1. Introduction
1.1. Neural tissUE ENGINEERing and the iN VITRO GAP
Neurological disorders remain a leading cause of disability, partly because laboratory models still lag behind the intricate architecture of the brain. Conventional two-dimensional monolayers confine neurons to a flat plane and supply only rudimentary extracellular-matrix cues, which markedly limits their physiological relevance for mechanistic or therapeutic studies [
1,
2,
3,
4,
5,
6,
7,
8,
9,
10,
11,
12,
13,
14]. Three-dimensional spheroids and unguided brain organoids increase fidelity, yet the absence of intrinsic vasculature drives hypoxia, central necrosis, and persistent expression of progenitor markers such as SOX2, features that undermine their value for late-stage or degenerative disease modeling [
12]. Regionalized or chimeric strategies that transplant human neural lineages into rodent brains alleviate these shortcomings: the host vasculature improves perfusion and accelerates electrophysiological maturation, although inter-species signaling and ethical issues constrain widespread deployment [
8].
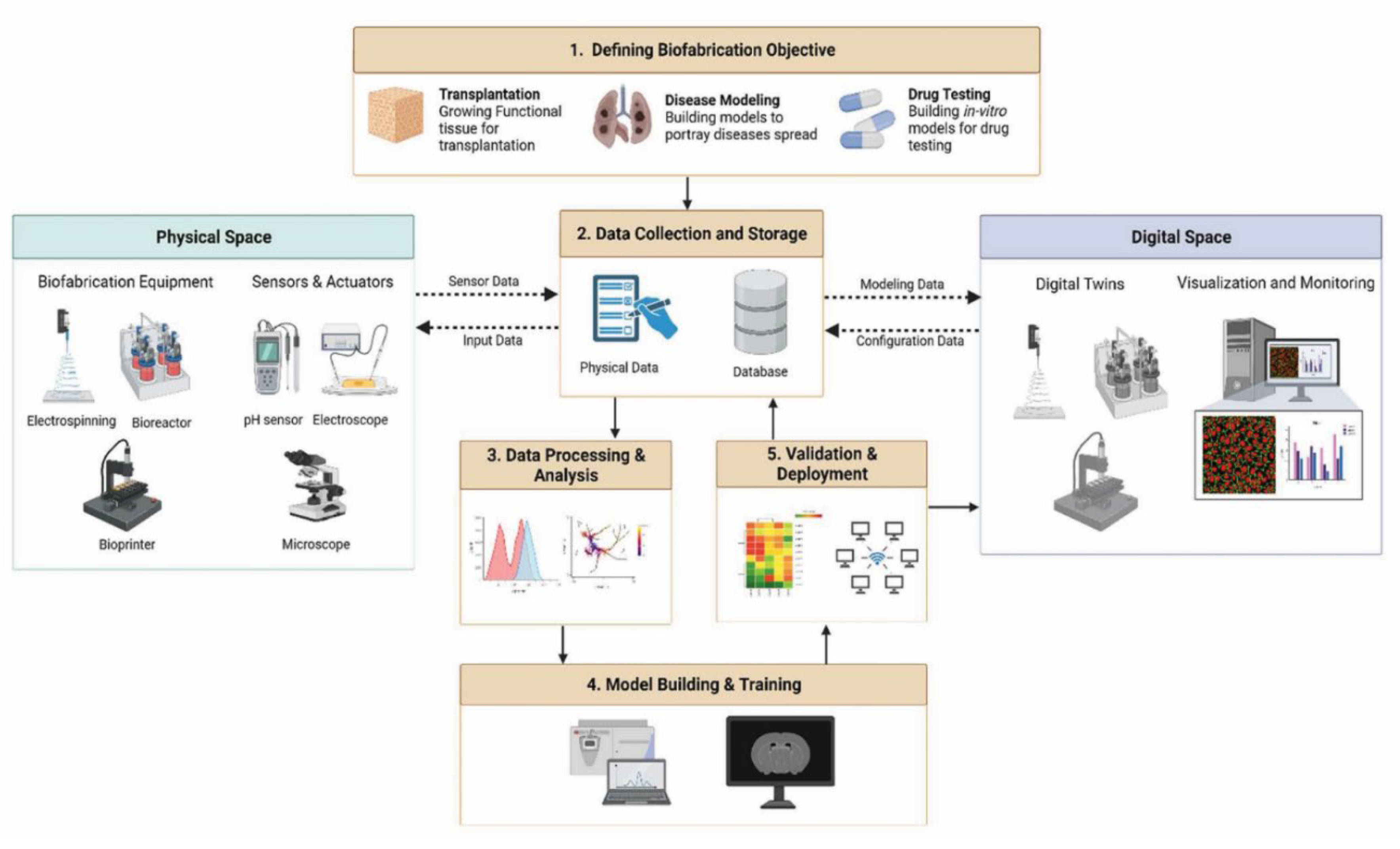
Organ-on-chip (OOC) technology offers a modular alternative (
Figure 1). Microfluidic perfusion recreates shear stress and nutrient gradients, whereas 3D-printing enables the precise placement of multiple cell types within perfusable channels. Despite notable success in liver and gut models, many neural chips still rely on passive hydrogels with limited capacity for matrix remodeling, which remains a persistent barrier to extended culture performance [
3].
Four-dimensional (4D) bioprinting adds time-programmed shape change to this microfluidic precision. Reviews catalog pH-responsive poly(2-vinyl-pyridine) constructs, ion-triggered alginate networks, and electro-responsive polypyrrole hydrogels that reversibly fold, swell, or contract, providing conceptual paths toward sulcus-like or layered architectures after printing [
7]. Complementing these material advances, recent analyses underscore how 4D approaches can be tuned for personalized therapeutic applications [
4]. When such inks are integrated with sacrificial microchannels, brain-on-chip platforms could, in principle, reproduce dynamic phenomena such as tissue swelling or neurovascular coupling in vitro.
Collectively, the field is moving from static replicas toward living constructs that mature alongside their resident cells. Closing the remaining in-vitro gap will demand scaffolds capable of adjusting their mechanical and biochemical landscapes as neural circuits evolve.
1.2. Role of Composite Materials and AI
Realizing that vision depends on substrates that feel to neurons such as the native cortex. Multiphase composites, soft, ionically conductive matrices interwoven with mechanically robust lattices, permit independent tuning of elasticity, porosity, and charge transport. Interpenetrating-phase lattice architectures efficiently distribute stress and exhibit high specific strength and energy absorption, qualities essential for long-term culture stability [
13]. Topographic patterning, grooves, aligned fibers, or multichannel conduits further bias stem cells toward neuronal lineages and guide axons into ordered trajectories reminiscent of cortical columns [
6]. Conductive coatings such as polypyrrole enhance electrical conductivity and promote neurite extension under electrical stimulation without compromising scaffold integrity [
6].
Navigating this expansive chemical-topographical design space increasingly relies on artificial intelligence. Statistics-encoded neural networks can reconstruct three-dimensional multiphase microstructures from sparse two-dimensional images, enabling rapid poro-mechanical simulations that replace exhaustive imaging pipelines [
1]. Deep- learning classifiers predict scaffold biocompatibility directly from slice images, flagging cytotoxic phase ratios before a single droplet of bio-ink is dispensed [
9]. Complementary 4D-bioprinting frameworks outline prospective closed-loop control strategies that can adjust the nozzle temperature, extrusion speed, and cross-linker dosing in real time to maintain user-defined mechanical and electrical targets [
4].
Multimodal AI pushes personalization further. By integrating patient--specific imaging, genomic, and histopathological data, emerging models can suggest biomaterial parameters that can be used to mitigate adverse glial responses while preserving functional conductivity [
14]. Conceptual studies likewise propose the use of such predictive engines to position shape-memory domains within stimuli-responsive inks so that future scaffolds might swell, stiffen, or release trophic factors in response to pathological cues [
7].
The convergence of bioadaptive composites and data-driven intelligence frames the bioinspired NeuroPod: an AI-enhanced, 4D-printed microenvironment designed to evolve alongside its neural inhabitants. By uniting materials that replicate the brain’s form with algorithms that anticipate its function, these systems promise disease models and regenerative scaffolds of unprecedented fidelity.
2. Bioinspiration in Neural Architecture
2.1. Hierarchical Brain Structures & Functional Cues
The mammalian cortex is stratified not only by neurons but also by gradients in glia, extracellular- matrix (ECM) mechanics and metabolic support. Systematic surveys of 3-dimensional (3D) bioprinted neural models have shown that excitatory and inhibitory neurons populate gray- matter- such as the periphery, whereas astrocytes, oligodendrocytes and microglia congregate in an interior zone that mirrors white matter, establishing the complementary biochemical and mechanical niches necessary for circuit assembly [
15,
16,
17,
18,
19,
20,
21,
22,
23,
24,
25,
26,
27,
28,
29,
30,
31].
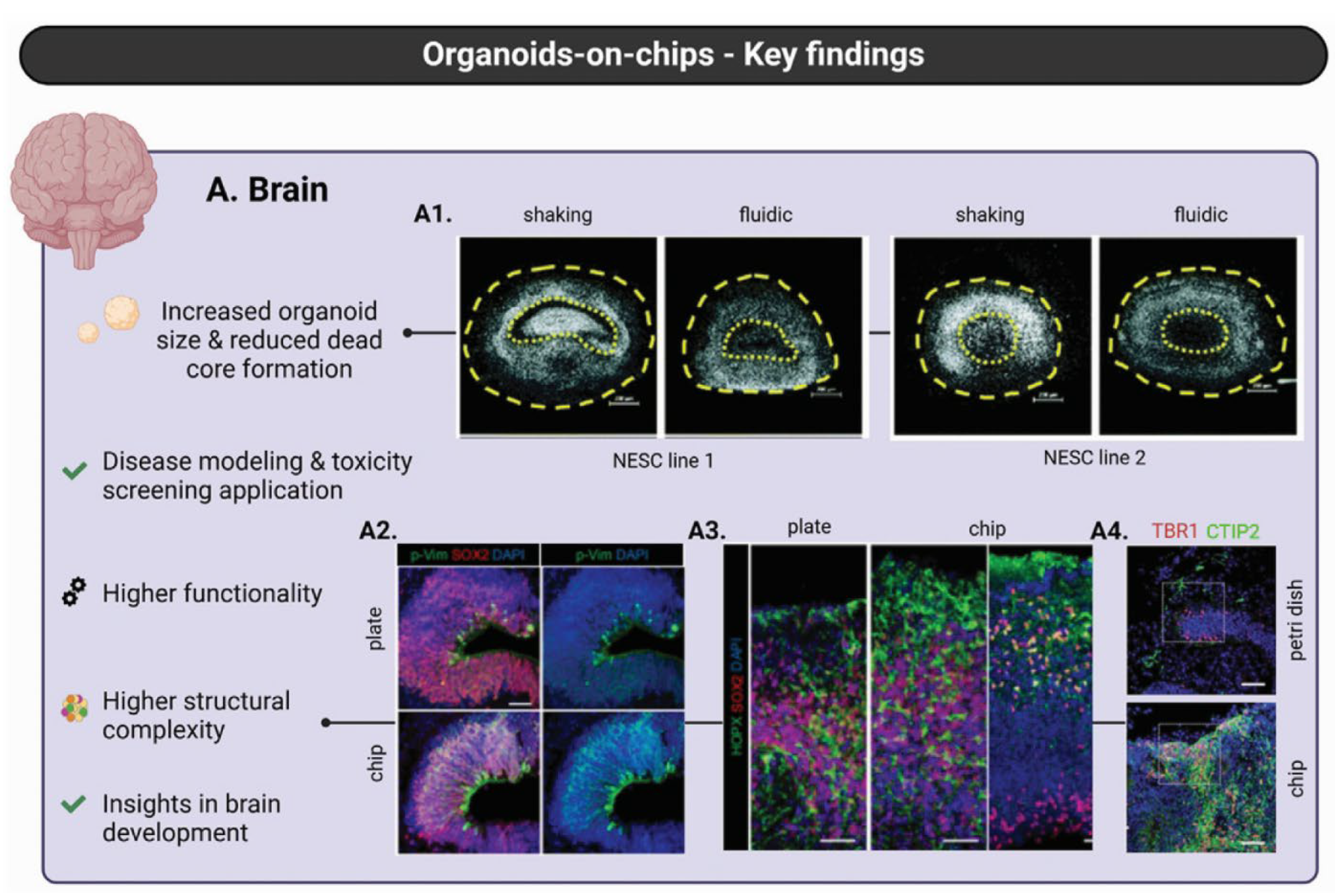
Lamination itself provides a developmental compass. Brain-like prints that mimic the six cortical layers, each 200 to 1 000 µm thick, must reproduce the orthogonal ECM and soluble-factor gradients that steer radial migration and axonal guidance. A gelatin–alginate ink tuned to a modulus close to that of the adult cortex (~1 kPa) and engineered with a controlled pore gradient (30–150 µm) supports balanced nutrient diffusion and sustained neurite extension over four weeks [
23]. The same study reported 94.5 % cell viability for human neural progenitors, demonstrating that gentle rheology can coexist with structural fidelity.
The functional cues are superimposed on this physical scaffold. Printing neurons in alternating 50 µm “bands” recreates the architectural rhythm of cortical minicolumns; within these stacked layers, a fibrin–hyaluronic-acid bioink supports rapid maturation as MAP2-positive neurons develop vGlut1–synapsin puncta by day 35 [
26].
Supportive glial meshes are active modulators rather than passive fillers. Soft, conductive hydrogels coated with ECM proteins transmit electrical cues while sharply reducing the astrogliosis that plagues rigid implants, an effect attributed to a matched modulus and bioactive coatings [
30]. Printing astrocytes alongside polypyrrole-- or PEDOT:PSS--based networks therefore enables stimulation or recording without compromising trophic support.
Engineering translation of these motifs has produced the first modular “NeuroPods.” Each pod houses layered cell strata within a collagen–alginate shell whose density increases from the outside, guiding axons toward less-crowded cores and encouraging dendrites to remain in synapse-rich outer layers. This gradient design reproduces cortical porosity, drives axonal penetration across interfaces within five days and preserves high viability [
23]. PDMS micro-compartment chips that employ guidance channels approximately 10 µm wide and 2.5 µm high fluidically isolate somata from axons to study direction-selective growth [
31], whereas a reversible variant enables long-term culture and network reuse across experiments [
19].
Key geometric and biophysical design targets abstracted from the studies above, spanning laminar thickness constraints, fine-pitch cellular banding, gradient porosity, modulus matching, and compartmental guidance scales, are compiled in (
Table 1) to guide layered NeuroPod engineering.
By fusing stiffness gradients, microcolumn-level synaptic niches and glia–neuron synergy inside centimeter-scale NeuroPods, investigators now capture key hierarchical rules of the brain in a format compatible with high-throughput drug screens. These constructs mark a turning point where structural fidelity and emergent electrophysiology coexist, laying the foundation for AI-guided optimization and regenerative translation.
2.2. Biomimetic Principles for Regenerative Neuroscience
Repairing cortical or spinal lesions demands implants that reinstate layered identity and long-range circuitry rather than merely filling voids. Bioprinting strategies therefore deposit 50 µm neuronal sheets sequentially so that an orthogonal view reveals a six-layer slab whose cytoarchitecture echoes ontogenetic migration patterns. Within these laminates, interneurons synchronize with projection neurons, and astrocytes generate glutamate-responsive calcium waves, confirming that geometric fidelity restores synaptic reciprocity [
26].
The hippocampal circuitry supplies a second template. Bilayer conductive hydrogels, electroactive polypyrrole- or PEDOT:PSS--based bases married to a degradable fibrin top, support dentate-like neurogenesis while delivering patterned electrical stimuli that preserve impedance stability and minimize glial scarring. The design emulates the dentate-CA axis: a cell-laden degradable layer fosters neurogenesis, whereas the electroactive substrate channels trisynaptic activity, thereby encouraging regeneration of the perforant and Schaffer pathways [
30].
Artificial-intelligence pipelines now compress iteration cycles. The BrAIn platform segments and classifies cerebral organoids grown under static, orbital-shaker or microfluidic conditions. Transfer-learning models achieve an area-under-curve of 92.7–96.8 % in distinguishing budding or abnormal morphologies while tracking area, Feret diameter and circularity over time [
20]. When paired with organoid-on-chip systems perfused with decellularized brain ECM, these metrics reveal that dynamic flow homogenizes growth and enhances neuron–glia interplay, information that directly feeds into layer--specific printing protocols [
27].
Nature-inspired gradients to further refine differentiation. Self-assembling peptide hydrogels present RGD and other bioactive motifs while offering tunable stiffness from 100 to 500 Pa, spanning embryonic to adult brain values, and promoting both neural adhesion and axonal elongation [
17]. When combined with neural stem cells or exosome-laden matrices, such gels simultaneously steer lineage commitment and temper immune responses, delivering dual benefits for regeneration [
24].
Temporal plasticity is introduced through 4D printing. Shape-memory or enzyme-responsive inks can swell or degrade, dynamically widening axon channels as neurite bundles thicken or curving scaffolds to match gyrification patterns seen during in vivo recovery; early prototypes show curvature changes of ~40 % upon matrix-metalloproteinase exposure, capturing the evolving microarchitecture of healing tissue [
16].
Finally, vascular integration remains paramount. Blood-brain- barrier (BBB)- on-chip platforms that co-cultivate endothelial cells and astrocytes achieve physiological tightness and selectively gate metabolites. When positioned upstream of NeuroPods, they recreate glucose and oxygen gradients that influence synaptic maturation, yielding a closed-loop model that couples circulation and the parenchyma [
22].
Together, these biomimetic strategies weave geometric layering, region-specific cell programming, AI-guided morphology tracking and adaptive materials into unified blueprints for neural repair. By honoring the brain’s intrinsic design rules, lamination, graded signals and glia–neuron synergy, printed NeuroPods grow, fire and remodel much like their in vivo counterparts do. These living constructs are poised to transform the brain-on-chip analytics, accelerate personalized drug testing and, ultimately, enable patient-tailored neural regeneration.
3. Advanced Composites & 4D Bioprinting for NeuroPods
3.1. Smart Nanocomposites & Hybrid Bioinks
Electroconductive nanocomposites act as “spark-plugs” that allow NeuroPods to exchange ionic and electronic cues with native brain tissue (
Figure 2). Gelatin–gelMA (GG) inks, which are known for their biocompatibility, were originally electrically inert; dispersing trace graphene-oxide (GO) sheets created continuous π-electron pathways, markedly increasing conductivity while slightly reinforcing the hydrogel network and preserving high cell viability during UV curing owing to the intrinsic light-shielding capacity of GO [
32,
33,
34,
35,
36,
37,
38,
39,
40,
41,
42,
43,
44]. The same addition of GO maintained shear-thinning behavior, ensuring smooth extrusion and shape fidelity in the printed lattices [
37].
A complementary strategy embeds carbon-nanotube (CNT) bundles inside PEG hydrogels. Ionic-liquid exfoliation followed by in- situ polymerization locks pristine CNTs into a percolating mesh that encourages neurite extension but keeps the nanotubes physically distant from neuronal membranes, a configuration shown to sustain homeostatic calcium oscillations in primary hippocampal networks rather than the hyper-excitability sometimes observed on bare CNT films [
38].
Two-dimensional MXene (Ti₃C₂Tₓ) flakes increase the conductivity further while remaining hydrophilic. In silk-methacrylate/pectin/MXene-soy phospholipid (SilMA/Pec/MX-SP) inks, the flakes bestow shear-thinning rheology and low charge-transfer resistance; daily 300 µA electrical pulses through printed neural-stem- cell (NSC) spheroids accelerate proliferation and significantly enhance neuronal differentiation, illustrating how matched impedance can amplify endogenous signaling [
44].
Hybrid bioinks also serve as molecular carriers. Reference [
44] summarizes earlier work in which a Morpho-butterfly-inspired nerve conduit combining reduced GO with brain-derived neurotrophic factor (BDNF) guided NSCs toward neuronal fates and supported regeneration across a 10 mm sciatic- nerve gap, an example of how topography, conductivity and growth- factor release can be co-engineered within one platform [
44].
The formulation of such multi-component inks is moving from an empirical trial-and-error to a data-driven design. A survey of extrusion bioprinting highlights machine-learning (ML) pipelines that correlate viscosity, nozzle gauge and pressure with filament quality, enabling rapid identification of printable windows while safeguarding cell viability [
36]. The vision-guided feedback described in the same review halts printing when surface defects emerge, allowing-- fly adjustment instead of wholesale scaffold rejection [
42]. For droplet-based systems, multilayer-perceptron models predict droplet volume for cell-laden inks with high precision, paving the way for uniform organoid arrays, although real-time in-line control remains a near-term goal [
39]. Collectively, ML augments nanocomposite chemistry by shrinking the experimental space needed to converge on inks that print cleanly, protect cells, and conduct bioelectric signals.
3.2. Stimuli-Responsive & Shape-Memory Approaches
Time-programmed materials give NeuroPods a fourth dimension, allowing printed constructs to be reshaped in concert with neural development (
Figure 3). This paper reviews programmed dynamic tissue catalog scaffolds that self-fold into conduits, stiffen selectively or elongate axially to direct axonal alignment, behaviors encoded through the distribution of responsive segments inside the print [
33].
Shape-memory polymers (SMPs) are the canonical workhorses. A 2025 survey dissects macro- versus micro-scale actions: bulk recovery enables minimally invasive deployment of neural electrodes, whereas micro-wrinkling surfaces impart cyclic mechanical cues that bias stem-cell lineage commitment [
34]. Thermo-responsive cyanate-ester SMPs can lock three discrete configurations, a catheter-friendly rod, a sulcus-hugging helix and a planar recording sheet, each triggered by step-wise warming to body temperature [
34]. Water-triggered PEG-PCL networks, meanwhile, cycle between extension and contraction as the local ionic strength shifts, opening avenues for axon stretch- growth without auxiliary hardware [
34].
Nano-fillers enrich these SMPs with extra functions. GO-grafted soybean-oil acrylate scaffolds produced by stereolithography self-roll into nerve guides when gently heated, marrying conductivity with shape morphing for peripheral- nerve repair [
33]. Embedding Fe₃O₄ particles adds magnetic programmability: alternating-field heating bends the scaffold in situ, circumventing systemic temperature increases [
34].
Predictive analytics underpin safe deployment. Artificial-neural-network models trained on hydrogel-swelling datasets forecast equilibrium volume with approximately 11 % error, allowing designers to calculate forces exerted on neighboring axons before fabrication begins [
40] . This integration of material informatics ensures that temporal transformations guide, rather than injure, delicate neural processes, converting 4D printing from imaginative origami into quantitative neuro-mechanical engineering.
3.3. Fabrication Methodologies & Programming
4D fabrication for NeuroPods follows two complementary logics. Ex situ printing produces constructs that are programmed after printing, through heat or solvent exchange, before implantation. In situ schemes deliver shape-memory inks directly into defects; physiological temperature or ionic gradients then activate morphogenesis on site, trimming surgical footprints [
35].
Whether NeuroPods are printed ex situ and later programmed, or delivered in situ as thermo- or ion-responsive inks, closed-loop performance depends on real-time data that report filament fidelity, evolving rheology, conductive network formation, thermal and light dose histories, post-print cell viability, swelling-driven shape change, and mechanical convergence toward neural tissue targets. Recent machine vision droplet studies, ML-empowered printability frameworks, thermo-responsive injectable hydrogels, photocurable conductive nanocomposites, and swelling-prediction models collectively define a practical instrumentation stack for AI supervision of 4D bioprinting. The principal sensor/data modalities and their control roles are summarized in (
Table 2).
Extrusion bioprinting dominates cell-laden applications because its moderate pressures accommodate fragile neural progenitors. ML-assisted parameter mapping identifies regions where the filament width and height track the G-code path closely while maintaining cell viability, shifting optimization from artisanal to algorithmic methods [
36]. Computer-vision feedback loops, highlighted in the same literature, detect sagging or under-extrusion layer by layer and adjust pressure or speed in near real time, maintaining print integrity even in soft hydrogel systems [
42].
When sub-100-micron vascular or synaptic geometries are needed, stereolithography (SLA) and digital-light processing (DLP) excel. The AI-driven bioprinting review describes voxel-level material assignment combined with finite-element/ML simulations that predict how composite voxels collaboratively deform over time, enabling multi-material prints whose global actuation is pre-calculated rather than empirically tuned [
32].
Bioelectronic programming blends deposition with circuitry. In SilMA/Pec/MX-SP scaffolds, conductive MXene traces are printed alongside insulating hydrogel walls; once cross-linked, micro-ampere pulses both stimulate maturing neurons and modulate the resistance of electro-responsive polymer segments, gradually harmonizing scaffold impedance with the evolving neural network [
44].
Finally, biochemical timing can be layered into prints without invoking external AI predictions. Gradients of neurotrophic factors such as BDNF are co-printed within conductive nanocomposites; their diffusion naturally synchronizes with the scaffold softening or unfolding windows described above [
44]. This multi-modal choreography, mechanical, electrical and biochemical, transforms a static print into a dynamically instructive micro-environment where cells are guided through sequential developmental milestones.
4. AI-Driven Modeling and Digital Twins
4.1. Predictive Multiscale Modeling
Creating NeuroPods that truly emulate brain micro-environments requires models able to converse across mechanics, mass transport, and cell-level decision making. Finite-element (FE) solvers map perfusion forces, strain gradients, and endogenous electric fields at micrometre resolution; agent-based or phase-field agents then interpret those maps to decide when a growth cone pauses, branches, or retracts.
A landmark in vascular tissue engineering has already fused a lattice-free smooth-muscle-cell agent-based model with dynamic FE mechanics. Each virtual cell “reads” local cyclic strain and interstitial flow before migrating, proliferating, or secreting collagen. The hybrid platform reproduced intimal hyperplasia patterns and showed how threshold-based collagen deposition progressively stiffened the scaffold, shifting compliance gradients that further steered cell behavior [
45,
46,
47,
48,
49,
50,
51,
52,
53,
54,
55,
56,
57,
58,
59,
60,
61,
62,
63]. This handshake, which exports tensor fields from the FE to cellular agents and feeds updated material moduli back, offers a ready template for neurite-scale mechanobiology.
For neural tissue, a multiphysics FE formulation of dendritic growth couples reaction–diffusion biochemistry with advection driven by membrane polarization. Simulations recover space-filling arborization and cathodal turning; a convolutional neural network then inversely estimates hidden kinetic constants directly from neuron images, closing a data-poor image-to-mechanism loop that is vital for early-stage brain-on-chip work [
48]. Phase-field work that blends isogeometric collocation captures lamellipodia emergence, axon specification, and dendrite elaboration while cutting the number of FE degrees of freedom owing to spline-based geometry [
60].
At the network level, the digital twin brain (DTB) framework states that a valid twin must preserve three nested layers: a structural atlas, neuro-physics, and functional applications. This hierarchy maps neatly onto NeuroPods, where the printed microarchitecture, scaffold mechanics, and emergent electrophysiology must remain coherent [
50].
Surrogate meta-models then accelerate design space exploration. The GAD-MALL workflow started with fewer than 100 fully solved FE samples yet searched for an 18,000-candidate lattice library. Within five to six active-learning iterations, it uncovered “treasure” scaffolds exhibiting elastic moduli of 2.5 GPa and 5 GPa, far beyond the uniform baseline, while holding FE calls below 100 per cycle [
63]. Similar loops can scan bio-ink composition, filament spacing, or cyclic-strain schedules for NeuroPods without brute-force sweeps.
Together, these advances outline a predictive stack: the FE delivers spatial cue fields, agent-based or phase-field solvers grow neurites in response, and active-learning meta-models mine the simulation archive to guide the next experiment. The result is a quantitative canvas on which AI can paint adaptive, brain-mimetic NeuroPods.
4.2. Continuous Optimization and Learning
The prediction is only half the story; NeuroPods must keep learning as materials, stimuli, and phenotypes drift. Data-efficient AI design- of-experiments (DoE) frameworks confront chronic data scarcity. Active-learning schedules have already reduced the number of physical scaffold prints while converging on optimal combinations of print fidelity, cell viability, and degradation kinetics (
Figure 4) [
63]. Bayesian optimization has outperformed random and genetic searches when tuning bioreactor variables; multi-objective reinforcement learning now targets simultaneous gains in stiffness and metabolic health [
63].
Hybrid model--based reinforcement learning (RL) pushes this further. A probabilistic knowledge graph overlays first-principles kinetics with data-driven residuals, whereas Bayesian RL selects robust control trajectories under batch-to-batch uncertainty in cell-therapy manufacture [
49]. When applied to NeuroPods, an internal twin can predict how tweaking stimulation duty cycles or growth-factor gradients will modulate neurite density; the RL agent updates printing or perfusion parameters in silico before committing in vitro.
Sensor fusion closes the loop. Micro-electrode arrays, piezoresistive strain gauges, and chemotaxis probe stream signals that single-modality controllers cannot parse. Deep RL agents trained on microbial co-cultures maintain target population ratios and maximize product yield even with sparse sampling, outperforming proportional-integral control under bang-bang actuation [
53]. A comparable agent could raster electrical pulses, fluid shear, and compressive loading across NeuroPods while maximizing a composite reward drawn from electrophysiological synchrony, calcium-imaging metrics, and scaffold modulus evolution.
The internet of Bio-Nano Things (IoBNT) sensor networks still promise finer granularity. The IoBNT specification supports nanoscale transceivers with bit-error rates between 0.01 % and 0.1 % and integrates federated learning (FL) to minimize bandwidth demands [
45]. In the reported CNN–FL framework, training bacterial classifiers across 33 species reached 98.7 % accuracy while cutting bandwidth by more than 99 % relative to centralized learning [
45]. Deploying the same FL-enabled DoE across thousands of parallel NeuroPods would let each construct refined local policies while sharing only encrypted weight updates.
4.3. Digital Twins and Real-Time Adaptation
A digital twin ceases to be a mere mirror when it starts prescribing interventions. The IoBNT–CNN–FL architecture supplies the infrastructure: physical NeuroPods, nano-sensor gateways, distributed learners, and an immersive dashboard where engineers nudge virtual parameters and observe physical correlates in real time [
45]. Edge computation streams electrophysiological movies, impedance spectra, and strain maps without saturating links, whereas FL aggregation ensures that the consolidated twin reflects population-level regularities.
The staged workflow in (
Table 3) distills how a NeuroPod digital twin evolves from initial design intent capture through process alignment, mechanistic initialization, live data assimilation, predictive scenario testing, closed loop control, and outcome-driven retraining (
Figure 5).
Digital twins already assist patient-specific tumor-resection planning by simulating how lesions perturb functional networks. The same principle can guide patient-derived NeuroPods engineered for injury repair, provided that the twin accommodates maladaptive rewiring as well as recovery trajectories, an imperative stressed in twin-centric neuroscience reviews [
55].
Real-time adaptation hinges on the multiscale propagation of perturbations. A complex- systems perspective on precision-medicine twins emphasizes that a molecular-scale tweak, CRISPR knock-in, mRNA burst, and ionic micro-gradient must ripple through cellular, tissue, and cognitive scales inside the twin before any intervention is approved [
61]. NeuroPod twins can apply this principle to predict how a modest increase in hydrogel- stiffness reshapes local mechanotransduction, alters branching-probability distributions, and shifts network-level oscillations.
Forecasting scaffold evolution is equally critical. The dendrite-electric FE model forecasts how evolving membrane polarization re-orients branches, allowing the twin to suggest polarity reversals if the arbor density plateaus [
48]. Moreover, the ABM-FEM vascular platform demonstrates how threshold-based collagen deposition stiffens a scaffold, and information from the twin translates into updated boundary conditions for subsequent simulations [
51]. Coupling such predictive engines with RL controllers yields a closed loop: sensors feed the twin, the twin anticipates drift, the policy optimizer proposes corrective stimuli, and actuators implement them in the living NeuroPod.
Bioprocess twins already rely on process-alytical- technology (PAT) streams, Raman spectra, and dielectric spectroscopy to recalibrate mammalian-cell cultures in real time [
62]. NeuroPod research can adopt analogous -line impedance tomography or optical- coherence probes as PAT surrogates that retune the twin every few seconds.
Translating the above architecture into regulated research and eventual clinical use requires staged verification and validation across code, parameters, biology, predictive utility, systems integration, and real-time performance. (
Table 4) maps these levels to evidence types, quantitative metrics, example acceptance criteria, and likely regulatory or translational milestones.
By merging predictive multiscale models, continuously learning optimizers, and sensor-driven digital twins, engineers craft a living, thinking replica for every NeuroPod. These replicas do more than monitor. They negotiate futures, guiding bioprinted neural constructs toward robust connectivity, adaptive plasticity, and regenerative potential under routine culture conditions.
5. Integration into the Brain-on-Chip Platforms
5.1. Microfluidics & Organ-Level Synergy
Brain-on-chip (BoC) platforms have progressed from static endothelial monolayers to perfusable microphysiological systems that sustain coordinated neuronal, glial, vascular and immune interactions (
Figure 6A). 4D-printed NeuroPods, defined here as shape-morphing neural constructs fabricated from stimuli-responsive bio-inks, fit naturally into this trajectory because their architecture adapts to precisely programmed microfluidic cues. A three-dimensional neurovascular-unit chip has already shown that a single device can carry a perfused endothelial channel adjacent to hiPSC-derived neurons and astrocytes; axons extend toward the vessel, and tumor-necrosis- factor-α (TNF-α) perfusion triggers a reversible drop in trans-endothelial electrical resistance, effectively modeling barrier disruption [
64,
65,
66,
67,
68,
69,
70,
71,
72,
73,
74,
75,
76,
77,
78]. Placing a NeuroPod into the extracellular-matrix channel of such a device allows its programmed folding to position dendrites closer to the vessel lumen, thereby heightening neurovascular coupling while preserving the electrical isolation needed for tight- junction integrity (
Figure 6B).
Immune-competent BoC formats provide an additional layer of realism. Neuroinflammation-on-a-chip systems for multiple-sclerosis modeling involve iPSC-derived microglia, controlled TNF-α perfusion and an endothelial interface; the resulting cytokine cascade reproducibly compromises barrier tightness [
67]. Integrating NeuroPods into this setting supplies a matrix whose stiffness can be tuned in real-time, enabling investigators to examine how mechanical cues influence leukocyte transmigration or oligodendrocyte remyelination. Because the bending behavior of 4D composites modulates local strain fields, NeuroPods transform a linear chemotaxis assay into a three-dimensional neuro-immune dialog without requiring additional hardware. Recent 4D bioprinting studies underline the feasibility of such mechanically adaptive scaffolds by demonstrating that polymeric materials are capable of reversible shape changes under physiological stimuli [70, 71].
Cross-organ microfluidic communication represents the next frontier. Multifunctional manifolds have already linked cerebral chips with cardiac constructs, providing an avenue to interrogate neuro-cardiac interactions such as stress-induced arrhythmic signaling [
76]. Comparable links to the intestinal epithelium are used to explore gut–brain axes. NeuroPods can be printed in each organ compartment with tissue-specific inks and then connected to an artificial-intelligence-controlled recirculating loop that continuously balances shear stress, nutrient supply and metabolite removal across all compartments, limiting the dominance of any one tissue in serial perfusion schemes.
Engineered flow is more common than passive plumbing; it is an active variable in neuropathophysiology. Precision control requires automation beyond conventional syringe pumps. A recent review on the fusion of microfluidics with artificial intelligence described closed-loop controllers that couple pressure sensors or optical probes to machine-learning algorithms, thereby optimizing flow rates, oxygen tension and solute concentrations without manual intervention [
73]. Parallel work on blood–brain-barrier chips shows that spatial or temporal gradients of inflammatory cytokines can regionally modulate tight-junction protein expression and that hypoxic episodes produce distinct patterns of barrier failure [
75]. Using the feedback schemes described above, the same microfluidic layout can alternate rapidly between homeostatic and inflammatory states, exposing NeuroPods to cycles of ischemia or cytokine stress that would be impractical to orchestrate manually.
Coupling NeuroPods with vascular and immune modules also enhances drug-delivery studies. The neurovascular-unit chip tracks real-time transcytosis of fluorescent nanobodies across the endothelial barrier, thereby supplying kinetic permeability data [
66]. Positioning a responsive NeuroPod downstream enables direct observation of how these cargoes affect synaptic plasticity minutes after crossing the barrier, effectively closing the pharmacokinetic–pharmacodynamic loop on-chip. When the same chip contains an immune channel, investigators can quantify how inflammatory mediators alter barrier integrity and cytokine profiles, a prerequisite to linking these cues to downstream neuronal responses, yielding mechanistic insights unavailable in static Transwell assays [
67].
Collectively, these advances transform the BoC into a holistic neuro-physiological system. Microfluidic perfusion maintains metabolic equilibrium, immune streams recreate inflammatory milieus, and 4D-printed NeuroPods actively remodel their mechanical micro-niches. AI-directed flow control unites the components, permitting rapid toggling between healthy and disease-like conditions without changing hardware.
5.2. Real-Time Sensing, Actuation & Drug Testing
The full potential of an integrated NeuroPod platform emerges when continuous, multimodal sensing closes the feedback loop between mechanics, electrophysiology and biochemical flux. Comprehensive surveys of sensor-integrated BoC devices catalog technologies such as micro-impedance tomography for cell motion, optical pH or oxygen beads for metabolic stress and complementary-metal-oxide-semiconductor multi-electrode arrays (CMOS MEAs) for network spiking, all of which are embedded within layered polydimethylsiloxane structures [
64].
NeuroPods complement these sensors because their 4D matrices can incorporate piezoresistive or graphene-based strain gauges without impairing cell viability. A self-folding graphene interface has been shown to envelop three-dimensional brain-like tissue and record action potentials deep within the construct, something that planar MEAs cannot achieve [
69]. Embedding such arrays as the conductive skeleton of a NeuroPod yields concurrent maps of mechanical deformation and neuronal firing during every shape-change cycle. Electrochemical microsensors, reviewed in the context of in vitro neurochemical detection, can quantify transient dopamine or glutamate release with sub-second resolution [
74]. The resulting datasets allow algorithms to correlate strain spikes with neurotransmitter surges in real time.
Artificial-intelligence analytics convert these data streams into actionable control signals. The BrAIn platform employs convolutional neural networks to analyze bright-field videos of organoids, extracting metrics such as area, circularity and budding rate that predict developmental stage or pathology [
78]. Feeding those metrics back into the microfluidic controller enables closed-loop interventions: if swelling suggests emerging edema, flow can increase to wash out cytokines; if burst frequency declines, oxygen tension can be elevated to rescue network activity. Digital-twin frameworks, proposed as a bridge between the silicon and carbon worlds, further support this paradigm by forecasting drug responses and mechanical stresses before any physical intervention [
72].
Personalized disease modeling benefits directly from these capabilities. Organoid-on-chip systems already employ patient-derived iPSCs to reproduce individual genotypes while supporting high-throughput drug screening [
65]. Printing NeuroPods from the same cellular source subjects maturing neurons to graded stiffness and shear reminiscent of early developmental niches, thereby increasing phenotypic fidelity. High-density MEAs within the chip can stratify patient-specific electrophysiological phenotypes, for example, the hyper-synchronous bursts characteristic of certain epileptic genotypes, and deliver quantitative end points for machine-learning-guided compound selection. The sensor-integrated review emphasizes that impedance electrodes and optical oxygen probes can be multiplexed without cross-talk, ensuring that parallel NeuroPod chips each receive reliable readouts [
64].
Mechanical cues are equally traceable. A nanotechnology-oriented review highlights transparent cantilever MEAs that protrude approximately 200 µm into the culture chamber, recording intratissue spikes while simultaneously acting as micro-strain gauges [
68]. Linking these signals to piezoelectric actuators beneath the NeuroPod allows haptic feedback, for example, pulsatile compression that mimics intracranial pressure or uniaxial stretching that models traumatic injury, with neuronal adaptation monitored in real time.
Drug-testing pipelines gain a distinctive edge when sensor data guide adaptive dosing. If a kinase inhibitor suppresses glutamate excitotoxicity yet slows axonal conduction, machine-learning algorithms can recognize the divergence, pause perfusion, allow recovery and resume dosing at a lower concentration, all within a single experiment. Because every variable, from shear stress to cytokine flux, is logged automatically, reproducibility surpasses that of manual protocols, an essential milestone for regulatory acceptance [
64].
High-throughput learning loops are already operational. Deep-learning pipelines track hundreds of patient-derived midbrain organoids, flagging samples that deviate from expected growth trajectories for immediate rescue or replacement [
78]. Coupled with flow-through electrochemical sensors that quantify extracellular lactate and glutamate, the system predicts neurotoxicity days before morphological deterioration becomes visible. The same logic triages candidate drugs, eliminating compounds that increase oxidative stress in early perfusate fractions.
In summary, embedding multimodal sensors into AI-orchestrated microfluidic BoC platforms converts 4D-printed NeuroPods into self-reports, self-adapting testbeds. Electrodes map spikes, graphene ribbons register strain, optical beads monitor metabolism, and electrochemical probes detect neurotransmitters, whereas artificial- intelligence algorithms fuse the streams into digital twins that forecast failure and prescribe intervention. This closed-loop architecture accelerates personalized drug discovery and simultaneously exposes the intricate interplay between biomechanics, electrophysiology and immunometabolism that underlies brain health and disease.
6. Key Challenges & Future Directions
6.1. Scalability & Manufacturing Bottlenecks
Early 4D-printed NeuroPods demonstrated that photo-, thermo-, and ion-responsive lattices can morph on cues, but pushing these demonstrations toward industrial volumes is still complicated by the classic speed–resolution dilemma. The latest review of 4D bioprinting highlights that as soon as layer thicknesses are driven toward the sub-100 µm regime, build times stretch into multiple-hour runs that jeopardize nutrient diffusion and hence cell viability (
Figure 7) [
79,
80,
81,
82,
83,
84,
85,
86,
87,
88,
89,
90,
91,
92,
93,
94,
95].
Reproducibility is the next gatekeeper. A vision-guided tool-path compensation scheme slashes filament-width variation on curved substrates and effectively halves the cumulative path-error area, turning previously manual “watch-and-tweak” steps into software-defined corrections [
91]. Complementary robotic-arm platforms already integrate adaptive force feedback and collision- avoidance routines; current roadmaps foresee multi-axis manipulators that remain enclosed in GMP-grade cleanrooms [
82].
Artificial- intelligence–centered quality-by-design (QbD) frameworks now weave these hardware advances into a single data fabric (
Figure 8). By fusing in-line multispectral imaging with digital twins, an AI controller detects thermal or rheological drifts in real time and retunes nozzle duty cycles without operator intervention, maintaining uniform strand diameters across multi-day campaigns [
81].
The logistics are scaled in parallel. A finite-state-machine algorithm driving a 4-axis robot achieved 83 % success for raw-chip pick-up, 100 % success for chip placement, but only 67 % success for protective-foil removal, still high enough to reduce operator attendance compared with manual workflows [
86]. When this handling system feeds a dual drop-on-a demand line, three tissue chambers can be fully loaded, cooled, sealed, and perfused- in approximately 70 s, with micro-vessels remaining patent for 14 days [
87].
Throughput at the tissue-block level is also surging. The HITS-Bio platform lifts spheroids with >90 % success and deposits hundreds of units in minutes, stitching centimeter-scale constructs while on-board cameras verify each placement [
88]. Together, vision-assisted robotics, AI-driven QbD, and multi-array deposition sketch a credible “smart-factory” blueprint capable of forecasting material consumption, flagging anomalies before they cascade, and propelling NeuroPod production toward GMP-compliant, patient-specific manufacture.
6.2. Long-Term Stability & Translation
Inside neural tissue, even micron-scale mismatches provoke astroglial scarring and signal loss. Flexible mesh electrodes coated with silk-fibroin sacrificial layers dissolve within days, minimizing chronic foreign-body burden while maintaining a conformable polyimide backbone [
89]. The same review illustrates a microporous drug-releasing coating in which cyclosporine A–loaded PLGA microspheres are embedded in a PEG hydrogel matrix, suggesting a route to on-board immunomodulation [
89].
Standardizing “what counts as stable” is equally vital. Clinical-trial guidance now recommends harmonized dashboards that track impedance, signal-to-noise, and electro-chemical interface markers, opening the door to cross-study meta-analysis and machine-learning prediction of early failure modes [
85]. Parallel policy papers for next-generation brain–computer interfaces insist on algorithmic transparency and mental-privacy safeguards from the first line of code [
92].
Regulators are moving in concert. The US FDA’s Total-Product-Lifecycle memorandum for generative-AI-enabled devices demands that developers pre-specify learning scopes, install real-time performance monitoring, and define drift-mitigation triggers, rather than freezing software at clearance [
95]. Such provisions dovetail with proposed AI risk-assessment engines for NeuroPods, where off-nominal impedance or cytokine signatures would trigger cloud alerts and the same-day recalibration.
Translation pathways are already being shortened. Hybrid testbeds now pair 4D-printed brain-on-chip arrays with pilot animal implants to track mechanical fatigue, biodegradable- layer thinning, and immune signatures under matched electrical loads, forming an iterative bench-to-bedside loop [
89]. Cloud-shared datasets from these rigs will feed broader predictive models, accelerating convergence on consensus standards and first-in-human timelines.
6.3. Visionary Concepts
Beyond today’s bottlenecks lies the convergence of synthetic biology, AI, and 4D printing. Smart-responsive hydrogels that switch from insulating to conductive with modest pH shifts have already rerouted ionic currents around inflamed regions and cut reactive- oxygen species in vitro [
93]. Embedding programmable gene circuits into these matrices could allow NeuroPods to secrete neurotrophic factors on demand, drive axonal regrowth, and then silence expression once synaptic density normalizes [
94].
4D architectures supply the morphing scaffold. The gradient- programmed bilayers unfurl into spiral micro-channels after implantation, steering axonal extension along chemotactic corridors and closing as lesions contract, and no revision surgery is needed [
80]. AI scheduling of thermal, electrical, and chemokine cues refines these shape changes, emulating developmental morphogenesis within adult brain tissue [
81].
Bio-inspired electronics push the interface toward “living synthelectronics.” Ultra-soft electrodes encapsulated in collagen–alginate blends achieve single-digit-kilopascal moduli and maintain low-kilohm impedance during ten-thousand-cycle strain tests while supporting spontaneous neuronal firing [
84]. Future designs envisage transient conductors that vanish once regeneration is complete, erasing the foreign-body footprint.
Manufacturing technologies are catching up. Multi-nozzle spheroid printers now co-deposit vascular and parenchymal spheroids at kilohertz rates, assembling centimeter-scale tissues in under 40 min with high viability [
88]. Coupling such heads to the robotic organ-on-chip line that seals and stores devices in a minute-scale cycle [
87] sketches a genuine “print-to-surgery” workflow.
Embedding low-power neuromorphic co-processors closes the loop between sensing and actuation, compressing spikes on-board and adjusting stimulation in real time, capabilities flagged as near-term deliverables in neuromorphic algorithm roadmaps [
90]. Regulatory scientists already anticipate such adaptive systems, aligning conditional approvals with live cloud-based audits under FDA lifecycle guidance [
95].
Finally, “living bridges” printed from patient-derived cerebral organoids on conductive hydrogel rails are gaining traction. Bio-hybrid interfaces that co-cultivate stem-cell-derived neurons with flexible electronics have sustained spontaneous activity for months without fibrotic encapsulation [
84]. As the organoids mature, embedded AI agents could modulate growth-factor release, synchronize oscillations with host networks and taper stimulation as connectivity is restored, transforming NeuroPods from passive scaffolds into actively self-healing neural surrogates.
Collectively, these advances delineate a credible trajectory from laboratory prototypes to clinic-ready, AI-enhanced 4D NeuroPods capable of high-volume manufacture, decades-long reliability, and seamless partnership with the human brain.
7. Conclusions
The interdisciplinary convergence that culminates in the bioinspired NeuroPod concept signals a decisive inflection point for neural-tissue engineering, brain-on-a-chip analytics, and regenerative medicine. Throughout this Review, we traced the field’s progression from static collagen or alginate hydrogels toward AI-curated, four-dimensional composite prints that can morph, conduct, and metabolically dialog with living neural networks. That journey rests on three reinforcing pillars. First, smart nanocomposites marry soft ion-permissive matrices with percolating conductive fillers, graphene oxide, MXenes, and carbon-nanotube bundles to sustain millivolt signaling while remaining cytocompatible. Second, stimuli-responsive or enzyme-labile domains provide temporal plasticity, allowing printed conduits to unfold, stiffen, or degrade in synchrony with neuronal development or inflammatory cues. Third, artificial-intelligence pipelines compress empirical trial-and-error cycles: surrogate meta-models mine vast design libraries in silico; computer-vision feedback corrects filament deviation in real time; and hybrid model-based reinforcement learning converts perfusion, stimulation, and dosing schedules from fixed set-points into adaptive trajectories that protect viability and enhance maturation.
Mater, NeuroPods integrate spatial and biochemical gradients that echo cortical lamination, hippocampal circuitry, or lesion-specific topology. Sacrificial pores introduce vascular access; stiffness gradients corral glia and orient axons; and co-printed neurotrophic-factor reservoirs release BDNF or GDNF in step with mechanical remodeling. Examples already demonstrate >90 percent progenitor viability, accelerated synaptogenesis under low-impedance stimulation, and centimeter-scale axonal bridging across transected spinal cords. Shape-memory polymers further extend capability: cyanate-ester electrodes self-unfurl to contact deep sulci after minimally invasive deployment, and hydro-ionic PEG–PCL lattices rhythmically contract to encourage axonal stretch growth without motors.
Computationally, digital-twin frameworks recruit multiscale finite-element solvers, agent-based cell models, and federated-learning controllers to forecast scaffold evolution hours ahead. Bright-field morphology extracted by convolutional networks, impedance tomography that maps mechanical strain, and electrochemical sensors that track real-time neurotransmitter flux feed these twins, allowing predictive re-tuning of flow rates, electrical pulse trains, or local stiffness before failure arises. In parallel, the active- learning design- of- experimental loops has already cut material-screening burdens by an order of magnitude, identifying printable inks that satisfy rheological, electrical, and biological constraints with fewer than 100 physical trials.
The practical impact is tangible. In pharmacology, NeuroPod-equipped brain-on-chip platforms correlate barrier permeability, burst-rate synchrony, and cytokine dynamics inside the same microfluidic loop, producing pharmacokinetic–pharmacodynamic fingerprints unattainable in siloed assays. In neurotrauma, patient-derived organoids embedded within shape-adaptive conduits have restored conduction across centimeter-scale lesions in rodent models, whereas self-folding graphene interfaces record signals deep inside three-dimensional constructs for months without gliosis. Looking forward, multi-organ chips are beginning to connect NeuroPods to cardiac and gut modules, enabling the study of neuro-cardiac arrhythmias or gut–brain inflammatory axes under closed-loop AI control.
However, translation hinges on multiscale harmonization. Conductive domains must retain impedance coherence as they biodegrade and as neurons myelinate; immune responses to mechanically dynamic polymers need baseline maps across species, ages and disease states; and lifelong data stewardship strategies must secure the intimate electrophysiological signatures that sensor-rich implants generate. On the computational front, high-fidelity twins must evolve from episodic recalibration toward continuous learning that detects drift, mitigates algorithmic bias, and guards against adversarial manipulation. Regulators already anticipate this need: total-product-lifecycle guidance for generative-AI-enabled devices demands transparent learning scopes, real-time performance monitoring, drift-mitigation triggers, and requirements that will shape every future NeuroPod.
Despite these hurdles, the architecture outlined here offers a coherent blueprint for the next decade. Artificial intelligence collapses the combinatorial design space; four-dimensional printing furnishes time-aware fabrication; composite bioelectronics enable bidirectional communication; and microfluidic perfusion maintains metabolic equilibrium. Together, they orchestrate mechanobiology, electrophysiology, and system control in a single living construct. The NeuroPod thus stands poised to transform drug discovery, accelerate personalized regenerative therapy, and redefine our experimental grasp of the human brain. In the same way, multi-core processors revolutionized computing by distributing workloads, NeuroPods herald a future in which distributed neuromorphic constructs shoulder the heavy lifting of disease modeling and repair, moving neuroscience from observation toward programmable intervention with precision once reserved for semiconductor engineering.
8. Unresolved Questions
While the NeuroPod framework sketches an ambitious trajectory, several foundational riddles remain and invite daring exploration. How might we create a single composite ink whose degradation rate, swelling amplitude, electrical impedance, and trophic-factor release all couple directly to real-time transcriptional or metabolic signatures, yielding an authentic self-regulating scaffold that dispenses with external controllers? Can we define universal geometric invariants, perhaps rooted in differential-topology metrics, that guarantee stable oscillatory dynamics across scales, from dendritic polarity to mesoscale synchrony, independent of patient-specific anatomy? What molecular and mechanical tipping points mark the transition where an adaptive material intended for regeneration triggers maladaptive fibrosis, and can predictive immuno-digital twins forecast that switch days in advance? Will it become feasible to embed neuromorphic co-processors fabricated from biocompatible semiconducting peptides or conductive polysaccharides directly inside the NeuroPod so that computation, sensing, and actuation share a common metabolic currency with neurons? How can federated-learning architectures be shielded against adversarial attacks when the training data are continuous neurochemical and electrophysiological streams that expose an individual’s cognitive fingerprint? At the regulatory frontier, what new ethical frameworks and audit algorithms will certify implants that autonomously rewrite their own control policies in response to unpredictable developmental trajectories or disease relapses? Can we devise cross-platform benchmarking assays that compare the functional output of NeuroPods, brain organoids and in vivo grafts on equal footing, clarifying when an in-vitro surrogate has matured enough to guide clinical decision-making? Equally pressing is the quest for energy autonomy: can printable biofuel cells or optogenetically driven ion pumps harvest endogenous metabolites or ambient light to power embedded electronics for months without a thermal burden? Finally, as multi-organ chips link the gut, heart, and brain NeuroPods into closed circulatory loops, how do we model and mitigate emergent cross-organ instabilities, coupled metabolic oscillations or synchronized inflammatory flares that have not precedent in single-organ studies? Addressing these questions will not only refine NeuroPod engineering but also illuminate the fundamental principles governing self-organizing neural systems in health and disease.
Ethics approval and consent to participate
Not Applicable.
Competing interests
Not Applicable.
Availability of data and material
Not Applicable.
Consent for publication
Not Applicable.
Declaration Regarding the Use of AI-Assisted Readability Enhancement
I hereby affirm that the utilization of AI-assisted tools in the refinement of the manuscript was strictly limited to enhancing its readability. At no point were AI technologies employed to supplant essential authorial responsibilities, including the generation of scientific, pedagogic, or medical insights; the formulation of scientific conclusions; or the issuance of clinical recommendations. The implementation of AI for readability enhancement was rigorously supervised under the discerning eye of human oversight and control.
References
- Fu, J. , & Tan, W. Stochastic reconstruction of multiphase composite microstructures using statistics-encoded neural network for poro/micromechanical modeling. Computer Methods in Applied Mechanics and Engineering 2025, 441, 117986. [Google Scholar] [CrossRef]
- Yan, Y. , Li, X., Gao, Y., Mathivanan, S., Kong, L., Tao, Y.,... & Zhang, S.C. 3D bioprinting of human neural tissues with functional connectivity. Cell Stem Cell 2024, 31, 260–274. [Google Scholar] [PubMed]
- Wu, X. , Shi, W., Liu, X., & Gu, Z. Recent advances in 3D-printing-based organ-on-a-chip. EngMedicine 2024, 1, 100003. [Google Scholar] [CrossRef]
- Kennedy, S.M. , M, S., A, V., & K, A. 4D bioprinting for personalized medicine, innovations in implant fabrication and regenerative therapies. Polymer-Plastics Technology and Materials 2025, 1–26. [Google Scholar] [CrossRef]
- Salmina, A.B. , Alexandrova, O.P., Averchuk, A.S., Korsakova, S.A., Saridis, M.R., Illarioshkin, S.N., & Yurchenko, S.O. Current progress and challenges in the development of brain tissue models: How to grow up the changeable brain in vitro? Journal of Tissue Engineering 2024, 15, 20417314241235527. [Google Scholar]
- Han, S. , Zhao, X., Cheng, L., & Fan, J. Recent progresses in neural tissue engineering using topographic scaffolds. American Journal of Stem Cells 2024, 13, 1. [Google Scholar] [PubMed]
- Arif, Z.U. , Khalid, M.Y., Ahmed, W., & Arshad, H. A review on four-dimensional (4D) bioprinting in pursuit of advanced tissue engineering applications. Bioprinting 2022, 27, e00203. [Google Scholar] [CrossRef]
- Papetti, A.V. , Jin, M., Ma, Z., Stillitano, A.C., & Jiang, P. (2025). Chimeric brain models: Unlocking insights into human neural development, aging, diseases, and cell therapies. Neuron.
- Oncu, E. , Ayanoglu, K.Y.U., & Ciftci, F. Comparative analysis of deep learning models for predicting biocompatibility in tissue scaffold images. Computers in Biology and Medicine 2025, 192, 110281. [Google Scholar] [CrossRef]
- Papamichail, L. , Koch, L.S., Veerman, D., Broersen, K., & van der Meer, A.D. Organoids-on-a-chip: microfluidic technology enables culture of organoids with enhanced tissue function and potential for disease modeling. Frontiers in bioengineering and biotechnology 2025, 13, 1515340. [Google Scholar] [CrossRef]
- Aili, Y. , Maimaitiming, N., Wang, Z., & Wang, Y. Brain organoids: A new tool for modeling of neurodevelopmental disorders. Journal of Cellular and Molecular Medicine 2024, 28, e18560. [Google Scholar] [CrossRef]
- Urrestizala-Arenaza, N. , Cerchio, S., Cavaliere, F., & Magliaro, C. Limitations of human brain organoids to study neurodegenerative diseases: a manual to survive. Frontiers in Cellular Neuroscience 2024, 18, 1419526. [Google Scholar] [CrossRef]
- Mu, Y. , Jin, Y., Ji, H., Luo, J., Li, G., Xu, M.,... & Du, J. Mechanical performance of interpenetrating phase composites with multisheet lattice structures. International Journal of Mechanical Sciences 2024, 276, 109369. [Google Scholar] [CrossRef]
- Parvin, N. , Joo, S.W., Jung, J.H., & Mandal, T.K. Multimodal AI in Biomedicine: Pioneering the Future of Biomaterials, Diagnostics, and Personalized Healthcare. Nanomaterials 2025, 15, 895. [Google Scholar] [CrossRef] [PubMed]
- Orr, A. , Kalantarnia, F., Nazir, S., Bolandi, B., Alderson, D., O’Grady, K.,... & Willerth, S.M. Recent advances in 3D bioprinted neural models: A systematic review on the applications to drug discovery. Advanced Drug Delivery Reviews 2025, 115524. [Google Scholar] [CrossRef]
- Lai, J. , Liu, Y., Lu, G., Yung, P., Wang, X., Tuan, R.S., & Li, Z.A. 4D bioprinting of programmed dynamic tissues. Bioactive Materials 2024, 37, 348–377. [Google Scholar] [CrossRef] [PubMed]
- Xie, C. , Chen, Y., Wang, L., Liao, K., Xue, B., Han, Y.,... & Jiang, Q. Recent research of peptide-based hydrogel in nervous regeneration. Bioactive Materials 2024, 40, 503–523. [Google Scholar] [CrossRef]
- Ferreira, M.J. , Colombani, S., Bernardin, A., Lacampagne, A., Pasquié, J.L., Costa, P.F.,... & Meli, A.C. Advancing organ-on-chip systems: the role of microfluidics in neuro-cardiac research. Current Research in Pharmacology and Drug Discovery 2025, 100227. [Google Scholar] [CrossRef]
- Moreau, S. , Flores-Berdines, R., Jalkh, T.E., Simon, A., Taret, G., Fomina, A.,... & Salmon, H. Reversible and reusable compartmentalized microfluidic chip for coculture of dorsal root ganglion neurons. bioRxiv.
- Kahveci, B. , Polatli, E., Evranos, A.E., Güner, H., Bastanlar, Y., Karakülah, G., & Güven, S. BrAIn: A comprehensive artificial intelligence-based morphology analysis system for brain organoids and neuroscience. bioRxiv 2025, 2025–02. [Google Scholar]
- Akcay, G. , & Luttge, R. Microenvironments matter: advances in brain-on-chip. Biosensors 2023, 13, 551. [Google Scholar] [CrossRef]
- da Silva, G.G. , Sacomani, D.P., de Carvalho, B.G., Porcionatto, M.A., Gobbi, A., Lima, R.S., & de la Torre, L.G. Microfluidic Systems to Mimic the Blood–Brain Barrier: from Market to Engineering Challenges and Perspectives. ACS Biomaterials Science & Engineering 2025. [Google Scholar]
- Pei, N. , Hao, Z., Wang, S., Pan, B., Fang, A., Kang, J.,... & Wang, L. 3D printing of layered gradient pore structure of brain-like tissue. International Journal of Bioprinting 2021, 7, 359. [Google Scholar] [CrossRef]
- Rybachuk, O. , Nesterenko, Y., & Zhovannyk, V. Modern advances in spinal cord regeneration: hydrogel combined with neural stem cells. Frontiers in Pharmacology 2024, 15, 1419797. [Google Scholar] [CrossRef] [PubMed]
- Parvin, N. , Joo, S.W., Jung, J.H., & Mandal, T.K. Multimodal AI in Biomedicine: Pioneering the Future of Biomaterials, Diagnostics, and Personalized Healthcare. Nanomaterials 2025, 15, 895. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y. , Li, X., Gao, Y., Mathivanan, S., Kong, L., Tao, Y.,... & Zhang, S.C. 3D bioprinting of human neural tissues with functional connectivity. Cell Stem Cell 2024, 31, 260–274. [Google Scholar] [CrossRef]
- Castiglione, H. , Vigneron, P.A., Baquerre, C., Yates, F., Rontard, J., & Honegger, T. Human brain organoids-on-chip: advances, challenges, and perspectives for preclinical applications. Pharmaceutics 2022, 14, 2301. [Google Scholar]
- Carvalho, V. , Rodrigues, R.O., Shin, S.R., Lima, R., & Teixeira, S.F. Advancing blood–brain barrier-on-a-chip models through numerical simulations. BioChip Journal 2024, 18, 546–565. [Google Scholar] [CrossRef]
- Amartumur, S. , Nguyen, H., Huynh, T., Kim, T.S., Woo, R.S., Oh, E.,... & Heo, C. Neuropathogenesis-on-chips for neurodegenerative diseases. Nature Communications 2024, 15, 2219. [Google Scholar] [CrossRef] [PubMed]
- Boufidis, D. , Garg, R., Angelopoulos, E., Cullen, D.K., & Vitale, F. Bioinspired electronics: Soft, biohybrid, and “living” neural interfaces. Nature Communications 2025, 16, 1861. [Google Scholar]
- De Vitis, E.; La Pesa, V.; Gervaso, F.; Romano, A.; Quattrini, A.; Gigli, G.; Moroni, L.; Polini, A. A microfabricated multi-compartment device for neuron and Schwann cell differentiation. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhou, X.; Fang, Y.; Xiong, Z.; Zhang, T. AI-driven 3D bioprinting for regenerative medicine: From bench to bedside. Bioact. Mater. 2025, 45, 201–230. [Google Scholar] [CrossRef]
- Lai, J.; Liu, Y.; Lu, G.; Yung, P.; Wang, X.; Tuan, R.S.; Li, Z.A. 4D bioprinting of programmed dynamic tissues. Bioact. Mater. 2024, 37, 348–377. [Google Scholar] [CrossRef]
- You, D.; Lin, L.; Dong, M.; Wu, Y.; Hu, Y.; Hu, X.; Shao, Y.; Xie, Y.; Xu, M.; Chen, G.; et al. Recent Advances in Shape Memory Polymers for Biomedical Applications: Bridging Macro- and Micro-Scale Effects. Smart Mater. Med. 2025. [Google Scholar] [CrossRef]
- Aizarna-Lopetegui, U.; Bittinger, S.C.; Álvarez, N.; Henriksen-Lacey, M.; de Aberasturi, D.J. Stimuli-responsive hybrid materials for 4D in vitro tissue models. Mater. Today Bio 2025, 33, 102035. [Google Scholar] [CrossRef] [PubMed]
- Krishna, D.V.; Sankar, M.R. Machine learning-assisted extrusion-based 3D bioprinting for tissue regeneration applications. Ann. 3D Print. Med. 2023, 12, 100132. [Google Scholar] [CrossRef]
- Lai, J. , Chen, X. , Lu, H.H., & Wang, M. 3D bioprinting of graphene oxide-incorporated hydrogels for neural tissue regeneration. 3D Printing and Additive Manufacturing 2024, 11, e2022–e2032. [Google Scholar] [PubMed]
- Ye, L.; Ji, H.; Liu, J.; Tu, C.; Kappl, M.; Koynov, K.; Vogt, J.; Butt, H. Carbon Nanotube–Hydrogel Composites Facilitate Neuronal Differentiation While Maintaining Homeostasis of Network Activity. Adv. Mater. 2021, 33, 2102981. [Google Scholar] [CrossRef]
- Shin, J.; Kang, R.; Hyun, K.; Li, Z.; Kumar, H.; Kim, K.; Park, S.S.; Kim, K. Machine Learning-Enhanced Optimization for High-Throughput Precision in Cellular Droplet Bioprinting. Adv. Sci. 2025, 12, e2412831. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wallmersperger, T.; Ehrenhofer, A. Prediction of hydrogel swelling states using machine learning methods. Eng. Rep. 2024, 6, e12893. [Google Scholar] [CrossRef]
- Esworthy, T.J.; Miao, S.; Lee, S.-J.; Zhou, X.; Cui, H.; Zuo, Y.Y.; Zhang, L.G. Advanced 4D-bioprinting technologies for brain tissue modeling and study. Int. J. Smart Nano Mater. 2019, 10, 177–204. [Google Scholar] [CrossRef]
- Freeman, S.; Calabro, S.; Williams, R.; Jin, S.; Ye, K. Bioink Formulation and Machine Learning-Empowered Bioprinting Optimization. Front. Bioeng. Biotechnol. 2022, 10, 913579. [Google Scholar] [CrossRef]
- Mofrad, Y.M.; Shamloo, A. The effect of conductive aligned fibers in an injectable hydrogel on nerve tissue regeneration. Int. J. Pharm. 2023, 645, 123419. [Google Scholar] [CrossRef]
- Yeh, Y.-C.; Chen, P.-Y.; Chen, K.-T.; Lee, I.-C. Innovative MXene/SilMA-Based Conductive Bioink for Three Dimensional Bioprinting of Neural Stem Cell Spheroids in Neural Tissue Engineering. ACS Appl. Mater. Interfaces 2025, 17, 10402–10416. [Google Scholar] [CrossRef] [PubMed]
- Jamshidi, M.B. , Hoang, D. T., Nguyen, D.N., Niyato, D., & Warkiani, M.E. Revolutionizing biological digital twins: Integrating internet of bionano things, convolutional neural networks, and federated learning. Computers in Biology and Medicine 2025, 189, 109970. [Google Scholar]
- Gangwal, A.; Lavecchia, A. Artificial intelligence in preclinical research: enhancing digital twins and organ-on-chip to reduce animal testing. Drug Discov. Today 2025, 30, 104360. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Narayanan, L.K.; Shirwaiker, R. A framework for digital twin integration in biofabrication and a scaffold 3D bioplotting case study. Manuf. Lett. 2024, 41, 1182–1191. [Google Scholar] [CrossRef]
- Wang, S.; Wang, X.; Holland, M.A. Multi-physics modeling and finite-element formulation of neuronal dendrite growth with electrical polarization. Brain Multiphysics 2023, 4, 100071. [Google Scholar] [CrossRef]
- Zheng, H. , Xie, W., Wang, K., & Li, Z. Opportunities of Hybrid Model-based Reinforcement Learning for Cell Therapy Manufacturing Process Control. arXiv 2022, arXiv:2201.03116. [Google Scholar]
- Xiong, H.; Chu, C.; Fan, L.; Song, M.; Zhang, J.; Ma, Y.; Zheng, R.; Zhang, J.; Yang, Z.; Jiang, T. The Digital Twin Brain: A Bridge between Biological and Artificial Intelligence. Intell. Comput. 2023, 2. [Google Scholar] [CrossRef]
- Zahedmanesh, H.; Lally, C. A multiscale mechanobiological modelling framework using agent-based models and finite element analysis: application to vascular tissue engineering. Biomech. Model. Mechanobiol. 2011, 11, 363–377. [Google Scholar] [CrossRef]
- Ramesh, S.; Deep, A.; Tamayol, A.; Kamaraj, A.; Mahajan, C.; Madihally, S. Advancing 3D bioprinting through machine learning and artificial intelligence. Bioprinting 2024, 38, e00331. [Google Scholar] [CrossRef]
- Treloar, N.J.; Fedorec, A.J.H.; Ingalls, B.; Barnes, C.P.; You, L. Deep reinforcement learning for the control of microbial co-cultures in bioreactors. PLOS Comput. Biol. 2020, 16, e1007783. [Google Scholar] [CrossRef]
- Xiong, H.; Chu, C.; Fan, L.; Song, M.; Zhang, J.; Ma, Y.; Zheng, R.; Zhang, J.; Yang, Z.; Jiang, T. The Digital Twin Brain: A Bridge between Biological and Artificial Intelligence. Intell. Comput. 2023, 2, 0055. [Google Scholar] [CrossRef]
- Fekonja, L.S.; Schenk, R.; Schröder, E.; Tomasello, R.; Tomšič, S.; Picht, T. The digital twin in neuroscience: from theory to tailored therapy. Front. Neurosci. 2024, 18, 1454856. [Google Scholar] [CrossRef]
- An, J.; Chua, C.K.; Mironov, V. Application of Machine Learning in 3D Bioprinting: Focus on Development of Big Data and Digital Twin. Int. J. Bioprinting 2021, 7, 342. [Google Scholar] [CrossRef]
- E Wang, H.; Triebkorn, P.; Breyton, M.; Dollomaja, B.; Lemarechal, J.-D.; Petkoski, S.; Sorrentino, P.; Depannemaecker, D.; Hashemi, M.; Jirsa, V.K. Virtual brain twins: from basic neuroscience to clinical use. Natl. Sci. Rev. 2024, 11, nwae079. [Google Scholar] [CrossRef]
- Malashin, I.; Masich, I.; Tynchenko, V.; Gantimurov, A.; Nelyub, V.; Borodulin, A.; Martysyuk, D.; Galinovsky, A. Machine Learning in 3D and 4D Printing of Polymer Composites: A Review. Polymers 2024, 16, 3125. [Google Scholar] [CrossRef] [PubMed]
- Akbarialiabad, H.; Seyyedi, M.S.; Paydar, S.; Habibzadeh, A.; Haghighi, A.; Kvedar, J.C. Bridging silicon and carbon worlds with digital twins and on-chip systems in drug discovery. npj Syst. Biol. Appl. 2024, 10, 150. [Google Scholar] [CrossRef] [PubMed]
- Qian, K.; Pawar, A.; Liao, A.; Anitescu, C.; Webster-Wood, V.; Feinberg, A.W.; Rabczuk, T.; Zhang, Y.J. Modeling neuron growth using isogeometric collocation based phase field method. Sci. Rep. 2022, 12, 8120. [Google Scholar] [CrossRef] [PubMed]
- De Domenico, M.; Allegri, L.; Caldarelli, G.; D’aNdrea, V.; Di Camillo, B.; Rocha, L.M.; Rozum, J.; Sbarbati, R.; Zambelli, F. Challenges and opportunities for digital twins in precision medicine from a complex systems perspective. npj Digit. Med. 2025, 8, 37. [Google Scholar] [CrossRef]
- Park, S.-Y.; Park, C.-H.; Choi, D.-H.; Hong, J.K.; Lee, D.-Y. Bioprocess digital twins of mammalian cell culture for advanced biomanufacturing. Curr. Opin. Chem. Eng. 2021, 33, 100702. [Google Scholar] [CrossRef]
- Wu, Y.; Ding, X.; Wang, Y.; Ouyang, D. Harnessing the power of machine learning into tissue engineering: current progress and future prospects. Burn. Trauma 2024, 12, tkae053. [Google Scholar] [CrossRef]
- Spitz, S. , Schobesberger, S. , Brandauer, K., & Ertl, P. Sensor-integrated brain-on-a-chip platforms: improving the predictive validity in neurodegenerative research. Bioengineering & Translational Medicine 2024, 9, e10604. [Google Scholar]
- Zhang, H.; Huang, N.; Bian, S.; Sawan, M. Brain organoids-on-chip for neural diseases modeling: History, challenges and trends. J. Pharm. Anal. 2025, 101323. [Google Scholar] [CrossRef]
- Qiu, B. , Pompe, S., Xenaki, K.T., van Bergen en Henegouwen, P.M., Oliveira, S., Mastrobattista, E., & Caiazzo, M. Generation of a Three-dimensional Human Neurovascular Unit Model in a Microfluidic Chip. bioRxiv 2025, 2025-04. [Google Scholar]
- Berjaoui, C.; Kachouh, C.; Joumaa, S.; Ghayyad, M.H.; Bekele, B.A.; Ajirenike, R.; Al Maaz, Z.; Awde, S.; Wojtara, M.; Nazir, A.; et al. Neuroinflammation-on-a-chip for multiple sclerosis research: a narrative review. Ann. Med. Surg. 2024, 86, 4053–4059. [Google Scholar] [CrossRef]
- Rodrigues, R.O.; Shin, S.-R.; Bañobre-López, M. Brain-on-a-chip: an emerging platform for studying the nanotechnology-biology interface for neurodegenerative disorders. J. Nanobiotechnology 2024, 22, 573. [Google Scholar] [CrossRef] [PubMed]
- Sakai, K. , Teshima, T. F., Goto, T., Nakashima, H., & Yamaguchi, M. Self-Folding Graphene-Based Interface for Brain-Like Modular 3D Tissue. Advanced Functional Materials 2023, 33, 2301836. [Google Scholar]
- Wang, Z.; Ma, D.; Liu, J.; Xu, S.; Qiu, F.; Hu, L.; Liu, Y.; Ke, C.; Ruan, C. 4D printing polymeric biomaterials for adaptive tissue regeneration. Bioact. Mater. 2025, 48, 370–399. [Google Scholar] [CrossRef]
- Lai, J.; Liu, Y.; Lu, G.; Yung, P.; Wang, X.; Tuan, R.S.; Li, Z.A. 4D bioprinting of programmed dynamic tissues. Bioact. Mater. 2024, 37, 348–377. [Google Scholar] [CrossRef] [PubMed]
- Akbarialiabad, H.; Seyyedi, M.S.; Paydar, S.; Habibzadeh, A.; Haghighi, A.; Kvedar, J.C. Bridging silicon and carbon worlds with digital twins and on-chip systems in drug discovery. npj Syst. Biol. Appl. 2024, 10, 150. [Google Scholar] [CrossRef]
- A Shah, P.; Shrivastav, P.S.; Ghate, M.; Chavda, V. The fusion of microfluidics and artificial intelligence: a novel alliance for medical advancements. Bioanalysis 2024, 16, 927–930. [Google Scholar] [CrossRef]
- Xu, X.; Zuo, Y.; Chen, S.; Hatami, A.; Gu, H. Advancements in Brain Research: The In Vivo/In Vitro Electrochemical Detection of Neurochemicals. Biosensors 2024, 14, 125. [Google Scholar] [CrossRef]
- Yu, S.; Jiang, L.; Song, M.; Yang, T.; Yuan, M.; Chen, X.; Shu, H. Regulation of BBB function and pathological evolution of PD by microenvironment “spatiotemporal gradient”: unique advantages of microfluidic chips. Front. Aging Neurosci. 2025, 17, 1599509. [Google Scholar] [CrossRef]
- Ferreira, M.J.; Colombani, S.; Bernardin, A.; Lacampagne, A.; Pasquié, J.-L.; Costa, P.F.; Charlot, B.; Meli, A.C. Advancing organ-on-chip systems: the role of microfluidics in neuro-cardiac research. Curr. Res. Pharmacol. Drug Discov. 2025, 9, 100227. [Google Scholar] [CrossRef]
- Yan, Y.; Li, X.; Gao, Y.; Mathivanan, S.; Kong, L.; Tao, Y.; Dong, Y.; Li, X.; Bhattacharyya, A.; Zhao, X.; et al. 3D bioprinting of human neural tissues with functional connectivity. Cell Stem Cell 2024, 31, 260–274. [Google Scholar] [CrossRef]
- Kahveci, B. , Polatli, E. bioRxiv 2025, 2025-02. [Google Scholar]
- Dhar, S.; Ahmad, F.; Deshpande, A.; Rana, S.S.; A, T.A.; Priyadarsini, S. 3-Dimensional printing and bioprinting in neurological sciences: applications in surgery, imaging, tissue engineering, and pharmacology and therapeutics. J. Mater. Sci. Mater. Med. 2025, 36, 32. [Google Scholar] [CrossRef]
- Lai, J.; Liu, Y.; Lu, G.; Yung, P.; Wang, X.; Tuan, R.S.; Li, Z.A. 4D bioprinting of programmed dynamic tissues. Bioact. Mater. 2024, 37, 348–377. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhou, X.; Fang, Y.; Xiong, Z.; Zhang, T. AI-driven 3D bioprinting for regenerative medicine: From bench to bedside. Bioact. Mater. 2025, 45, 201–230. [Google Scholar] [CrossRef]
- Li, K.; Huang, W.H.; Guo, H.T.; Liu, Y.Y.; Chen, S.; Liu, H.; Gu, Q. Advancements in robotic arm-based 3D bioprinting for biomedical applications. Life Med. 2023, 2, lnad046. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.; Deep, A.; Tamayol, A.; Kamaraj, A.; Mahajan, C.; Madihally, S. Advancing 3D bioprinting through machine learning and artificial intelligence. Bioprinting 2024, 38, e00331. [Google Scholar] [CrossRef]
- Boufidis, D. , Garg, R. , Angelopoulos, E., Cullen, D.K., & Vitale, F. Bioinspired electronics: Soft, biohybrid, and “living” neural interfaces. Nature Communications 2025, 16, 1861. [Google Scholar]
- Ienca, M.; Valle, G.; Raspopovic, S. Clinical trials for implantable neural prostheses: understanding the ethical and technical requirements. Lancet Digit. Heal. 2025, 7, e216–e224. [Google Scholar] [CrossRef]
- Lindner, N.; Mejia-Wille, A.; Fritschen, A.; Blaeser, A. Development of a robotic-assisted handling and manipulation system for the high-scale bioproduction of 3D-bioprinted organ-on-a-chip devices. HardwareX 2024, 19, e00572. [Google Scholar] [CrossRef]
- Fritschen, A. , Lindner, N. , Scholpp, S., Richthof, P., Dietz, J., Linke, P.,... & Blaeser, A. High-Scale 3D-Bioprinting Platform for the Automated Production of Vascularized Organs-on-a-Chip. Advanced Healthcare Materials 2024, 13, 2304028. [Google Scholar]
- Kim, M.H.; Singh, Y.P.; Celik, N.; Yeo, M.; Rizk, E.; Hayes, D.J.; Ozbolat, I.T. High-throughput bioprinting of spheroids for scalable tissue fabrication. Nat. Commun. 2024, 15, 1–21. [Google Scholar] [CrossRef]
- Lv, S.; Xu, Z.; Mo, F.; Wang, Y.; Duan, Y.; Liu, Y.; Jing, L.; Shan, J.; Jia, Q.; Wang, M.; et al. Long-term stability strategies of deep brain flexible neural interface. npj Flex. Electron. 2025, 9, 1–17. [Google Scholar] [CrossRef]
- Pawlak, W.A.; Howard, N. Neuromorphic algorithms for brain implants: a review. Front. Neurosci. 2025, 19, 1570104. [Google Scholar] [CrossRef] [PubMed]
- Barjuei, E.S.; Shin, J.; Kim, K.; Lee, J. Precision improvement of robotic bioprinting via vision-based tool path compensation. Sci. Rep. 2024, 14, 17764. [Google Scholar] [CrossRef] [PubMed]
- Sirbu, R. , Morley, J., Schroder, T., Taddeo, M., Pothukuchi, R.P., Ugur, M.,... & Floridi, L. Regulating next-generation implantable brain-computer interfaces: Recommendations for ethical development and implementation. arXiv 2025, arXiv:2506.12540. [Google Scholar]
- Gao, H.; Shen, H.; Zhang, X.; Liu, Y.; Shang, Y.; Sun, S.; Guan, W.; Gu, X.; Yang, Y.; Li, G. Revolutionizing neural regeneration with smart responsive materials: Current insights and future prospects. Bioact. Mater. 2025, 52, 393–421. [Google Scholar] [CrossRef]
- Groff-Vindman, C.S.; Trump, B.D.; Cummings, C.L.; Smith, M.; Titus, A.J.; Oye, K.; Prado, V.; Turmus, E.; Linkov, I. The convergence of AI and synthetic biology: the looming deluge. npj Biomed. Innov. 2025, 2, 20. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. (2024, –21). Executive summary for the Digital Health Advisory Committee meeting: Total product lifecycle considerations for generative AI-enabled devices [PDF]. U.S. Food and Drug Administration. https://www.fda. 20 November 1828.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).