Submitted:
09 July 2025
Posted:
10 July 2025
You are already at the latest version
Abstract

Keywords:
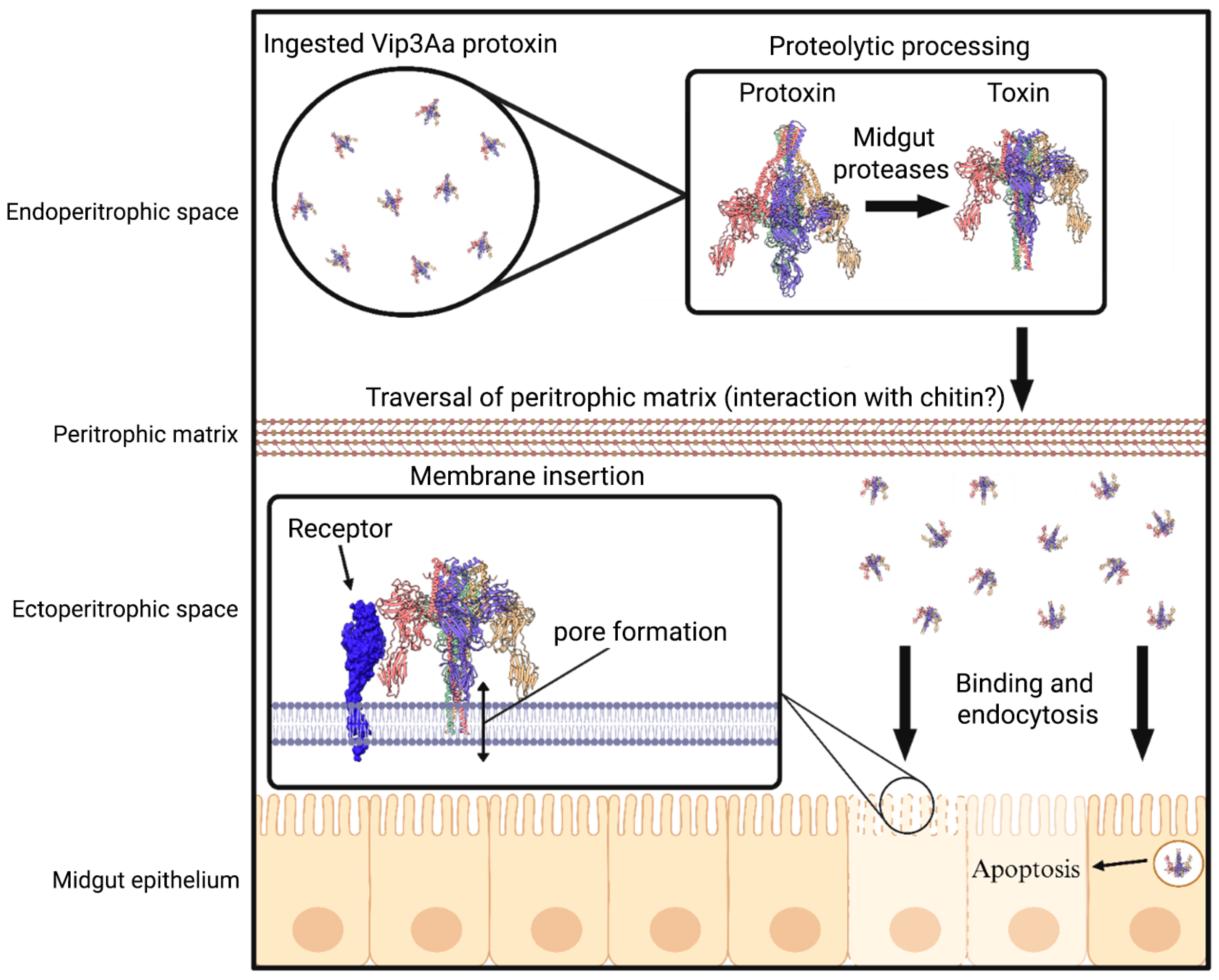
1. Introduction
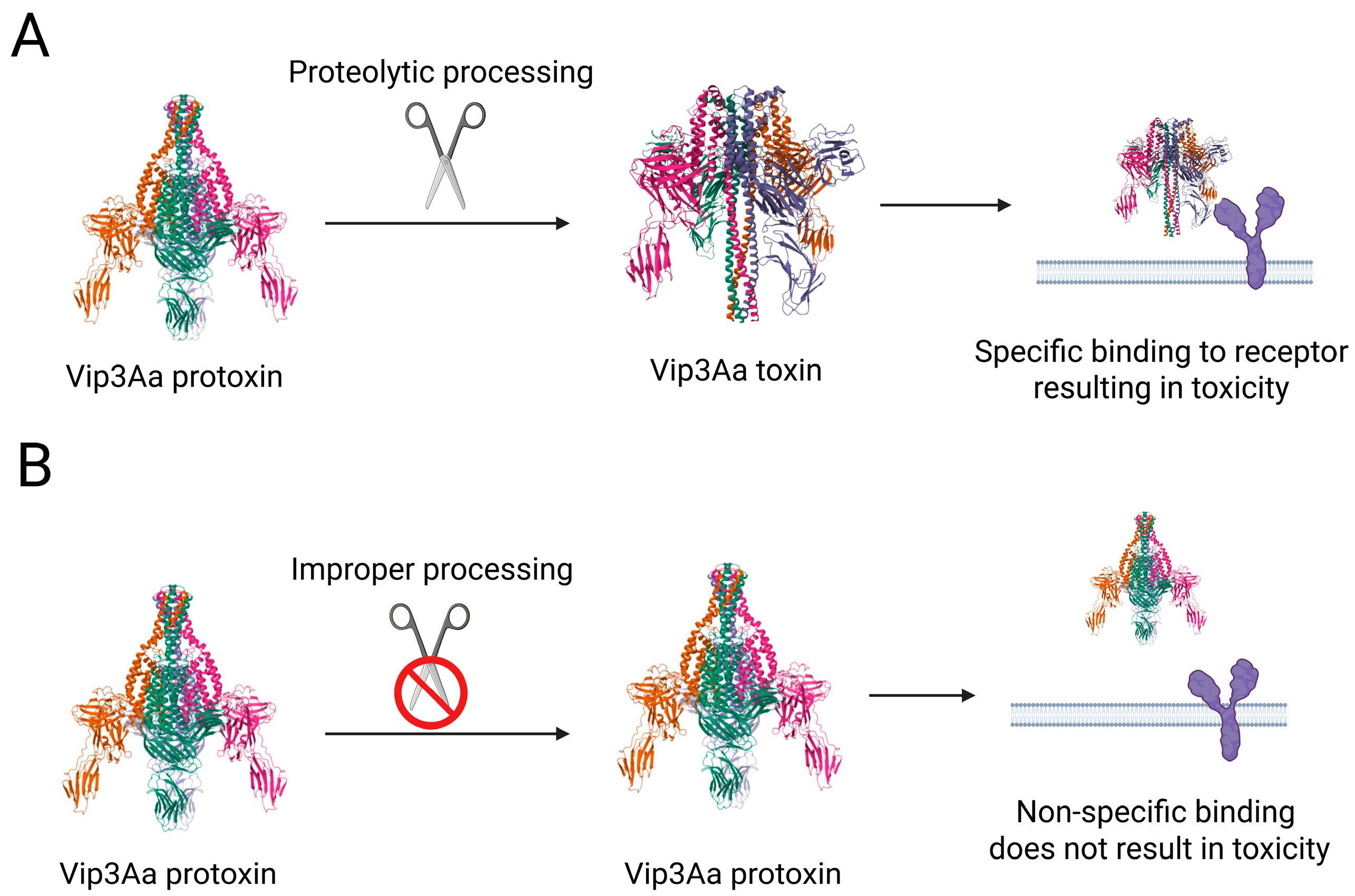
2. Altered Protoxin Processing
3. Interactions with the Peritrophic Matrix
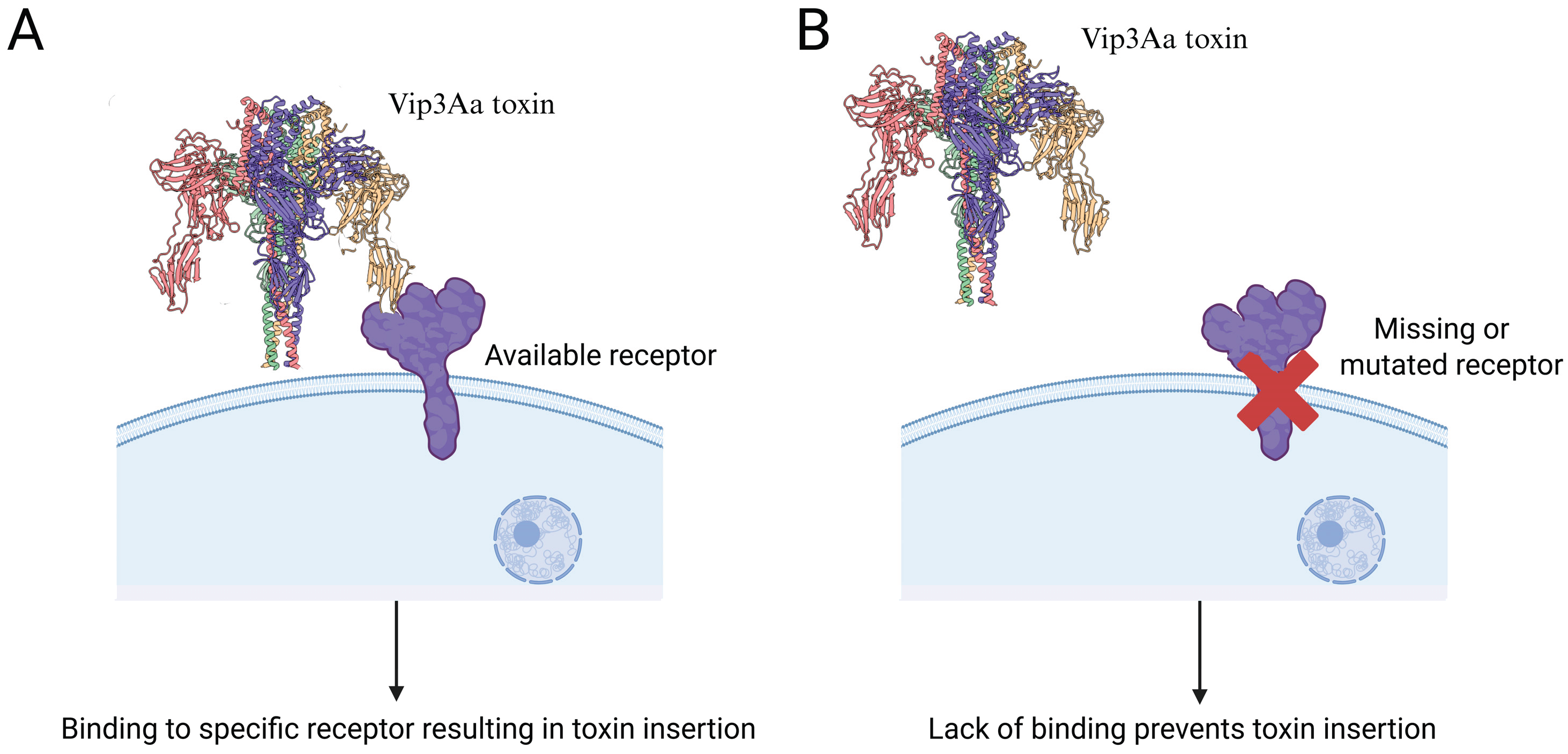
4. Binding to Receptors in the Midgut
5. Mechanism of Cytotoxicity
6. Other Critical Genes Involved in Resistance to Vip3Aa
7. Current Status of Field Resistance to Vip3Aa
| Species | Origin | RR | Inheritance | CRR* | Fitness cost | Refs |
| Chloridea virescens | Field Mississippi 2006 |
2,040 | Paternal influence Polygenic | 6.7 Cry1Ab 1.0 Cry1Ac |
Reduced survival to adult, egg viability, and mating success | [90] |
| Helicoverpa armigera | F2 screen Australia 2009-2010 |
>232 | Maternal influence Recessive Monogenic |
1.7 Cry1Ac 0.3 Cry2Ab |
NA | [42,63,70] |
| Helicoverpa punctigera | F2 screen Australia 2009-2010 |
>215 | Autosomal Recessive Monogenic |
3.2 Cry1Ac 1.7 Cry2Ab |
NA | [70] |
| Helicoverpa zea | F2 screen Snook (TX) 2019 |
45,194 | Autosomal Recessive Monogenic |
0.2 Cry1A.105 1.6 Cry1Ac 0.5 Cry2Ab |
NA | [87] |
| F2 screen Alexandria (LA) 2019 |
>909 | Autosomal Recessive Monogenic |
NA | NA | [81] | |
| F2 screen Winnsboro (LA) 2020 |
>909 | Autosomal Recessive Monogenic |
NA | NA | [81] | |
| F2 screen Stoneville (MS) 2020 |
>909 | Autosomal Recessive Monogenic |
NA | NA | [81] | |
| Lab selection Georgia (USA) 2023 |
267 | Autosomal Recessive |
NA | Resistance reduced in heterogeneous strains without selection | [76] | |
| Spodoptera frugiperda | F2 screen Brazil 2016 |
>3,200 | Autosomal Recessive Monogenic |
Survives Cry1Ab + Vip3Aa20 corn | Reduced fecundity, survival, and reproductive rate | [71,77] |
| F2 screen Rapides (LA) 2016 |
>632 | Autosomal Recessive Monogenic |
0.5 Cry1F 1.4 Cry2Ab 0.4 Cry2Ae |
Reduced pupal weight and longer pupal development | [41,85] | |
| F2 screen Snook (TX) 2018 |
>395 | Autosomal Recessive Monogenic |
2.9 Cry2Ab 1.9 Cry1F |
NA | [80,86] | |
| Field Tifton, (GA) 2012 |
>9,800 | NA | NA | Slower growth and lower pupation rate | [75] | |
| Lab selection Yunan (China) 2023 |
5,562 | Autosomal Recessive Monogenic |
2.0 Cry1Ab 0.9 Cry1Ac 2.0 Cry1F 1.8 Cry2Ab |
NA | [38] | |
| Lab selection Yunan (China) 2023 |
206 | Autosomal Recessive |
1.2 Cry1Ab 2.0 Cry1F 1.6 Cry2Ab |
NA | [69] | |
| Mythimna separata | Lab selection Jilin (China) 2021 |
>3,061 | NA | 2.8 Cry1Ab 0.6 Cry1F |
NA | [43] |
| Lab selection Jilin (China) 2022 |
>634 | Maternal influence Incomplete dominant Polygenic |
NA | Longer larval development, lower pupation rate, adult emergence, and reproductive rate | [79] |
8. Conclusions and Prospects
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| Bt | Bacillus thuringiensis |
| PIP | Plant incorporated protectant |
| BBMV | Brush border membrane vesicle |
References
- Crickmore, N.; Berry, C.; Panneerselvam, S.; Mishra, R.; Connor, T.R.; Bonning, B.C. A structure-based nomenclature for Bacillus thuringiensis and other bacteria-derived pesticidal proteins. J. Invert. Path. 2020, 107438. [CrossRef]
- Dively, G.P.; Kuhar, T.P.; Taylor, S.V.; Doughty, H.; Holmstrom, K.; Gilrein, D.O.; Nault, B.A.; Ingerson-Mahar, J.; Huseth, A.; Reisig, D.; et al. Extended sentinel monitoring of Helicoverpa zea resistance to Cry and Vip3Aa toxins in Bt sweet corn: Assessing changes in phenotypic and allele frequencies of resistance. Insects 2023, 14(7), 577. [CrossRef]
- International Service for the Acquisition of Agri-biotech Applications (ISAAA), Brief 55: Global Status of Commercialized Biotech/GM Crops. 2019. Ithaca, NY.
- Fleming, D.; Musser, F.; Reisig, D.; Greene, J.; Taylor, S.; Parajulee, M.; Lorenz, G.; Catchot, A.; Gore, J.; Kerns, D.; et al. Effects of transgenic Bacillus thuringiensis cotton on insecticide use, heliothine counts, plant damage, and cotton yield: A meta-analysis, 1996-2015. PLoS ONE 2018, 13, e0200131. [CrossRef]
- Bueno, A.d.F.; Braz-Zini, E.C.; Horikoshi, R.J.; Bernardi, O.; Andrade, G.; Sutil, W.P. Over 10 years of Bt soybean in Brazil: Lessons, benefits, and challenges for its use in integrated pest management (IPM). Neotrop. Entomol. 2025, 54, 61. [CrossRef]
- Wu, F. Mycotoxin risks are lower in biotech corn. Curr. Opin. Biotechnol. 2022, 78, 102792. [CrossRef]
- Lee, M.K.; Walters, F.S.; Hart, H.; Palekar, N.; Chen, J.S. The mode of action of the Bacillus thuringiensis vegetative insecticidal protein Vip3A differs from that of Cry1Ab delta-endotoxin. Appl. Environ. Microbiol. 2003, 69, 4648-4657. [CrossRef]
- Gilreath, R.T.; Kerns, D.L.; Huang, F.; Yang, F. No positive cross-resistance to Cry1 and Cry2 proteins favors pyramiding strategy for management of Vip3Aa resistance in Spodoptera frugiperda. Pest Manag. Sci. 2021, 77, 1963-1970. [CrossRef]
- Tabashnik, B.E.; Carriere, Y. Surge in insect resistance to transgenic crops and prospects for sustainability. Nat. Biotechnol. 2017, 35, 926-935. [CrossRef]
- Reisig, D.; Buntin, G.D.; Greene, J.K.; Paula-Moraes, S.V.; Reay-Jones, F.; Roberts, P.; Smith, R.; Taylor, S.V. Magnitude and extent of Helicoverpa zea resistance levels to Cry1Ac and Cry2Ab2 across the Southeastern USA. Insects 2023, 14(3), 262. [CrossRef]
- Dively, G.P.; Venugopal, P.D.; Finkenbinder, C. Field-evolved resistance in corn earworm to Cry proteins expressed by transgenic sweet corn. PLoS One 2016, 11, e0169115. [CrossRef]
- Gassmann, A.J.; Reisig, D.D. Management of insect pests with Bt crops in the United States. Annu. Rev. Entomol. 2023, 68, 31-49. [CrossRef]
- Yang, F.; Kerns, D.L.; Little, N.S.; Santiago González, J.C.; Tabashnik, B.E. Early warning of resistance to Bt toxin Vip3Aa in Helicoverpa zea. Toxins 2021, 13, 618. [CrossRef]
- Gottems, L. IMA reveals increased insecticide use in Viptera biotechnology. AgNews 2025. https://news.agropages.com/News/NewsDetail---53498.htm (accessed 04/15/2025).
- Ferré, J.; Bel, Y.; Lázaro-Berenguer, M.; Hernández-Martínez, P. Chapter Three - Vip3 insecticidal proteins: Structure and mode of action. In Advances in Insect Physiology, Jurat-Fuentes, J.L., Ed.; Academic Press: 2023; Volume 65, pp. 93-122. [CrossRef]
- Lee, M.K.; Miles, P.; Chen, J.S. Brush border membrane binding properties of Bacillus thuringiensis Vip3A toxin to Heliothis virescens and Helicoverpa zea midguts. Biochem. Biophys. Res. Commun. 2006, 339, 1043-1047. [CrossRef]
- Tabashnik, B.E.; Carrière, Y. Evaluating cross-resistance between Vip and Cry toxins of Bacillus thuringiensis. J. Econ. Entomol. 2020, 113, 553-561. [CrossRef]
- Yu, C.G.; Mullins, M.A.; Warren, G.W.; Koziel, M.G.; Estruch, J.J. The Bacillus thuringiensis vegetative insecticidal protein Vip3A lyses midgut epithelium cells of susceptible insects. Appl. Environ. Microbiol. 1997, 63, 532-536. [CrossRef]
- Núñez-Ramírez, R.; Huesa, J.; Bel, Y.; Ferré, J.; Casino, P.; Arias-Palomo, E. Molecular architecture and activation of the insecticidal protein Vip3Aa from Bacillus thuringiensis. Nat. Commun. 2020, 11, 3974. [CrossRef]
- Caccia, S.; Chakroun, M.; Vinokurov, K.; Ferré, J. Proteolytic processing of Bacillus thuringiensis Vip3A proteins by two Spodoptera species. J. Insect Physiol. 2014, 67, 76-84. [CrossRef]
- Zhang, J.; Pan, Z.Z.; Xu, L.; Liu, B.; Chen, Z.; Li, J.; Niu, L.Y.; Zhu, Y.J.; Chen, Q.X. Proteolytic activation of Bacillus thuringiensis Vip3Aa protein by Spodoptera exigua midgut protease. Int. J. Biol. Macromol. 2018, 107, 1220-1226. [CrossRef]
- Chakroun, M.; Bel, Y.; Caccia, S.; Abdelkefi-Mesrati, L.; Escriche, B.; Ferré, J. Susceptibility of Spodoptera frugiperda and S. exigua to Bacillus thuringiensis Vip3Aa insecticidal protein. J. Invert. Pathol. 2012, 110, 334-339. [CrossRef]
- Jiang, K.; Mei, S.-q.; Wang, T.-t.; Pan, J.-h.; Chen, Y.-h.; Cai, J. Vip3Aa induces apoptosis in cultured Spodoptera frugiperda (Sf9) cells. Toxicon 2016, 120, 49-56. [CrossRef]
- Jiang, K.; Hou, X.-Y.; Tan, T.-T.; Cao, Z.-L.; Mei, S.-Q.; Yan, B.; Chang, J.; Han, L.; Zhao, D.; Cai, J. Scavenger receptor-C acts as a receptor for Bacillus thuringiensis vegetative insecticidal protein Vip3Aa and mediates the internalization of Vip3Aa via endocytosis. PLOS Pathogens 2018, 14, e1007347. [CrossRef]
- Hou, X.; Han, L.; An, B.; Zhang, Y.; Cao, Z.; Zhan, Y.; Cai, X.; Yan, B.; Cai, J. Mitochondria and lysosomes participate in Vip3Aa-induced Spodoptera frugiperda Sf9 cell apoptosis. Toxins 2020, 12, 116. [CrossRef]
- Nimsanor, S.; Srisaisup, M.; Jammor, P.; Promdonkoy, B.; Boonserm, P. Intracellular localization and cytotoxicity of Bacillus thuringiensis Vip3Aa against Spodoptera frugiperda (Sf9) cells. J. Invert. Pathol 2020, 171, 107340. [CrossRef]
- Lázaro-Berenguer, M.; Ferré, J.; Hernández-Martínez, P. Receptor interactions of protoxin and activated Vip3Aa structural conformations in Spodoptera exigua. Pest Manag. Sci. 2024, 80, 6142-6149. [CrossRef]
- Barkhade, U.; Thakare, A. Protease mediated resistance mechanism to Cry1C and Vip3A in Spodoptera litura. Egypt. Acad. J. Biol. Sci. A Entomol. 2010, 3, 43-50. [CrossRef]
- Chakroun, M.; Banyuls, N.; Walsh, T.; Downes, S.; James, B.; Ferre, J. Characterization of the resistance to Vip3Aa in Helicoverpa armigera from Australia and the role of midgut processing and receptor binding. Sci. Rep. 2016, 6, 24311. [CrossRef]
- Ayra-Pardo, C.; Ochagavía, M.E.; Raymond, B.; Gulzar, A.; Rodríguez-Cabrera, L.; Rodríguez de la Noval, C.; Morán Bertot, I.; Terauchi, R.; Yoshida, K.; Matsumura, H.; et al. HT-SuperSAGE of the gut tissue of a Vip3Aa-resistant Heliothis virescens (Lepidoptera: Noctuidae) strain provides insights into the basis of resistance. Insect Sci. 2019, 26, 479-498. [CrossRef]
- Roy, R.; Abdelgaffar, H.; Kerns, D.; Huff, M.; Staton, M.; Yang, F.; Huang, F.; Jurat-Fuentes, J.L. Reduced processing and toxin binding associated with resistance to Vip3Aa in a strain of fall armyworm (Spodoptera frugiperda) from Louisiana. Pest Manag. Sci. 2025, 81(7), 4097-4107. [CrossRef]
- Infante, O.; Gómez, I.; Pélaez-Aguilar, A.E.; Verduzco-Rosas, L.A.; García-Suárez, R.; García-Gómez, B.I.; Wang, Z.; Zhang, J.; Guerrero, A.; Bravo, A.; et al. Insights into the structural changes that trigger receptor binding upon proteolytic activation of Bacillus thuringiensis Vip3Aa insecticidal protein. PLoS Pathogens 2024, 20, e1012765. [CrossRef]
- Lázaro-Berenguer, M.; Paredes-Martínez, F.; Bel, Y.; Núñez-Ramírez, R.; Arias-Palomo, E.; Casino, P.; Ferré, J. Structural and functional role of Domain I for the insecticidal activity of the Vip3Aa protein from Bacillus thuringiensis. Microb. Biotechnol. 2022, 15, 2607-2618. [CrossRef]
- Qi, L.; Dai, H.; Jin, Z.; Shen, H.; Guan, F.; Yang, Y.; Tabashnik, B.E.; Wu, Y. Evaluating cross-resistance to Cry and Vip toxins in four strains of Helicoverpa armigera with different genetic mechanisms of resistance to Bt toxin Cry1Ac. Front. Microbiol. 2021, Volume 12 - 2021. [CrossRef]
- Wang, M.; Zhang, S.; Shi, Y.; Yang, Y.; Wu, Y. Global gene expression changes induced by knockout of a protease gene cluster in Helicoverpa armigera with CRISPR/Cas9. J. Insect Physiol. 2020, 122, 104023. [CrossRef]
- Sharath Chandra, G.; Asokan, R.; Manamohan, M.; Ellango, R.; Sharma, H.C.; Akbar, S.M.D.; Krishna Kumar, N.K. Double-stranded RNA-mediated suppression of trypsin-like serine protease (t-SP) triggers over-expression of another t-SP isoform in Helicoverpa armigera. Appl. Biochem. Biotechnol. 2018, 184, 746-761. [CrossRef]
- Jiang, K.; Chen, Z.; Zang, Y.; Shi, Y.; Shang, C.; Jiao, X.; Cai, J.; Gao, X. Functional characterization of Vip3Aa from Bacillus thuringiensis reveals the contributions of specific domains to its insecticidal activity. J. Biol. Chem. 2023, 299, 103000. [CrossRef]
- Liu, Z.; Liao, C.; Zou, L.; Jin, M.; Shan, Y.; Quan, Y.; Yao, H.; Zhang, L.; Wang, P.; Liu, Z.; et al. Retrotransposon-mediated variation of a chitin synthase gene confers insect resistance to Bacillus thuringiensis Vip3Aa toxin. PLoS Biol. 2024, 22(7), e3002704. [CrossRef]
- Jin, M.; Shan, Y.; Peng, Y.; Chen, S.; Zhou, X.; Liu, K.; Xiao, Y. CRISPR-mediated chromosome deletion facilitates genetic mapping of Vip3Aa resistance gene within complex genomic region in an invasive global pest. bioRxiv 2024, 2024.2007.2030.605831. [CrossRef]
- Wang, P.; Liu, Z.; Kang, Q.; Liao, C.; Zou, L.; Mao, K.; Yao, H.; Li, Y.; Xiao, Y. Functional loss of CHS2 confers high levels resistance to Bacillus thuringiensis Vip3Aa, but with significant fitness costs in five lepidopteran species. bioRxiv 2025, 2025.2006.2005.658017. [CrossRef]
- Chen, X.; Head, G.P.; Price, P.; Kerns, D.L.; Rice, M.E.; Huang, F.; Gilreath, R.T.; Yang, F. Fitness costs of Vip3A resistance in Spodoptera frugiperda on different hosts. Pest Manag. Sci. 2019, 75, 1074-1080. [CrossRef]
- Tandy, P.; Lamour, K.; Placidi de Bortoli, C.; Nagoshi, R.; Emrich, S.J.; Jurat-Fuentes, J.L. Screening for resistance alleles to Cry1 proteins through targeted sequencing in the native and invasive range of Spodoptera frugiperda (Lepidoptera: Noctuidae). J. Econ. Entomol. 2023, 116, 935-944. [CrossRef]
- Quan, Y.; Yang, J.; Wang, Y.; Hernández-Martínez, P.; Ferré, J.; He, K. The Rapid Evolution of resistance to Vip3Aa insecticidal protein in Mythimna separata (Walker) is not related to altered binding to midgut receptors. Toxins 2021, 13, 364. [CrossRef]
- Abdelkefi-Mesrati, L.; Boukedi, H.; Chakroun, M.; Kamoun, F.; Azzouz, H.; Tounsi, S.; Rouis, S.; Jaoua, S. Investigation of the steps involved in the difference of susceptibility of Ephestia kuehniella and Spodoptera littoralis to the Bacillus thuringiensis Vip3Aa16 toxin. J. Invert. Pathol. 2011, 107, 198-201. [CrossRef]
- Chakrabarty, S.; Jin, M.; Wu, C.; Chakraborty, P.; Xiao, Y. Bacillus thuringiensis vegetative insecticidal protein family Vip3A and mode of action against pest Lepidoptera. Pest Manag. Sci. 2020, 76, 1612-1617. [CrossRef]
- Syed, T.; Askari, M.; Meng, Z.; Li, Y.; Abid, M.; Wei, Y.; Guo, S.; Liang, C.; Zhang, R. Current insights on vegetative insecticidal proteins (Vip) as next generation pest killers. Toxins 2020, 12, 522. [CrossRef]
- Jurat-Fuentes, J.L.; Heckel, D.G.; Ferré, J. Mechanisms of resistance to insecticidal proteins from Bacillus thuringiensis. Annu. Rev. Entom. 2021, 66, 121-140. [CrossRef]
- Kerns, D.D.; Yang, F.; Kerns, D.L.; Stewart, S.D.; Jurat-Fuentes, J.L. Reduced toxin binding associated with resistance to Vip3Aa in the corn earworm (Helicoverpa zea). Appl. Environ. Microbiol. 2023, 89, e01644-23. [CrossRef]
- Singh, G.; Sachdev, B.; Sharma, N.; Seth, R.; Bhatnagar, R.K. Interaction of Bacillus thuringiensis vegetative insecticidal protein with ribosomal S2 protein triggers larvicidal activity in Spodoptera frugiperda. Appl. Environ. Microbiol. 2010, 76, 7202-7209. [CrossRef]
- An, B.; Zhang, Y.; Li, X.; Hou, X.; Yan, B.; Cai, J. PHB2 affects the virulence of Vip3Aa to Sf9 cells through internalization and mitochondrial stability. Virulence 2022, 13, 684-697. [CrossRef]
- Jiang, K.; Hou, X.; Han, L.; Tan, T.; Cao, Z.; Cai, J. Fibroblast growth factor receptor, a novel receptor for vegetative insecticidal protein Vip3Aa. Toxins 2018, 10, 546. [CrossRef]
- Ben Hamadou-Charfi, D.; Boukedi, H.; Abdelkefi-Mesrati, L.; Tounsi, S.; Jaoua, S. Agrotis segetum midgut putative receptor of Bacillus thuringiensis vegetative insecticidal protein Vip3Aa16 differs from that of Cry1Ac toxin. J. Invert. Pathol. 2013, 114, 139-143. [CrossRef]
- Osman, G.H.; Altaf, W.J.; Saleh, I.A.S.; Soltane, R.; Abulreesh, H.H.; Arif, I.A.; Ramadan, A.M.; Osman, Y.A. First report of detection of the putative receptor of Bacillus thuringiensis toxin Vip3Aa from black cutworm (Agrotis ipsilon). Saudi J. Biol. Sci. 2018, 25, 441-445. [CrossRef]
- Shan, Y.; Jin, M.; Chakrabarty, S.; Yang, B.; Li, Q.; Cheng, Y.; Zhang, L.; Xiao, Y. Sf-FGFR and Sf-SR-C are not the receptors for Vip3Aa to exert insecticidal toxicity in Spodoptera frugiperda. Insects 2022, 13, 547. [CrossRef]
- Liu, J.-G.; Yang, A.-Z.; Shen, X.-H.; Hua, B.-G.; Shi, G.-L. Specific binding of activated Vip3Aa10 to Helicoverpa armigera brush border membrane vesicles results in pore formation. J. Invert. Pathol. 2011, 108, 92-97. [CrossRef]
- Kunthic, T.; Watanabe, H.; Kawano, R.; Tanaka, Y.; Promdonkoy, B.; Yao, M.; Boonserm, P. pH regulates pore formation of a protease activated Vip3Aa from Bacillus thuringiensis. Biochim. Biophys. Acta - Biomembr. 2017, 1859, 2234-2241. [CrossRef]
- Pacheco, S.; Gómez, I.; Peláez-Aguilar, A.E.; Verduzco-Rosas, L.A.; García-Suárez, R.; Do Nascimento, N.A.; Rivera-Nájera, L.Y.; Cantón, P.E.; Soberón, M.; Bravo, A. Structural changes upon membrane insertion of the insecticidal pore-forming toxins produced by Bacillus thuringiensis. Front. Insect Sci. 2023, 3, 1188891. [CrossRef]
- Hernández-Martínez, P.; Gomis-Cebolla, J.; Ferré, J.; Escriche, B. Changes in gene expression and apoptotic response in Spodoptera exigua larvae exposed to sublethal concentrations of Vip3 insecticidal proteins. Sci. Rep. 2017, 7(1), 16245. [CrossRef]
- Tanaka, S.; Yoshizawa, Y.; Sato, R. Response of midgut epithelial cells to Cry1Aa is toxin-dependent and depends on the interplay between toxic action and the host apoptotic response. FEBS J. 2012, 279, 1071-1079. [CrossRef]
- Jurat-Fuentes, J.L.; Adang, M.J. Cry toxin mode of action in susceptible and resistant Heliothis virescens larvae. J. Invert. Pathol. 2006, 92, 166-171. [CrossRef]
- Bachler, A.; Padovan, A.; Anderson, C.J.; Wei, Y.; Wu, Y.; Pearce, S.; Downes, S.; James, B.; Tessnow, A.E.; Sword, G.A.; et al. Disruption of HaVipR1 confers Vip3Aa resistance in the moth crop pest Helicoverpa armigera. PLoS Biol. 2025, 23, e3003165. [CrossRef]
- Zhang, Z.; Wang, L.; Pang, X.; Tay, W.T.; Gordon, K.H.J.; Walsh, T.K.; Yang, Y.; Wu, Y. Knockout of the SfVipR1 gene confers high-level resistance to Bacillus thuringiensis Vip3Aa toxin in Spodoptera frugiperda. bioRxiv 2024, 2024.2009.2026.615236. [CrossRef]
- Bachler, A.; Padovan, A.; Anderson, C.J.; Wei, Y.; Wu, Y.; Pearce, S.; Downes, S.; James, B.; Tessnow, A.; Sword, G.A.; et al. Identification of a novel resistance gene which provides insight into Vip3Aa mode of action in Helicoverpa armigera. bioRxiv 2024, 2024.2008.2011.607516. [CrossRef]
- Pinos, D.; Chakroun, M.; Millán-Leiva, A.; Jurat-Fuentes, J.L.; Wright, D.J.; Hernández-Martínez, P.; Ferré, J. Reduced membrane-bound alkaline phosphatase does not affect binding of Vip3Aa in a Heliothis virescens resistant colony. Toxins 2020, 12, 409. [CrossRef]
- Jakka, S.R.K.; Knight, V.R.; Jurat-Fuentes, J.L. Spodoptera frugiperda (J.E. Smith) with field-evolved resistance to Bt maize are susceptible to Bt pesticides. J. Invert. Pathol. 2014, 122, 52-54. [CrossRef]
- Shabbir, M.Z.; Yang, X.; Batool, R.; Yin, F.; Kendra, P.E.; Li, Z.-Y. Bacillus thuringiensis and chlorantraniliprole trigger the expression of detoxification-related genes in the larval midgut of Plutella xylostella. Front. Physiol. 2021, 12, 780255. doi: 0.3389/fphys.2021.780255.
- Wu, Q.-L.; He, L.-M.; Shen, X.-J.; Jiang, Y.-Y.; Liu, J.; Hu, G.; Wu, K.-M. Estimation of the potential infestation area of newly-invaded fall armyworm Spodoptera frugiperda in the Yangtze River Valley of China. Insects 2019, 10(9), 298. [CrossRef]
- Jakka Siva, R.K.; Gong, L.; Hasler, J.; Banerjee, R.; Sheets Joel, J.; Narva, K.; Blanco Carlos, A.; Jurat-Fuentes Juan, L. Field-evolved mode 1 resistance of the fall armyworm to transgenic Cry1Fa-expressing corn associated with reduced Cry1Fa toxin binding and midgut alkaline phosphatase expression. Appl. Environ. Microbiol. 2016, 82, 1023-1034. [CrossRef]
- Jin, M.; Shan, Y.; Peng, Y.; Wang, W.; Zhang, H.; Liu, K.; Heckel, D.G.; Wu, K.; Tabashnik, B.E.; Xiao, Y. Downregulation of a transcription factor associated with resistance to Bt toxin Vip3Aa in the invasive fall armyworm. Proc. Natl. Acad. Ssci. USA 2023, 120(44), e2306932120. [CrossRef]
- Mahon, R.J.; Downes, S.J.; James, B. Vip3A resistance alleles exist at high levels in Australian targets before release of cotton expressing this toxin. PLoS ONE 2012, 7, e39192. [CrossRef]
- Bernardi, O.; Bernardi, D.; Ribeiro, R.S.; Okuma, D.M.; Salmeron, E.; Fatoretto, J.; Medeiros, F.C.L.; Burd, T.; Omoto, C. Frequency of resistance to Vip3Aa20 toxin from Bacillus thuringiensis in Spodoptera frugiperda (Lepidoptera: Noctuidae) populations in Brazil. Crop Protect. 2015, 76, 7-14. [CrossRef]
- Amaral, F.S.A.; Guidolin, A.S.; Salmeron, E.; Kanno, R.H.; Padovez, F.E.O.; Fatoretto, J.C.; Omoto, C. Geographical distribution of Vip3Aa20 resistance allele frequencies in Spodoptera frugiperda (Lepidoptera: Noctuidae) populations in Brazil. Pest Manag. Sci. 2020, 76, 169-178. [CrossRef]
- Yang, F.; Wang, Z.; Kerns, D.L. Resistance of Spodoptera frugiperda to Cry1, Cry2, and Vip3Aa proteins in Bt corn and cotton in the Americas: Implications for the rest of the world. J. Econ. Entomol. 2022, 115, 1752-1760. [CrossRef]
- Santiago-González, J.C.; Kerns, D.L.; Yang, F. Resistance allele frequency of Helicoverpa zea to Vip3Aa Bacillus thuringiensis protein in the Southeastern U.S. Insects 2023, 14, 161. [CrossRef]
- Wen, Z.; Conville, J.; Matthews, P.; Hootman, T.; Himes, J.; Wong, S.; Huang, F.; Ni, X.; Chen, J.S.; Bramlett, M. More than 10 years after commercialization, Vip3A-expressing MIR162 remains highly efficacious in controlling major Lepidopteran maize pests: laboratory resistance selection versus field reality. Pestic. Biochem. Physiol. 2023, 192, 105385. [CrossRef]
- Carrière, Y.; Degain, B.; Unnithan, G.C.; Tabashnik, B.E. Inheritance and fitness cost of laboratory-selected resistance to Vip3Aa in Helicoverpa zea (Lepidoptera: Noctuidae). J. Econ. Entomol. 2023, 116, 1804-1811. [CrossRef]
- Bernardi, O.; Bernardi, D.; Horikoshi, R.J.; Okuma, D.M.; Miraldo, L.L.; Fatoretto, J.; Medeiros, F.C.; Burd, T.; Omoto, C. Selection and characterization of resistance to the Vip3Aa20 protein from Bacillus thuringiensis in Spodoptera frugiperda. Pest Manag. Sci. 2016, 72, 1794-1802. [CrossRef]
- Pickett Brian, R.; Gulzar, A.; Ferré, J.; Wright D. J. Bacillus thuringiensis Vip3Aa toxin resistance in Heliothis virescens (Lepidoptera: Noctuidae). Appl. Environ. Microbiol. 2017, 83, e03506-03516. [CrossRef]
- Wang, Y.; Yang, J.; Zhang, T.; Bai, S.; Wang, Z.; He, K. Inheritance and fitness costs of Vip3Aa19 resistance in Mythimna separata. Toxins 2022, 14(6), 388. [CrossRef]
- Yang, F.; Williams, J.; Huang, F.; Kerns, D.L. Genetic basis and cross-resistance of Vip3Aa resistance in Spodoptera frugiperda (lepidoptera: Noctuidae) derived from Texas, USA. Crop Protection 2021, 147, 105702. [CrossRef]
- Yang, F.; Head, G.P.; Kerns, D.D.; Jurat-Fuentes, J.L.; Santiago-González, J.C.; Kerns, D.L. Diverse genetic basis of Vip3Aa resistance in five independent field-derived strains of Helicoverpa zea in the US. Pest Manag. Sci. 2024, 80(6), 2796-2803. [CrossRef]
- Fabrick, J.A.; Wu, Y. Chapter Four - Mechanisms and molecular genetics of insect resistance to insecticidal proteins from Bacillus thuringiensis. In Advances in Insect Physiology, Jurat-Fuentes, J.L., Ed.; Academic Press: 2023; Volume 65, pp. 123-183. [CrossRef]
- Tabashnik, B.E.; Fabrick, J.A.; Carrière, Y. Global patterns of insect resistance to transgenic Bt crops: The first 25 years. J. Econ. Entomol. 2023, 116, 297-309. [CrossRef]
- U. S. Environmental protection Agency (EPA) Biopesticides registration action document: Bacillus thuringiensis modified Cry1Ab (SYN-IR67B-1) and Vip3Aa19 (SYN-IR102-7) insecticidal proteins and the genetic material necessary for their production in COT102 X COT67B cotton. 2008. https://www3.epa.gov/pesticides/chem_search/reg_actions/registration/decision_PC-006529_12-Aug-08.pdf (accessed 04/15/2025).
- Yang, F.; Morsello, S.; Head, G.P.; Sansone, C.; Huang, F.; Gilreath, R.T.; Kerns, D.L. F2 screen, inheritance and cross-resistance of field-derived Vip3A resistance in Spodoptera frugiperda (Lepidoptera: Noctuidae) collected from Louisiana, USA. Pest Manag. Sci. 2018, 74, 1769-1778. [CrossRef]
- Yang, F.; Williams, J.; Porter, P.; Huang, F.; Kerns, D.L. F2 screen for resistance to Bacillus thuringiensis Vip3Aa51 protein in field populations of Spodoptera frugiperda (Lepidoptera: Noctuidae) from Texas, USA. Crop Prot. 2019, 126, 104915. [CrossRef]
- Yang, F.; Santiago González, J.C.; Sword, G.A.; Kerns, D.L. Genetic basis of resistance to the Vip3Aa Bt protein in Helicoverpa zea. Pest Manag. Sci. 2021, 77, 1530-1535. [CrossRef]
- Chae, H.; Wen, Z.; Hootman, T.; Himes, J.; Duan, Q.; McMath, J.; Ditillo, J.; Sessler, R.; Conville, J.; Niu, Y.; et al. eCry1Gb.1Ig, A novel chimeric Cry protein with high efficacy against multiple fall armyworm (Spodoptera frugiperda) strains resistant to different GM traits. Toxins 2022, 14(12), 852. [CrossRef]
- Wang, Y.; Wang, J.; Fu, X.; Nageotte Jeffrey, R.; Silverman, J.; Bretsnyder Eric, C.; Chen, D.; Rydel Timothy, J.; Bean Gregory, J.; Li Ke, S.; et al. Bacillus thuringiensis Cry1Da_7 and Cry1B.868 protein interactions with novel receptors allow control of resistant fall armyworms, Spodoptera frugiperda (J.E. Smith). Appl. Environ. Microbiol. 2019, 85, e00579-00519. [CrossRef]
- Gulzar, A.; Pickett, B.; Sayyed, A.H.; Wright, D.J. Effect of temperature on the fitness of a Vip3A resistant population of Heliothis virescens (Lepidoptera: Noctuidae). J. Econ. Entomol. 2012, 105, 964-970. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





