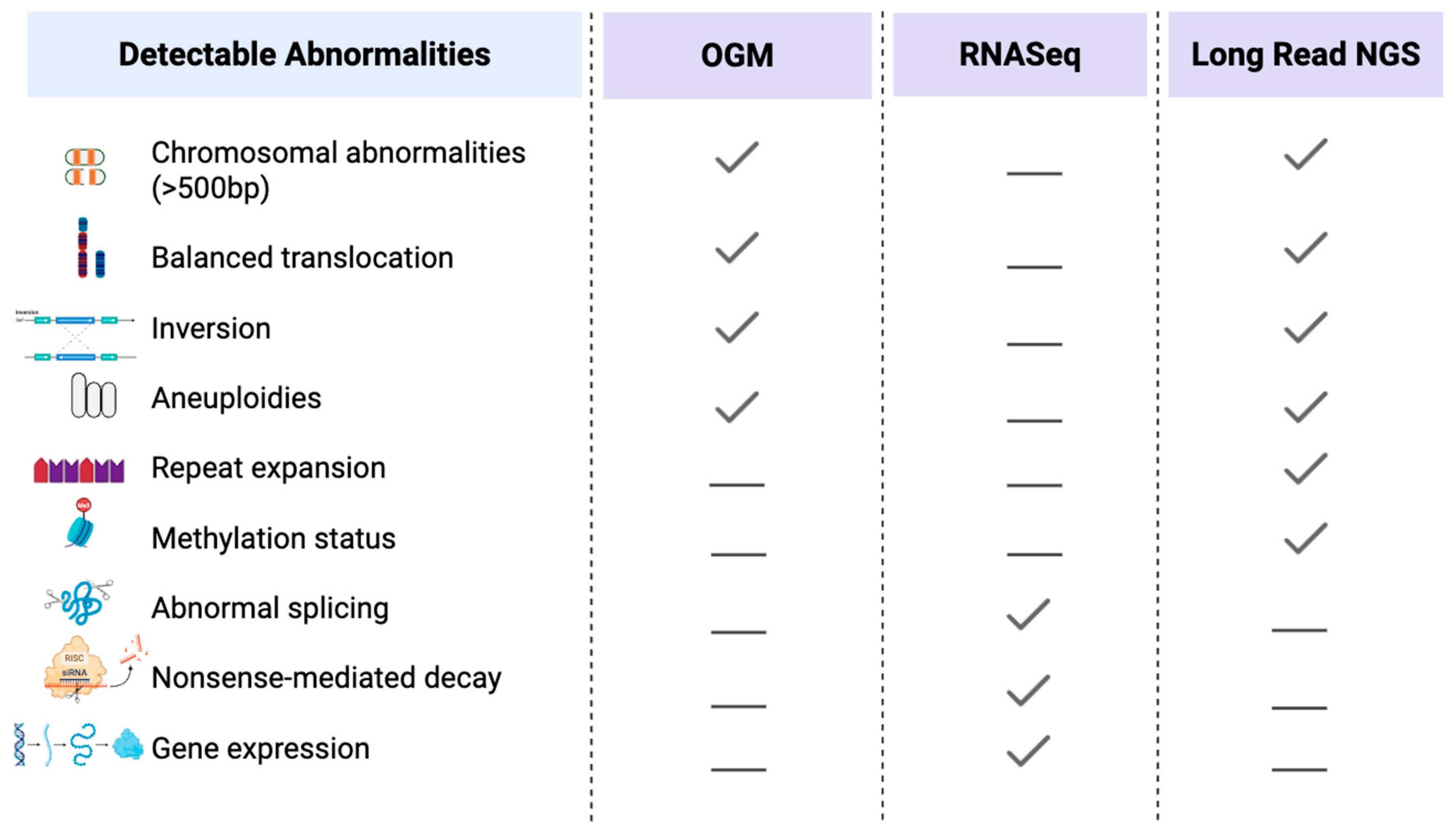
6. Emerging and Future Technologies
6.1. Optical Genome Mapping
6.2. RNA Sequencing
6.3. Long-Read Sequencing
6.4. Cell-Based Noninvasive Prenatal Testing
7. Conclusions
Acknowledgments
References
- Mardis, E.R. , Next-generation sequencing platforms. Annu Rev Anal Chem (Palo Alto Calif) 2013, 6, 287–303. [Google Scholar] [CrossRef]
- Gregg, A.R. , et al., Screening for autosomal recessive and X-linked conditions during pregnancy and preconception: a practice resource of the American College of Medical Genetics and Genomics (ACMG). Genet Med 2021, 23, 1793–1806. [Google Scholar] [CrossRef]
- Rose, N.C. , et al., Systematic evidence-based review: The application of noninvasive prenatal screening using cell-free DNA in general-risk pregnancies. Genet Med 2022, 24, 1379–1391. [Google Scholar] [CrossRef]
- van der Meij, K.R.M. , et al., TRIDENT-2: National Implementation of Genome-wide Non-invasive Prenatal Testing as a First-Tier Screening Test in the Netherlands. Am J Hum Genet 2019, 105, 1091–1101. [Google Scholar] [CrossRef] [PubMed]
- Dungan, J.S. , et al., Noninvasive prenatal screening (NIPS) for fetal chromosome abnormalities in a general-risk population: An evidence-based clinical guideline of the American College of Medical Genetics and Genomics (ACMG). Genet Med 2023, 25, 100336. [Google Scholar] [CrossRef]
- Mohan, P. , et al., Clinical experience with non-invasive prenatal screening for single-gene disorders. Ultrasound Obstet Gynecol 2022, 59, 33–39. [Google Scholar] [CrossRef]
- Wapner, R.J. , et al., Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med 2012, 367, 2175–2184. [Google Scholar] [CrossRef]
- Chau, M.H.K. , et al., Low-pass genome sequencing: a validated method in clinical cytogenetics. Hum Genet 2020, 139, 1403–1415. [Google Scholar] [CrossRef]
- Mellis, R. , et al., Diagnostic yield of exome sequencing for prenatal diagnosis of fetal structural anomalies: A systematic review and meta-analysis. Prenat Diagn 2022, 42, 662–685. [Google Scholar] [CrossRef]
- Richards, S. , et al., Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015, 17, 405–424. [Google Scholar] [CrossRef]
- American College of Obstetricians and Gynecologists, Carrier screening for genetic conditions. Committee opinion no. 691. Obstet Gynecol 2017, 129, e41–e55. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.G. , et al., Expanded carrier screening in reproductive medicine-points to consider: a joint statement of the American College of Medical Genetics and Genomics, American College of Obstetricians and Gynecologists, National Society of Genetic Counselors, Perinatal Quality Foundation, and Society for Maternal-Fetal Medicine. Obstet Gynecol 2015, 125, 653–662. [Google Scholar] [PubMed]
- Gregg, A.R. , Expanded Carrier Screening. Obstet Gynecol Clin North Am 2018, 45, 103–112. [Google Scholar] [CrossRef] [PubMed]
- National Human Genome Research Institute. The Cost of Sequencing a Human Genome. 2023 [cited 2025 ]; Available from: https://www.genome.gov/about-genomics/fact-sheets/Sequencing-Human-Genome-cost. 23 May.
- Guo M.H., G. A., Carrier Screening for Genetic Conditions, in Perinatal Genetics, M.E. Norton, Kuller, J.A., Dugoff, L., Editor. 2019, ELSEVIER.
- Rehder, C. , et al., Next-generation sequencing for constitutional variants in the clinical laboratory, 2021 revision: a technical standard of the American College of Medical Genetics and Genomics (ACMG). Genet Med 2021, 23, 1399–1415. [Google Scholar] [CrossRef]
- Fridman, H. , et al., Preconception carrier screening yield: effect of variants of unknown significance in partners of carriers with clinically significant variants. Genet Med 2020, 22, 646–653. [Google Scholar] [CrossRef]
- Beauchamp, K.A. , et al., Systematic design and comparison of expanded carrier screening panels. Genet Med 2018, 20, 55–63. [Google Scholar] [CrossRef]
- Ben-Shachar, R. , et al., A data-driven evaluation of the size and content of expanded carrier screening panels. Genet Med 2019, 21, 1931–1939. [Google Scholar] [CrossRef]
- Guo, M.H. and A.R. Gregg, Estimating yields of prenatal carrier screening and implications for design of expanded carrier screening panels. Genet Med 2019, 21, 1940–1947. [Google Scholar] [CrossRef]
- Chokoshvili, D., D. F. Vears, and P. Borry, Growing complexity of (expanded) carrier screening: Direct-to-consumer, physician-mediated, and clinic-based offers. Best Pract Res Clin Obstet Gynaecol 2017, 44, 57–67. [Google Scholar] [CrossRef]
- Haque, I.S. , et al., Modeled Fetal Risk of Genetic Diseases Identified by Expanded Carrier Screening. Jama 2016, 316, 734–742. [Google Scholar] [CrossRef]
- Lazarin, G.A. , et al., An empirical estimate of carrier frequencies for 400+ causal Mendelian variants: results from an ethnically diverse clinical sample of 23,453 individuals. Genet Med 2013, 15, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Westemeyer, M. , et al., Clinical experience with carrier screening in a general population: support for a comprehensive pan-ethnic approach. Genet Med 2020, 22, 1320–1328. [Google Scholar] [CrossRef] [PubMed]
- Rink, B.D. , Informed consent for expanded carrier screening: Past, present, and future. Prenatal Diagnosis 2023, 43, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Nussbaum, R.L., R. N. Slotnick, and N.J. Risch, Challenges in providing residual risks in carrier testing. Prenatal Diagnosis 2021, 41, 1049–1056. [Google Scholar] [CrossRef]
- Yang, Y. , et al., Applications of genome sequencing as a single platform for clinical constitutional genetic testing. Genet Med Open 2024, 2, 101840. [Google Scholar] [CrossRef]
- Punj, S. , et al., Preconception Carrier Screening by Genome Sequencing: Results from the Clinical Laboratory. Am J Hum Genet 2018, 102, 1078–1089. [Google Scholar] [CrossRef]
- Shamseldin, H.E. , et al., Molecular autopsy in maternal-fetal medicine. Genet Med 2018, 20, 420–427. [Google Scholar] [CrossRef]
- Fan, H.C. , et al., Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci U S A 2008, 105, 16266–16271. [Google Scholar] [CrossRef]
- Palomaki, G.E. , et al., DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genet Med 2011, 13, 913–920. [Google Scholar] [CrossRef]
- American College of Obstetricians and Gynecologists, Screening for Fetal Chromosomal Abnormalities: ACOG Practice Bulletin, Number 226. Obstet Gynecol 2020, 136, e48–e69. [CrossRef]
- Lo, Y.M. , et al., Presence of fetal DNA in maternal plasma and serum. Lancet 1997, 350, 485–487. [Google Scholar] [CrossRef] [PubMed]
- Wang, E. , et al., Gestational age and maternal weight effects on fetal cell-free DNA in maternal plasma. Prenat Diagn 2013, 33, 662–666. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, D.W. and R.W.K. Chiu, Sequencing of Circulating Cell-free DNA during Pregnancy. N Engl J Med 2018, 379, 464–473. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.J. , et al., Noninvasive prenatal diagnosis of fetal trisomy 21 by allelic ratio analysis using targeted massively parallel sequencing of maternal plasma DNA. PLoS One 2012, 7, e38154. [Google Scholar] [CrossRef]
- Nicolaides, K.H. , et al., Validation of targeted sequencing of single-nucleotide polymorphisms for non-invasive prenatal detection of aneuploidy of chromosomes 13, 18, 21, X, and Y. Prenat Diagn 2013, 33, 575–579. [Google Scholar] [CrossRef]
- Dugoff, L. , et al., Cell-free DNA screening for trisomy 21 in twin pregnancy: a large multicenter cohort study. Am J Obstet Gynecol 2023, 229, e1–e435. [Google Scholar] [CrossRef]
- Wang, Y. , et al., Noninvasive Evaluation of Fetal Zygosity in Twin Pregnancies Involving a Binary Analysis of Single-Nucleotide Polymorphisms. J Mol Diagn 2023, 25, 682–691. [Google Scholar] [CrossRef]
- van Riel, M. , et al., Performance and Diagnostic Value of Genome-Wide Noninvasive Prenatal Testing in Multiple Gestations. Obstet Gynecol 2021, 137, 1102–1108. [Google Scholar] [CrossRef]
- Ehrich, M. , et al., Genome-wide cfDNA screening: clinical laboratory experience with the first 10,000 cases. Genet Med 2017, 19, 1332–1337. [Google Scholar] [CrossRef]
- Grati, F.R. , et al., Prevalence of recurrent pathogenic microdeletions and microduplications in over 9500 pregnancies. Prenat Diagn 2015, 35, 801–809. [Google Scholar] [CrossRef]
- Dar, P. , et al., Cell-free DNA screening for prenatal detection of 22q11.2 deletion syndrome. Am J Obstet Gynecol 2022, 227, e1–e79. [Google Scholar] [CrossRef] [PubMed]
- Acreman, M.L. , et al., The predictive value of prenatal cell-free DNA testing for rare autosomal trisomies: a systematic review and meta-analysis. Am J Obstet Gynecol 2023, 228, 292–305.e6. [Google Scholar] [CrossRef] [PubMed]
- van Prooyen Schuurman, L. , et al., Clinical impact of additional findings detected by genome-wide non-invasive prenatal testing: Follow-up results of the TRIDENT-2 study. Am J Hum Genet 2022, 109, 1140–1152. [Google Scholar] [CrossRef] [PubMed]
- Eggenhuizen, G.M. , et al., Confined placental mosaicism and the association with pregnancy outcome and fetal growth: a review of the literature. Hum Reprod Update 2021, 27, 885–903. [Google Scholar] [CrossRef] [PubMed]
- Chitty, L.S., L. Hudgins, and M.E. Norton, Current controversies in prenatal diagnosis 2: Cell-free DNA prenatal screening should be used to identify all chromosome abnormalities. Prenat Diagn 2018, 38, 160–165. [Google Scholar] [CrossRef]
- Grati, F.R. , et al., Implications of fetoplacental mosaicism on cell-free DNA testing for sex chromosome aneuploidies. Prenat Diagn 2017, 37, 1017–1027. [Google Scholar] [CrossRef]
- Chitty, L.S. , Advances in the prenatal diagnosis of monogenic disorders. Prenat Diagn 2018, 38, 3–5. [Google Scholar] [CrossRef]
- Liao, J. , et al., Advances in Prenatal Cell-Free DNA Screening for Dominant Monogenic Conditions: A Review of Current Progress and Future Directions in Clinical Implementation. Prenat Diagn 2025, 45, 445–452. [Google Scholar] [CrossRef]
- Hayward, J. and L.S. Chitty, Beyond screening for chromosomal abnormalities: Advances in non-invasive diagnosis of single gene disorders and fetal exome sequencing. Semin Fetal Neonatal Med 2018, 23, 94–101. [Google Scholar] [CrossRef]
- Zhang, J. , et al., Non-invasive prenatal sequencing for multiple Mendelian monogenic disorders using circulating cell-free fetal DNA. Nat Med 2019, 25, 439–447. [Google Scholar] [CrossRef]
- Adams, S. , et al., Routine Prenatal cfDNA Screening for Autosomal Dominant Single-Gene Conditions. Clinical Chemistry 2025, 71, 129–140. [Google Scholar] [CrossRef]
- Rego, S. , et al., Cell-Free DNA Analysis for the Determination of Fetal Red Blood Cell Antigen Genotype in Individuals With Alloimmunized Pregnancies. Obstet Gynecol 2024, 144, 436–443. [Google Scholar] [CrossRef] [PubMed]
- Grace, M.R., B. Goodhue, and N.L. Vora, Rho(D) immune globulin shortage and fetal Rh(D) screening with cell-free DNA. Curr Opin Obstet Gynecol 2025, 37, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Hoskovec, J. , et al., Maternal carrier screening with single-gene NIPS provides accurate fetal risk assessments for recessive conditions. Genet Med 2023, 25, 100334. [Google Scholar] [CrossRef] [PubMed]
- Wynn, J. , et al., Performance of single-gene noninvasive prenatal testing for autosomal recessive conditions in a general population setting. Prenat Diagn 2023, 43, 1344–1354. [Google Scholar] [CrossRef]
- Lo, Y.M. , et al., Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med 2010, 2. [Google Scholar] [CrossRef]
- Yu, S.C. , et al., Size-based molecular diagnostics using plasma DNA for noninvasive prenatal testing. Proc Natl Acad Sci U S A 2014, 111, 8583–8588. [Google Scholar] [CrossRef]
- Yu, S.C.Y. , et al., Combined Count- and Size-Based Analysis of Maternal Plasma DNA for Noninvasive Prenatal Detection of Fetal Subchromosomal Aberrations Facilitates Elucidation of the Fetal and/or Maternal Origin of the Aberrations. Clinical Chemistry 2017, 63, 495–502. [Google Scholar] [CrossRef]
- Yang, Q. , et al., Size-selective separation and overall-amplification of cell-free fetal DNA fragments using PCR-based enrichment. Sci Rep 2017, 7, 40936. [Google Scholar] [CrossRef]
- Acevedo, A. , et al., Fetal fraction amplification within prenatal cfDNA screening enables detection of genome-wide copy-number variants at enhanced resolution. Genet Med 2025, 27, 101269. [Google Scholar] [CrossRef]
- Welker, N.C. , et al., High-throughput fetal fraction amplification increases analytical performance of noninvasive prenatal screening. Genet Med 2021, 23, 443–450. [Google Scholar] [CrossRef]
- Brand, H. , et al., High-Resolution and Noninvasive Fetal Exome Screening. N Engl J Med 2023, 389, 2014–2016. [Google Scholar] [CrossRef]
- Miceikaite, I. , et al., Comprehensive prenatal diagnostics: Exome versus genome sequencing. Prenat Diagn 2023, 43, 1132–1141. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, D.W. , et al., Noninvasive Prenatal Testing and Incidental Detection of Occult Maternal Malignancies. Jama 2015, 314, 162–169. [Google Scholar] [CrossRef]
- Ji, X. , et al., Identifying occult maternal malignancies from 1.93 million pregnant women undergoing noninvasive prenatal screening tests. Genet Med 2019, 21, 2293–2302. [Google Scholar] [CrossRef] [PubMed]
- Turriff Amy, E. , et al., Prenatal cfDNA Sequencing and Incidental Detection of Maternal Cancer. New England Journal of Medicine 2024, 391, 2123–2132. [Google Scholar] [CrossRef]
- Shao, L. , et al., Chromosomal microarray analysis, including constitutional and neoplastic disease applications, 2021 revision: a technical standard of the American College of Medical Genetics and Genomics (ACMG). Genet Med 2021, 23, 1818–1829. [Google Scholar] [CrossRef]
- Elron, E. , et al., The Diagnostic Yield of Chromosomal Microarray Analysis in Third-Trimester Fetal Abnormalities. Am J Perinatol 2024, 41, 2232–2242. [Google Scholar]
- Armour, C.M. , et al., Practice guideline: joint CCMG-SOGC recommendations for the use of chromosomal microarray analysis for prenatal diagnosis and assessment of fetal loss in Canada. J Med Genet 2018, 55, 215–221. [Google Scholar] [CrossRef]
- Mitrakos, A. , et al., Prenatal Chromosomal Microarray Analysis: Does Increased Resolution Equal Increased Yield? Genes (Basel).
- Dong, Z. , et al., Low-pass whole-genome sequencing in clinical cytogenetics: a validated approach. Genet Med 2016, 18, 940–948. [Google Scholar] [CrossRef]
- Monaghan, K.G. , et al., The use of fetal exome sequencing in prenatal diagnosis: a points to consider document of the American College of Medical Genetics and Genomics (ACMG). Genet Med 2020, 22, 675–680. [Google Scholar] [CrossRef]
- Wang, H. , et al., Low-pass genome sequencing versus chromosomal microarray analysis: implementation in prenatal diagnosis. Genet Med 2020, 22, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X. , et al., Prenatal diagnosis of skeletal dysplasias using a targeted skeletal gene panel. Prenat Diagn 2018, 38, 692–699. [Google Scholar] [CrossRef]
- Scott, A. , et al., When to test fetuses for RASopathies? Proposition from a systematic analysis of 352 multicenter cases and a postnatal cohort. Genet Med 2021, 23, 1116–1124. [Google Scholar] [CrossRef] [PubMed]
- Norton, M.E. , et al., Exome sequencing vs targeted gene panels for the evaluation of nonimmune hydrops fetalis. Am J Obstet Gynecol 2022, 226, 128–e1. [Google Scholar] [CrossRef]
- Yaldiz, B. , et al., Twist exome capture allows for lower average sequence coverage in clinical exome sequencing. Hum Genomics 2023, 17, 39. [Google Scholar] [CrossRef]
- Mellis, R. , et al., Fetal exome sequencing for isolated increased nuchal translucency: should we be doing it? BJOG 2022, 129, 52–61. [Google Scholar] [CrossRef]
- Pauta, M., R. J. Martinez-Portilla, and A. Borrell, Diagnostic yield of exome sequencing in fetuses with multisystem malformations: systematic review and meta-analysis. Ultrasound Obstet Gynecol 2022, 59, 715–722. [Google Scholar] [CrossRef]
- Mone, F. , et al., Should we offer prenatal exome sequencing for intrauterine growth restriction or short long bones? A systematic review and meta-analysis. Am J Obstet Gynecol 2023, 228, 409–417. [Google Scholar] [CrossRef]
- Lord, J. , et al., Prenatal exome sequencing analysis in fetal structural anomalies detected by ultrasonography (PAGE): a cohort study. Lancet 2019, 393, 747–757. [Google Scholar] [CrossRef]
- Petrovski, S. , et al., Whole-exome sequencing in the evaluation of fetal structural anomalies: a prospective cohort study. Lancet 2019, 393, 758–767. [Google Scholar] [CrossRef]
- Van den Veyver, I.B. , et al., International Society for Prenatal Diagnosis Updated Position Statement on the use of genome-wide sequencing for prenatal diagnosis. Prenat Diagn 2022, 42, 796–803. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, M.H. , et al., Genome Sequencing for Diagnosing Rare Diseases. N Engl J Med 2024, 390, 1985–1997. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J. , et al., Whole Genome Sequencing in the Evaluation of Fetal Structural Anomalies: A Parallel Test with Chromosomal Microarray Plus Whole Exome Sequencing. Genes (Basel).
- Westenius, E. , et al., Whole-genome sequencing in prenatally detected congenital malformations: prospective cohort study in clinical setting. Ultrasound Obstet Gynecol 2024, 63, 658–663. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, S. , et al., Exome sequencing for perinatal phenotypes: The significance of deep phenotyping. Prenat Diagn 2020, 40, 260–273. [Google Scholar] [CrossRef]
- Dhombres, F. , et al., Prenatal phenotyping: A community effort to enhance the Human Phenotype Ontology. Am J Med Genet C Semin Med Genet 2022, 190, 231–242. [Google Scholar] [CrossRef]
- Van den Veyver, I.B., Y. Yaron, and Z.C. Deans, International Society for Prenatal Diagnosis 2022 debate 3-Fetal genome sequencing should be offered to all pregnant patients. Prenat Diagn 2023, 43, 428–434. [Google Scholar] [CrossRef]
- Giordano, J.L. and R.J. Wapner, The fetal sequencing consortium: The value of multidisciplinary dialog and collaboration. Prenat Diagn 2022, 42, 807–810. [Google Scholar] [CrossRef]
- Jeanne, M. and W.K. Chung, Prenatal genomic sequencing: Navigating uncertainty. Semin Perinatol 2025, 49, 152058. [Google Scholar] [CrossRef]
- Lee, K. , et al., ACMG SF v3.3 list for reporting of secondary findings in clinical exome and genome sequencing: A policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med 2025, 27, 101454. [Google Scholar] [CrossRef]
- Dremsek, P. , et al., Optical Genome Mapping in Routine Human Genetic Diagnostics-Its Advantages and Limitations. Genes (Basel).
- Levy, B. , et al., Multisite Evaluation and Validation of Optical Genome Mapping for Prenatal Genetic Testing. J Mol Diagn 2024, 26, 906–916. [Google Scholar] [CrossRef]
- Liu, P. and L. Vossaert, Emerging technologies for prenatal diagnosis: The application of whole genome and RNA sequencing. Prenat Diagn 2022, 42, 686–696. [Google Scholar] [CrossRef]
- Truty, R. , et al., Spectrum of splicing variants in disease genes and the ability of RNA analysis to reduce uncertainty in clinical interpretation. Am J Hum Genet 2021, 108, 696–708. [Google Scholar] [CrossRef]
- Wai, H.A. , et al., Correction: Blood RNA analysis can increase clinical diagnostic rate and resolve variants of uncertain significance. Genet Med 2020, 22, 1129. [Google Scholar] [CrossRef]
- Mahmoud, M. , et al., Utility of long-read sequencing for All of Us. Nat Commun 2024, 15, 837. [Google Scholar] [CrossRef] [PubMed]
- Long, J. , et al., Evaluating the clinical efficacy of a long-read sequencing-based approach for carrier screening of spinal muscular atrophy. Hum Genomics 2024, 18, 110. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. , et al., Comprehensive SMN1 and SMN2 profiling for spinal muscular atrophy analysis using long-read PacBio HiFi sequencing. Am J Hum Genet 2023, 110, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y. , et al., Comprehensive Analysis of Hemophilia A (CAHEA): Towards Full Characterization of the F8 Gene Variants by Long-Read Sequencing. Thromb Haemost 2023, 123, 1151–1164. [Google Scholar]
- Wang, R. , et al., Long-Read Sequencing Solves Complex Structure of CYP21A2 in a Large 21-Hydroxylase Deficiency Cohort. J Clin Endocrinol Metab 2025, 110, 406–416. [Google Scholar] [CrossRef]
- Yu, S.C.Y., L. Y.L. Choy, and Y.M.D. Lo, ‘Longing’ for the Next Generation of Liquid Biopsy: The Diagnostic Potential of Long Cell-Free DNA in Oncology and Prenatal Testing. Mol Diagn Ther 2023, 27, 563–571. [Google Scholar] [CrossRef]
- Yu, S.C.Y. , et al., Single-molecule sequencing reveals a large population of long cell-free DNA molecules in maternal plasma. Proc Natl Acad Sci U S A.
- Vong, J.S.L. , et al., Enrichment of fetal and maternal long cell-free DNA fragments from maternal plasma following DNA repair. Prenat Diagn 2019, 39, 88–99. [Google Scholar] [CrossRef]
- Maktabi, M.A., L. Vossaert, and I.B. Van den Veyver, Cell-based Noninvasive Prenatal Testing (cbNIPT)-A Review on the Current Developments and Future Prospects. Clin Obstet Gynecol 2023, 66, 636–648. [Google Scholar] [CrossRef]
- Jeppesen, L.D. , et al., Clinical interpretation of cell-based non-invasive prenatal testing for monogenic disorders including repeat expansion disorders: potentials and pitfalls. Front Genet 2023, 14, 1188472. [Google Scholar] [CrossRef] [PubMed]
- Vossaert, L. , et al., Validation Studies for Single Circulating Trophoblast Genetic Testing as a Form of Noninvasive Prenatal Diagnosis. Am J Hum Genet 2019, 105, 1262–1273. [Google Scholar] [CrossRef] [PubMed]
- Imudia, A.N. , et al., Retrieval of trophoblast cells from the cervical canal for prediction of abnormal pregnancy: a pilot study. Hum Reprod 2009, 24, 2086–2092. [Google Scholar] [CrossRef] [PubMed]
- Kadam, L. , et al., Endocervical trophoblast for interrogating the fetal genome and assessing pregnancy health at five weeks. Eur J Med Genet 2019, 62, 103690. [Google Scholar] [CrossRef]
- Jou, H.J., P. H. Lo, and P.Y. Ling, Recent Advances of Microfluidic Platform for Cell Based Non-Invasive Prenatal Diagnosis. Int J Mol Sci.
- Li, X. , et al., Aptamer-Mediated Enrichment of Rare Circulating Fetal Nucleated Red Blood Cells for Noninvasive Prenatal Diagnosis. Anal Chem 2023, 95, 5419–5427. [Google Scholar] [CrossRef]
- Feng, C. , et al., The path winds along isolation and analyses of fetal nucleated red blood cells in maternal peripheral blood: Past, present, and future toward non-invasive prenatal diagnosis. Life Sci 2025, 369, 123530. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




