Submitted:
30 June 2025
Posted:
30 June 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Core Concepts or Technologies
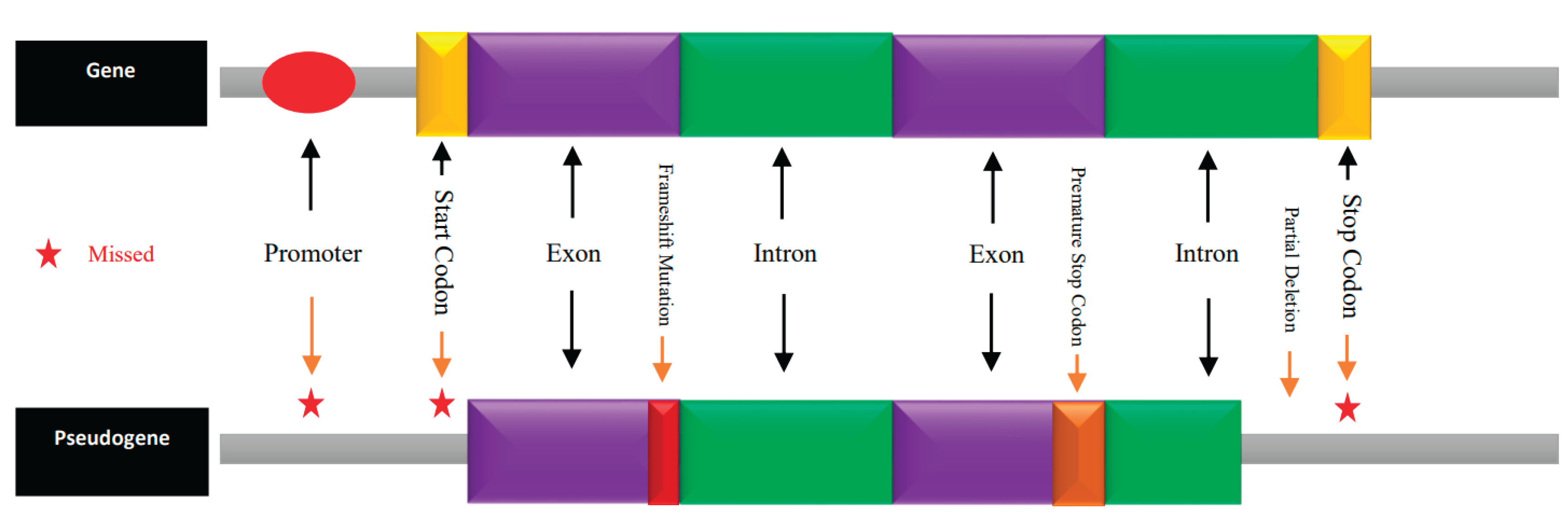
2.1. Pseudogenes: Evolutionary Relics with Functional Potential
- Processed pseudogenes: Resulting from retrotransposition events, lacking introns and often flanked by direct repeats.
- Unprocessed pseudogenes: Arise from gene duplication followed by deleterious mutations.
- Unitary pseudogenes: Formed when a functional gene becomes inactivated without duplication.
2.2. Long Non-Coding RNAs (lncRNAs)
- Chromatin remodeling by interacting with epigenetic modifiers like PRC2 (polycomb repressive complex 2) [3].
- Transcriptional interference by directly binding to transcription factors.
- Post-transcriptional regulation, including splicing and mRNA decay.
2.3. Small Regulatory RNAs: miRNAs and siRNAs
2.4. Enhancer RNAs (eRNAs) and Chromatin Architecture
2.5. Technologies Uncovering the Dark Genome
3. Applications Across Sectors
3.1. Healthcare and Precision Medicine
3.2. Agriculture and Crop Engineering
3.3. Environmental Science and Microbial Genomics
3.4. Industry and Synthetic Biology
4. Challenges and Limitations
4.1. Annotation and Functional Characterization
- The human genome contains over 15,000 pseudogenes and tens of thousands of lncRNAs, yet less than 5% have confirmed biological roles [21].
- Tools like ENCODE and FANTOM5 have improved mapping, but experimental validation remains limited.
- Develop standardized functional assays for non-coding RNA screening.
- Incorporate machine learning to predict function from sequence and structure [22].
4.2. Context-Dependent Activity and Tissue Specificity
- For instance, lncRNAs such as NEAT1 and MEG3 may be oncogenic in one tissue and tumor-suppressive in another [23].
- Employ single-cell RNA sequencing to resolve context-specific roles.
- Use conditional knockout models for in vivo validation.
4.3. Genetic Redundancy and Compensation
- Loss of a pseudogene may be buffered by the presence of homologous sequences, making loss-of-function phenotypes difficult to interpret [24].
- Use multiplex CRISPR systems to knock out entire gene families or ncRNA clusters simultaneously.
- Apply synthetic lethality screens to uncover dependencies.
4.4. Translational and Therapeutic Hurdles
- Instability in circulation.
- Off-target effects and poor tissue-specific delivery.
- Potential Solutions:
- Develop RNA stabilization chemistries and ligand-targeted delivery systems (e.g., aptamer-conjugates).
- Apply exosome-based delivery platforms for precision targeting [25].
4.5. Ethical and Regulatory Ambiguities
- Modifying enhancers or pseudogenes may have unintended long-range effects on gene expression [26].
- Potential Solutions:
- Introduce predictive modeling frameworks to simulate genome-wide effects before interventions.
- Promote international bioethical consensus on non-coding genome editing. See Table 3.
5. Future Directions
5.1. AI-Powered Functional Annotation
- Forecast the regulatory impact of non-coding variants.
- Infer enhancer-promoter interactions and ncRNA function from sequence alone [27].
5.2. CRISPR-Based Functional Genomics in Non-Coding Regions
- CRISPRi (interference) and CRISPRa (activation) allow targeted regulation of lncRNAs, pseudogenes, and enhancers without altering DNA sequence.
- CRISPR tiling screens offer high-resolution maps of functional non-coding elements in disease loci [28].
5.3. Single-Cell and Spatial Transcriptomics
- Single-cell RNA-seq (scRNA-seq) reveals lncRNA heterogeneity across individual cells.
- Spatial transcriptomics captures tissue-specific expression of regulatory elements, vital for developmental biology and cancer studies [29].
5.4. Multi-Omics and Systems Biology Approaches
- Dissecting complex regulatory networks.
- Modeling genotype-to-phenotype transitions driven by non-coding elements [30].
5.5. Synthetic Biology and ncRNA Engineering
- Engineered lncRNAs can act as scaffolds, decoys, or sponges in synthetic gene circuits.
- Riboregulators—RNA-based switches—are being used to control gene expression in response to environmental cues [31].
5.6. Clinical Translation and Personalized Medicine
- Routine inclusion of pseudogene and lncRNA panels in diagnostic assays.
- Personalized therapies targeting individual ncRNA profiles for precision oncology and neurology.
5.7. Ethical Frameworks and Governance
6. Conclusions
Funding
Data Availability Statement
Acknowledgments
AI Declaration
Conflict of Interest
References
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: what is functional and what is junk? Front Genet. 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Poliseno, L.; Salmena, L.; Zhang, J.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010, 465, 1033–1038. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu Rev Biochem.
- Esteller, M. Non-coding RNAs in human disease. Nat Rev Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Kim, T.K.; Shiekhattar, R. Architectural and functional commonalities between enhancers and promoters. Cell. 2015, 162, 948–959. [Google Scholar] [CrossRef]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012, 485, 376–380. [Google Scholar] [CrossRef]
- Poliseno, L.; Pandolfi, P.P. PTENP1 and the ceRNA hypothesis: an intricate balance. Methods.
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Arun, G.; Diermeier, S.; Spector, D.L. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol Med. 2018, 24, 257–277. [Google Scholar] [CrossRef]
- Liu, S.J.; Horlbeck, M.A.; Cho, S.W.; Birk, H.S.; Malatesta, M.; He, D.; et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017, 355, eaah7111. [Google Scholar] [CrossRef]
- Faghihi, M.A.; Modarresi, F.; Khalil, A.M.; Wood, D.E.; Sahagan, B.G.; Morgan, T.E.; et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat Med. 2008, 14, 723–730. [Google Scholar] [CrossRef]
- Si-Ammour, A.; Windels, D.; Arn-Bouldoires, E.; Kutter, C.; Ailhas, J.; Meins, F.; et al. miR393 and secondary siRNAs regulate expression of the auxin-related gene TIR1 and AFB2 in Arabidopsis. Plant J. 2011, 68, 452–461. [Google Scholar]
- Ariel, F.; Romero-Barrios, N.; Jégu, T.; Benhamed, M.; Crespi, M. Battles and hijacks: noncoding transcription in plants. Trends Plant Sci. 2015, 20, 362–371. [Google Scholar] [CrossRef]
- Lowder, L.G.; Zhang, D.; Baltes, N.J.; Paul, J.W.; Tang, X.; Zheng, X.; et al. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 2015, 169, 971–985. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Valera, F.; Martin-Cuadrado, A.B.; López-Pérez, M.; García-Heredia, I. Pseudogenes in microbial genomes: theory and practice. Res Microbiol. 2016, 167, 664–673. [Google Scholar]
- Odah, M.A.A. Temporal dynamics of environmental DNA (eDNA) as early-warning indicators of climate-driven ecosystem shifts in diverse Saudi habitats. Preprints. 2025. [Google Scholar]
- Odah, M.A.A. Ultra-short DNA satellites as environmental sensing elements in soil microbiomes: a frontier review. Preprints. 2025. [Google Scholar]
- Odah, M.A.A. Programmable DNA devices: the next generation of living sensors in agriculture, health, and the environment. Preprints. 2025. [Google Scholar]
- Odah, M.A.A. Photosynthetic reprogramming enhancing carbon fixation in crops through synthetic biology. Preprints. 2025. [Google Scholar]
- Odah, M.A.A. Microbiota-immune-brain crosstalk: synthetic biology solutions for neuroinflammatory disorders. Preprints. 2025. [Google Scholar]
- Pei, B.; Sisu, C.; Frankish, A.; Howald, C.; Habegger, L.; Mu, X.J.; et al. The GENCODE pseudogene resource. Genome Biol. 2012, 13, R51. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Troyanskaya, O.G. Predicting effects of noncoding variants with deep learning–based sequence model. Nat Methods. 2015, 12, 931–934. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Rice, K.; Wang, Y.; Chen, W.; Zhong, Y.; Nakayama, Y.; et al. Maternally expressed gene 3 (MEG3) noncoding RNA: isoform structure, expression, and functions. Nucleic Acids Res. 2010, 38, 4740–4751. [Google Scholar]
- Schmitz, J.F.; Zimmer, F.; Bornberg-Bauer, E. Mechanisms of transcription factor evolution in Metazoa. Nucleic Acids Res. 2016, 44, 6287–6297. [Google Scholar] [CrossRef]
- Li, Z.; Rana, T.M. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014, 13, 622–638. [Google Scholar] [CrossRef]
- Cwiklinska, M.; Kolb, G.; Michalakis, S. Non-coding RNAs in inherited retinal diseases. Cell Tissue Res. 2021, 386, 435–450. [Google Scholar]
- Kelley, D.R.; Reshef, Y.A.; Bileschi, M.; Belanger, D.; McLean, C.Y.; Snoek, J. Sequential regulatory activity prediction across chromosomes with convolutional neural networks. Genome Res. 2018, 28, 739–750. [Google Scholar] [CrossRef]
- Sanjana, N.E.; Wright, J.; Zheng, K.; Shalem, O.; Fontanillas, P.; Joung, J.; et al. High-resolution interrogation of functional elements in the noncoding genome. Science. 2016, 353, 1545–1549. [Google Scholar] [CrossRef]
- Stahl, P.L.; Salmen, F.; Vickovic, S.; Lundmark, A.; Navarro, J.F.; Magnusson, J.; et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016, 353, 78–82. [Google Scholar] [CrossRef]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef]
- Chappell, J.; Takahashi, M.K.; Lucks, J.B. Creating small transcription activating RNAs. Nat Chem Biol. 2015, 11, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Iyer, E.P.; Salvado, O.; Gaikwad, S. A policy framework for genome editing. Nat Biotechnol. 2017, 35, 484–486. [Google Scholar]
| Technology | Application |
|---|---|
| RNA-Seq | Transcriptome profiling of non-coding RNAs |
| ChIP-Seq | Identifying transcription factor binding sites |
| ATAC-Seq | Mapping chromatin accessibility |
| Hi-C/3C | Studying 3D genome architecture |
| CRISPR interference (CRISPRi) | Functional dissection of non-coding elements |
| Sector | Application Example | Key Molecule |
|---|---|---|
| Healthcare | PTENP1 in tumor suppression | Pseudogene |
| Agriculture | miR393 enhances drought resistance in rice | miRNA |
| Environment | eDNA-based stress markers in marine microbiomes | lncRNAs |
| Industry | RNA-based logic gates for biosensors | Synthetic lncRNAs |
| Challenge | Description | Proposed Solution |
|---|---|---|
| Poor functional annotation | Limited understanding of roles of ncRNAs/pseudogenes | Functional assays, ML-based prediction |
| Context specificity | Varying function across tissues | Single-cell transcriptomics |
| Redundancy and compensation | Masking of phenotypes by similar elements | Multiplexed CRISPR screens |
| Therapeutic delivery limitations | Instability and off-target effects | RNA modifications, targeted delivery vehicles |
| Ethical and regulatory uncertainties | Editing regulatory DNA with unknown consequences | Simulation models, ethical frameworks |
| Domain | Emerging Direction |
|---|---|
| AI & Bioinformatics | Predictive ncRNA function and variant annotation |
| CRISPR Technology | Targeted manipulation of non-coding elements |
| Single-Cell Biology | Context-specific ncRNA mapping |
| Multi-Omics | Integrated regulatory network modeling |
| Synthetic Biology | Engineered lncRNAs for smart applications |
| Clinical Translation | Diagnostic panels & RNA-targeting therapies |
| Ethics & Governance | Non-coding genome editing regulations |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





