Submitted:
28 July 2025
Posted:
30 July 2025
You are already at the latest version
Abstract
Interferon regulatory factor 5 (IRF5) plays a pivotal role in innate immune responses and macrophage polarization. Although its role in obesity-associated inflammation has been described, sex-specific differences in adipose IRF5 expression and its association with immune and metabolic markers remain poorly defined. To evaluate sex-specific associations between adipose tissue IRF5 expression and key inflammatory and metabolic markers in overweight and obese individuals. Subcutaneous adipose tissue samples from overweight/obese male and female subjects were analyzed for IRF5 expression using quantitative RT-PCR. Correlation and multiple linear regression analyses were performed to identify associations with inflammatory gene expression and metabolic parameters including insulin, glucose, HOMA-IR, and adipokines. RF5 gene and protein levels were significantly elevated in the AT of overweight/obese females compared to males (p < 0.0001), with expression increasing progressively with BMI in females but not in males. Despite these sex-dependent expression levels, IRF5 demonstrated consistent, sex-independent positive correlations with several core immune and inflammatory markers, including CCR5, CD11c, CD16, CD163, FOXP3, RUNX1, and MyD88. However, distinct sex-specific patterns emerged: in males, IRF5 correlated positively with classical pro-inflammatory markers such as IL-2, IL-6, IL-8, TNF-α, and IRAK1; whereas in females, IRF5 was associated with a broader array of immune markers, including chemokines (CCL7, CXCL11), pattern recognition receptors (TLR2, TLR8, TLR9), and macrophage markers (CD68, CD86), along with anti-inflammatory mediators such as IL-10 and IRF4. Notably, IRF5 expression in overweight/obese males, but not females, was significantly associated with metabolic dysfunction, showing positive correlations with fasting blood glucose, HbA1c, insulin, and HOMA-IR levels. Multiple regression analyses revealed sex-specific predictors of IRF5 expression, with metabolic (HOMA-IR) and inflammatory (IRAK1, MyD88) markers emerging in males, while immune-related genes (RUNX1, CD68, CCL7, MyD88) predominated in females. These findings underscore a sex-divergent role of IRF5 in AT, with implications for differential regulation of immune-metabolic pathways in obesity and its complications.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Anthropometric and Physio-Clinical Measurements
2.3. Adipose Tissue Collection and Processing
2.4. Real-Time Reverse-Transcription Polymerase Chain Reaction (RT-PCR)
2.5. Immunofluorescence Microscopy
2.6. Statistical Analysis
3. Results
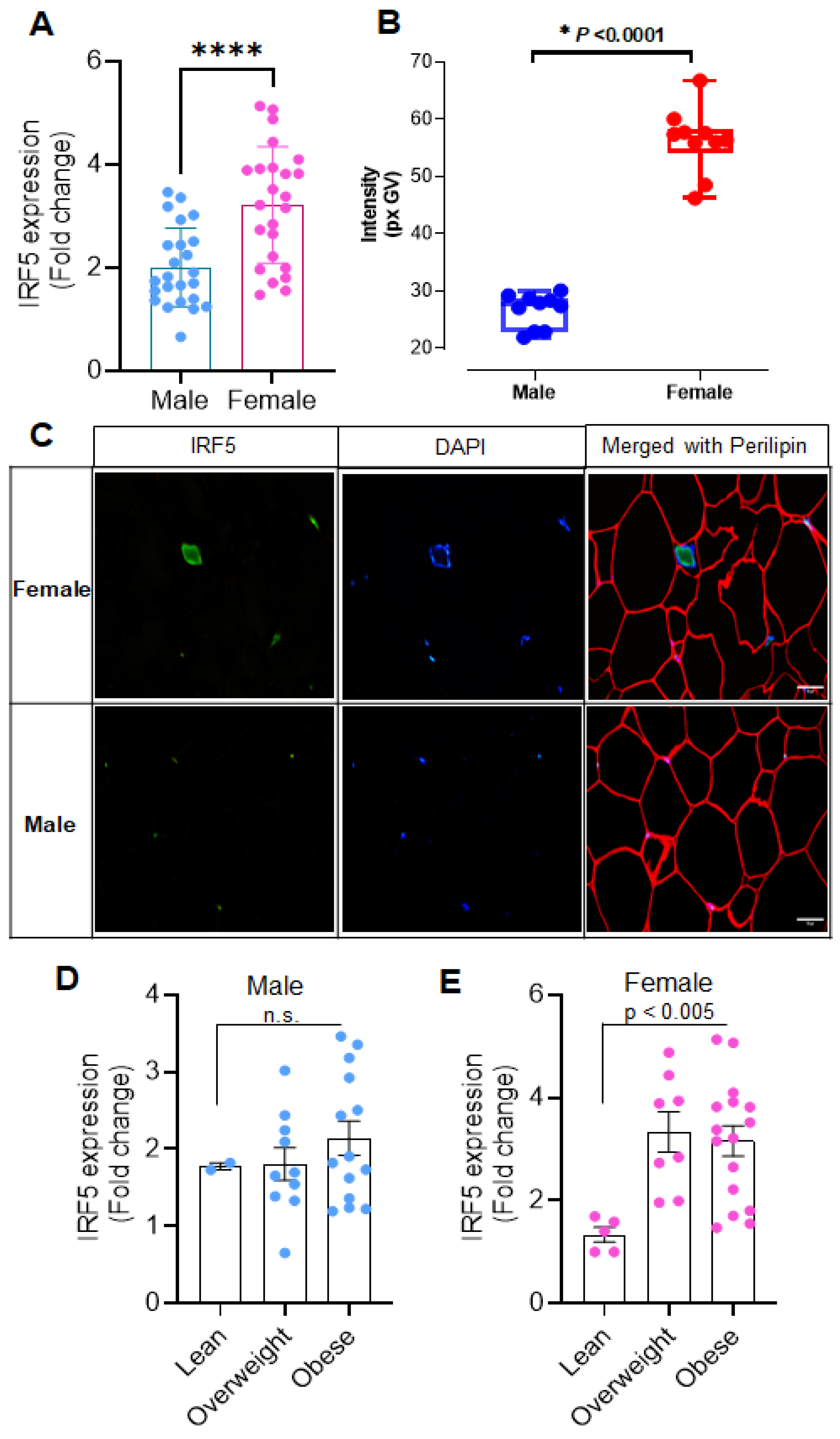
3.1. Sex-Related Differences in Adipose IRF5 Expression
3.2. Sex-Independent Correlation of IRF5 with Core Immune and Inflammatory Markers
3.3. Sex-Specific Associations of AT IRF5 with Inflammatory Markers
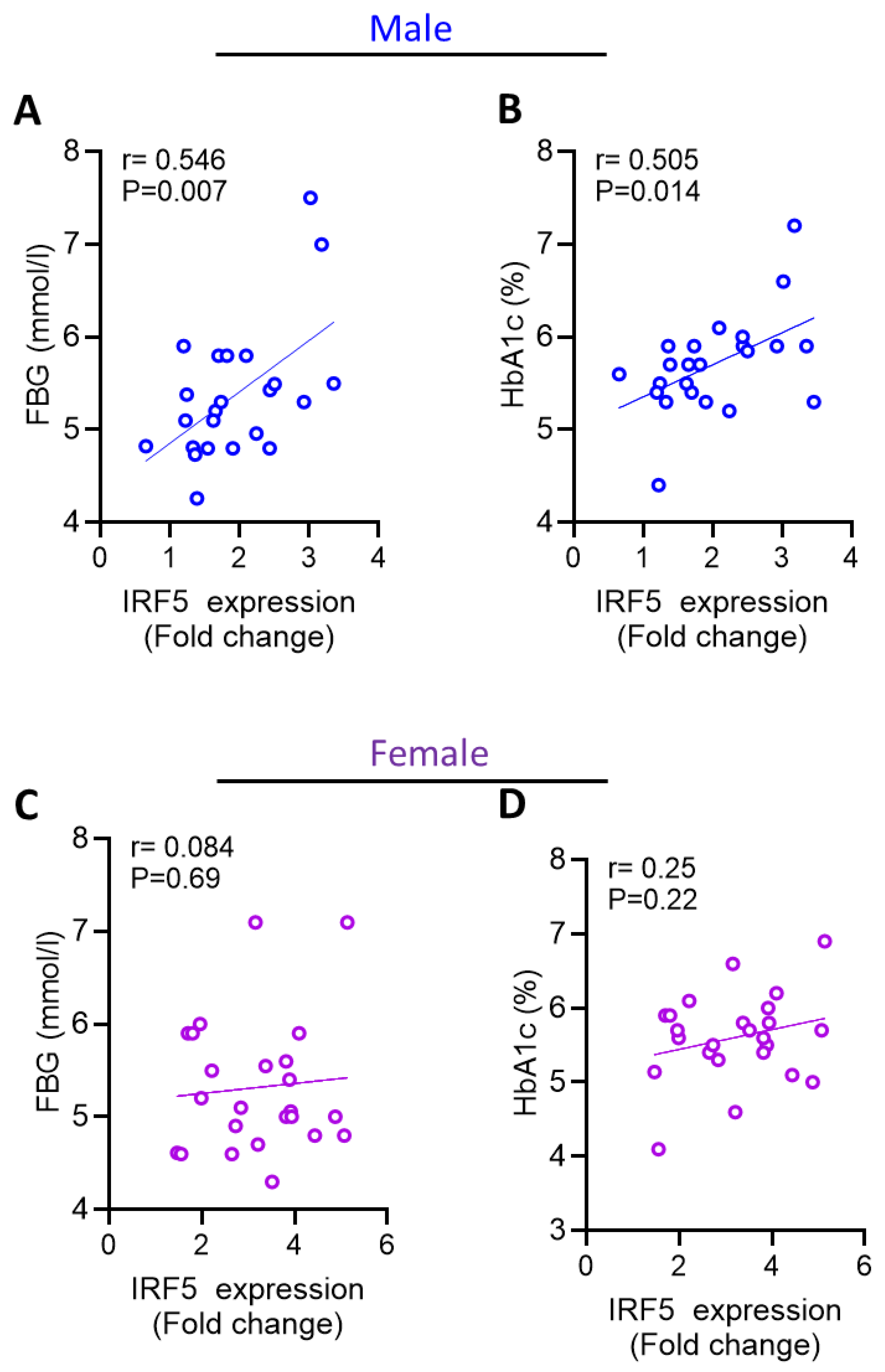
3.4. Increased AT IRF5 Gene Expression in Obesity Correlates with Diabetes Markers in males
3.5. Independent Predictors of IRF5 Expression
4. Discussion
5. Conclusion
6. Limitations and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alzaid, F.; Fagherazzi, G.; Riveline, J.P.; Bahman, F.; Al-Rashed, F.; Al-Mulla, F.; Ahmad, R. Immune cell-adipose tissue crosstalk in metabolic diseases with a focus on type 1 diabetes. Diabetologia 2025, 68, 1616–1631. [Google Scholar] [CrossRef]
- Tramunt, B.; Smati, S.; Grandgeorge, N.; Lenfant, F.; Arnal, J.F.; Montagner, A.; Gourdy, P. Sex differences in metabolic regulation and diabetes susceptibility. Diabetologia 2020, 63, 453–461. [Google Scholar] [CrossRef]
- Karastergiou, K.; Smith, S.R.; Greenberg, A.S.; Fried, S.K. Sex differences in human adipose tissues - the biology of pear shape. Biology of sex differences 2012, 3, 13. [Google Scholar] [CrossRef]
- Dalmas E, Toubal A, Alzaid F, Blazek K, Eames HL, Lebozec K, Pini M, Hainault I, Montastier E, Denis RG et al: Irf5 deficiency in macrophages promotes beneficial adipose tissue expansion and insulin sensitivity during obesity. Nature medicine 2015, 21, 610–618. [CrossRef]
- Krausgruber, T.; Blazek, K.; Smallie, T.; Alzabin, S.; Lockstone, H.; Sahgal, N.; Hussell, T.; Feldmann, M.; Udalova, I.A. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol 2011, 12, 231–238. [Google Scholar] [CrossRef]
- Griesbeck, M.; Ziegler, S.; Laffont, S.; Smith, N.; Chauveau, L.; Tomezsko, P.; Sharei, A.; Kourjian, G.; Porichis, F.; Hart Met, a.l. Sex Differences in Plasmacytoid Dendritic Cell Levels of IRF5 Drive Higher IFN-α Production in Women. The Journal of Immunology 2015, 195, 5327–5336. [Google Scholar] [CrossRef]
- Sindhu, S.; Thomas, R.; Shihab, P.; Sriraman, D.; Behbehani, K.; Ahmad, R. Obesity Is a Positive Modulator of IL-6R and IL-6 Expression in the Subcutaneous Adipose Tissue: Significance for Metabolic Inflammation. PLoS One 2015, 10, e0133494. [Google Scholar] [CrossRef]
- Kochumon, S.; Al-Rashed, F.; Abu-Farha, M.; Devarajan, S.; Tuomilehto, J.; Ahmad, R. Adipose tissue expression of CCL19 chemokine is positively associated with insulin resistance. Diabetes Metab Res Rev 2019, 35, e3087. [Google Scholar] [CrossRef]
- Albeloushi, S.; Hasan, A.; Arefanian, H.; Sindhu, S.; Al-Rashed, F.; Kochumon, S.; Abukhalaf, N.; Jacob, T.; Shenouda, S.; Al Madhoun Aet, a.l. Differential effects of fish-oil and cocoa-butter based high-fat/high-sucrose diets on endocrine pancreas morphology and function in mice. Frontiers in endocrinology 2024, 15, 1265799. [Google Scholar] [CrossRef]
- Albeloushi, S.M. Effect of Branched-Chain Amino Acid Supplements on Pancreatic Development in Preterm Lambs. Ph.D. Thesis, The University of Auckland, 2022. [Google Scholar]
- Cizkova, K.; Foltynkova, T.; Gachechiladze, M.; Tauber, Z. Comparative Analysis of Immunohistochemical Staining Intensity Determined by Light Microscopy, ImageJ and QuPath in Placental Hofbauer Cells. Acta histochemica et cytochemica 2021, 54, 21–29. [Google Scholar] [CrossRef]
- Duplancic, R.; Kero, D. Novel approach for quantification of multiple immunofluorescent signals using histograms and 2D plot profiling of whole-section panoramic images. Scientific reports 2021, 11, 8619. [Google Scholar] [CrossRef]
- Iyer, S.S.; Cheng, G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Critical reviews in immunology 2012, 32, 23–63. [Google Scholar] [CrossRef]
- Eguchi, J.; Kong, X.; Tenta, M.; Wang, X.; Kang, S.; Rosen, E.D. Interferon regulatory factor 4 regulates obesity-induced inflammation through regulation of adipose tissue macrophage polarization. Diabetes 2013, 62, 3394–3403. [Google Scholar] [CrossRef]
- Saraiva, M.; O'Garra, A. The regulation of IL-10 production by immune cells. Nature reviews Immunology 2010, 10, 170–181. [Google Scholar] [CrossRef]
- Klein, S.L.; Flanagan, K.L. Sex differences in immune responses. Nature Reviews Immunology 2016, 16, 626–638. [Google Scholar] [CrossRef]
- Ghosh, A.R.; Bhattacharya, R.; Bhattacharya, S.; Nargis, T.; Rahaman, O.; Duttagupta, P.; Raychaudhuri, D.; Liu, C.S.; Roy, S.; Ghosh Pet, a.l. Adipose Recruitment and Activation of Plasmacytoid Dendritic Cells Fuel Metaflammation. Diabetes 2016, 65, 3440–3452. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nature immunology 2010, 11, 373–384. [Google Scholar] [CrossRef]
- Ahmad, Z.; Kahloan, W.; Rosen, E.D. Transcriptional control of metabolism by interferon regulatory factors. Nat Rev Endocrinol 2024, 20, 573–587. [Google Scholar] [CrossRef]
- Lumeng, C.N.; Saltiel, A.R. Inflammatory links between obesity and metabolic disease. The Journal of clinical investigation 2011, 121, 2111–2117. [Google Scholar] [CrossRef]
- de Bruijn, M.; Dzierzak, E. Runx transcription factors in the development and function of the definitive hematopoietic system. Blood 2017, 129, 2061–2069. [Google Scholar] [CrossRef]
- Regitz-Zagrosek, V.; Oertelt-Prigione, S.; Seeland, U.; Hetzer, R. Sex and gender differences in myocardial hypertrophy and heart failure. Circulation journal : official journal of the Japanese Circulation Society 2010, 74, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Lovejoy, J.C.; Sainsbury, A. Sex differences in obesity and the regulation of energy homeostasis. Obesity reviews : an official journal of the International Association for the Study of Obesity 2009, 10, 154–167. [Google Scholar] [CrossRef]
- Gannon, M.; Kulkarni, R.N.; Tse, H.M.; Mauvais-Jarvis, F. Sex differences underlying pancreatic islet biology and its dysfunction. Molecular metabolism 2018, 15, 82–91. [Google Scholar] [CrossRef]
- Bastard, J.P.; Maachi, M.; Lagathu, C.; Kim, M.J.; Caron, M.; Vidal, H.; Capeau, J.; Feve, B. Recent advances in the relationship between obesity, inflammation, and insulin resistance. European cytokine network 2006, 17, 4–12. [Google Scholar]
| Inflammatory Markers | Male (Overweight + Obese) | Female (Overweight + Obese) | ||
|---|---|---|---|---|
| r-value | p-value | r-value | p-value | |
| Cytokines | ||||
| IL2 | 0.413 | 0.045* | 0.277 | 0.212 |
| IL6 | 0.37 | 0.075 | 0.068 | 0.765 |
| IL8 | 0.458 | 0.032* | 0.208 | 0.352 |
| IL10 | 0.107 | 0.635 | 0.542 | 0.008** |
| IL33 | 0.148 | 0.489 | 0.114 | 0.613 |
| TNFα | 0.474 | 0.019* | 0.359 | 0.101 |
| TGFβ | 0.207 | 0.343 | 0.079 | 0.727 |
| CC and CXC chemokine ligands and receptors | ||||
| CCL2 | 0.251 | 0.236 | 0.197 | 0.355 |
| CCL5 | 0.394 | 0.077 | 0.265 | 0.273 |
| CCL7 | 0.255 | 0.241 | 0.53 | 0.009** |
| CCL19 | 0.368 | 0.084 | 0.195 | 0.396 |
| CXCL10 | -0.007 | 0.975 | 0.41 | 0.052 |
| CXCL11 | 0.137 | 0.534 | 0.431 | 0.040* |
| Dectin1 | 0.328 | 0.126 | 0.441 | 0.040* |
| CCR1 | 0.031 | 0.895 | 0.369 | 0.083 |
| CCR2 | 0.403 | 0.07 | 0.375 | 0.085 |
| CCR5 | 0.434 | 0.034* | 0.44 | 0.032* |
| Macrophage markers | ||||
| CD11c | 0.677 | 0.000** | 0.639 | 0.001** |
| CD16 | 0.578 | 0.003** | 0.44 | 0.040* |
| CD68 | 0.394 | 0.057 | 0.701 | 0.000** |
| CD86 | 0.195 | 0.385 | 0.637 | 0.001** |
| CD141 | 0.464 | 0.026* | 0.032 | 0.883 |
| CD163 | 0.711 | 0.000** | 0.534** | 0.009** |
| Toll-like receptors (TLRs) signaling cascade | ||||
| TLR2 | 0.108 | 0.633 | 0.549 | 0.012* |
| TLR4 | 0.385 | 0.094 | 0.257 | 0.262 |
| TLR8 | 0.169 | 0.431 | 0.751 | 0.000** |
| TLR9 | 0.355 | 0.089 | 0.432 | 0.035* |
| TLR10 | 0.019 | 0.931 | 0.442 | 0.051 |
| MyD88 | 0.495 | 0.014* | .584** | 0.003 |
| IRAK1 | 0.548 | 0.006** | 0.304 | 0.158 |
| TRAF6 | 0.087 | 0.688 | 0.087 | 0.687 |
| Transcription factors | ||||
| FOXP3 | 0.548 | 0.006** | 0.464 | 0.030* |
| RUNX1 | 0.507 | 0.011* | 0.681 | 0.000** |
| IRF4 | 0.11 | 0.626 | 0.414 | 0.044* |
| IRF3 | 0.383 | 0.096 | 0.265 | 0.246 |
| Metabolic Markers | Male (Overweight + Obese) | Female (Overweight + Obese) | ||
|---|---|---|---|---|
| r-value | p-value | r-value | p-value | |
| Weight | 0.366 | 0.079 | -0.119 | 0.579 |
| Height | 0.347 | 0.097 | -0.158 | 0.461 |
| BMI | 0.207 | 0.333 | -0.012 | 0.955 |
| PBF | 0.179 | 0.463 | 0.002 | 0.992 |
| Waist | 0.412 | 0.071 | -0.075 | 0.741 |
| Chol | 0.121 | 0.573 | 0.136 | 0.525 |
| HDL | -0.171 | 0.424 | -0.203 | 0.341 |
| LDL | 0.014 | 0.947 | 0.23 | 0.279 |
| TGL | 0.304 | 0.148 | -0.257 | 0.226 |
| FBG | 0.546 | 0.007** | 0.084 | 0.696 |
| HbA1C | 0.505 | 0.014* | 0.255 | 0.229 |
| Insulin | 0.617 | 0.014* | -0.363 | 0.167 |
| HOMAIR | 0.657 | 0.008** | -0.283 | 0.289 |
| Adiponectin | -0.543 | 0.068 | -0.371 | 0.13 |
| RANTES | 0.515 | 0.086 | 0.073 | 0.774 |
| ANOVA (Sig) R2 = 082; p < 0.0001 | ||||
|---|---|---|---|---|
| Predictor Variable | Male | Female | ||
| Scandalized Confinement (β) | p-Value | Scandalized Confinement (β) | p-Value | |
| IRAK1 | 0.57 | 0.006 | _ | _ |
| HOMA-IR | 7.548 | 0.001 | _ | _ |
| MyD88 | 0.512 | 0.003 | 0.288 | 0.030 |
| RUNX1 | _ | _ | 0.399 | 0.005 |
| CD68 | _ | _ | 0.333 | 0.016 |
| CCL7 | _ | _ | 0.313 | 0.017 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





