Submitted:
19 June 2025
Posted:
20 June 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
- in samples of corticotroph tumors with visualization on MRI/more than 6 mm and without visualization of pituitary tumors on MRI/less than 6 mm for differential diagnosis of pituitary and ectopic localization of ACTH-secreting tumour.
- depending on the tumor volume (according to MRI data) to determine prognostic biomarkers.
- depending on the tumor cell subtype and proliferation level (according to immunohistochemical study) to determine the tumour prognosis.
- depending on the type of biological behavior of tumours - to identify indolent and aggressive corticotropinomas.
2. Materials and Methods
2.1. Patients and Samples
2.2. Description of Medical (Diagnostic) Intervention
2.3. Immunohistochemistry Imaging
2.4. Statistical Analysis
3. Results
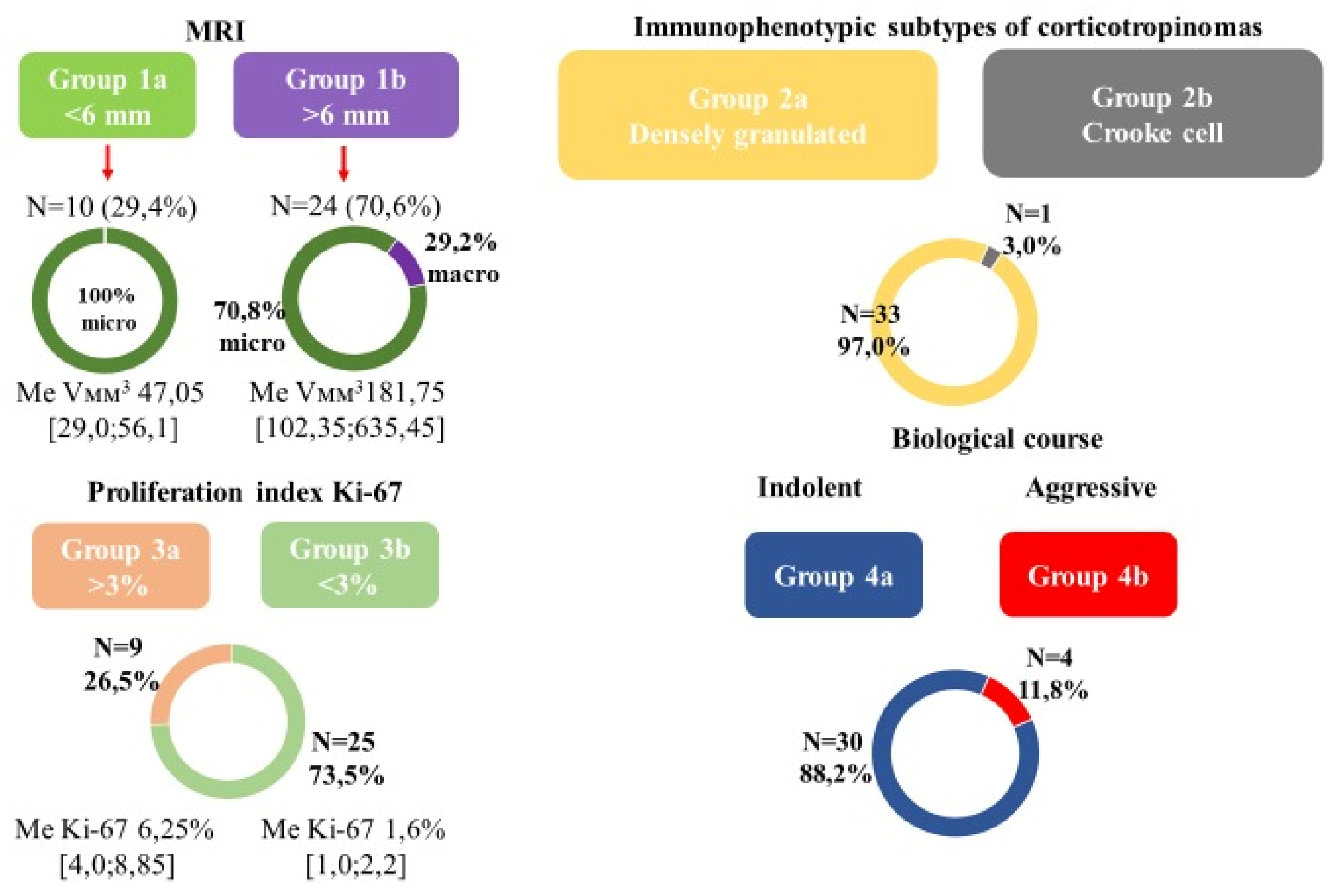
3.1. Clinical and Morphological Characteristics of Patients
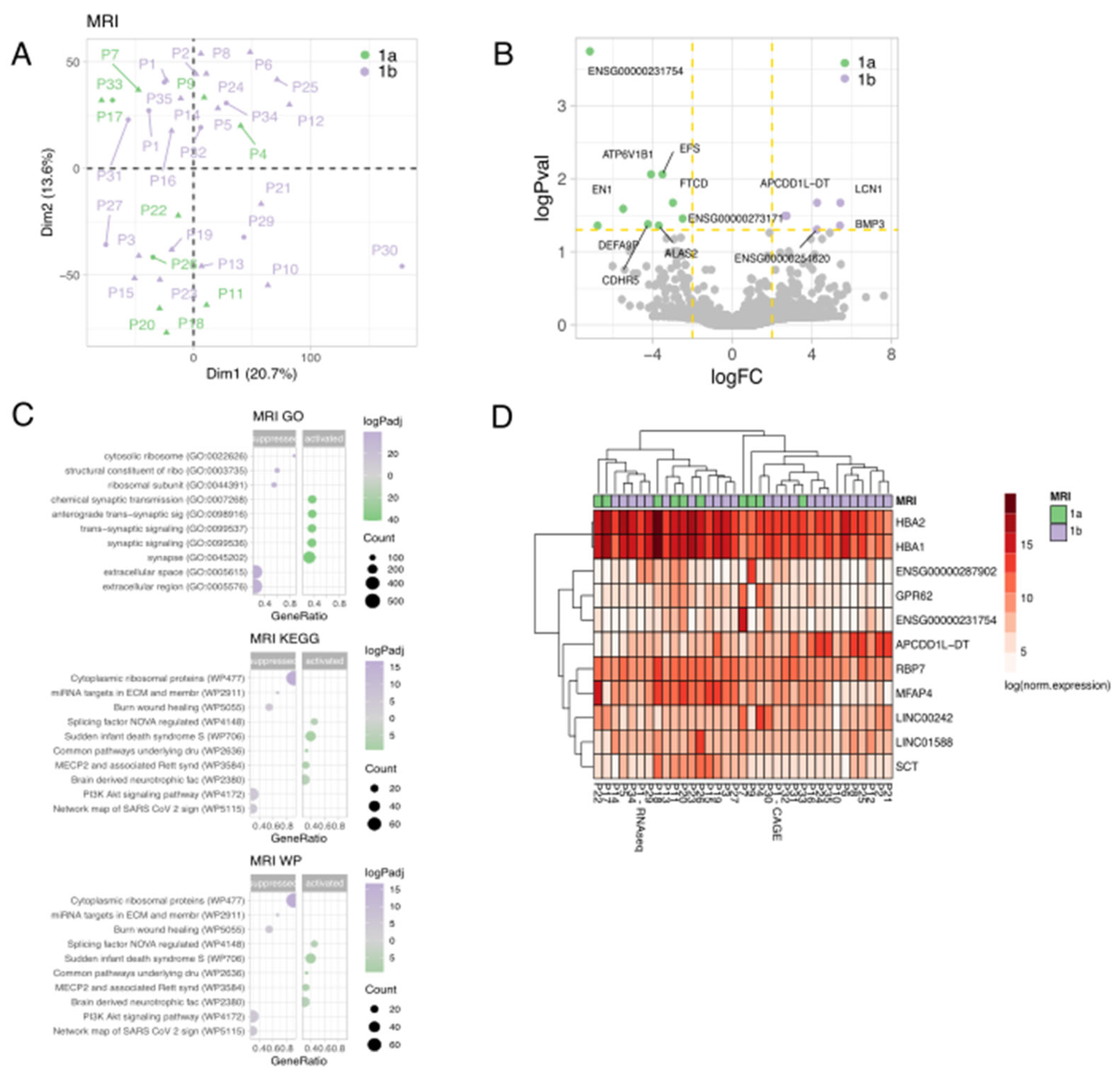
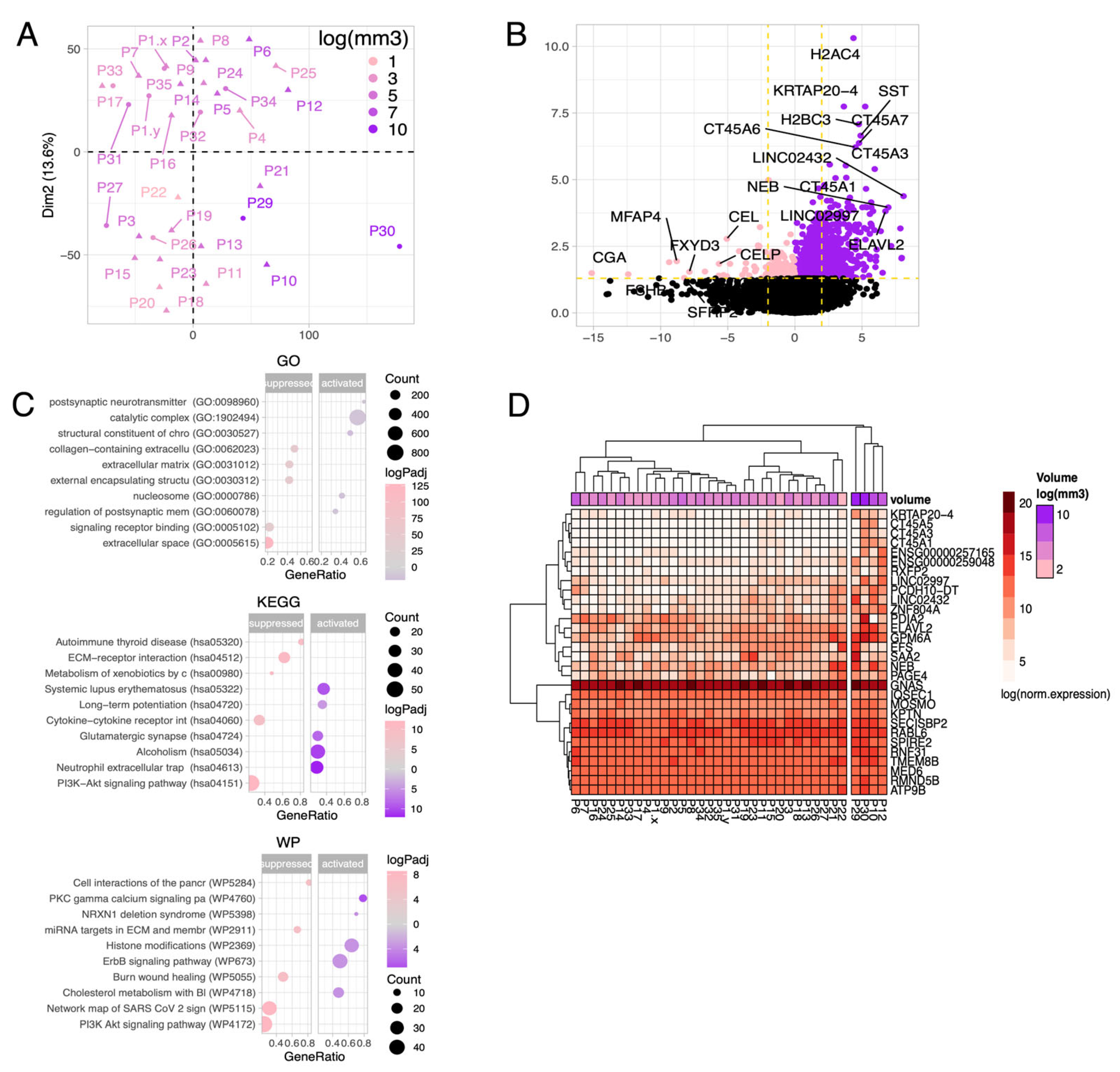
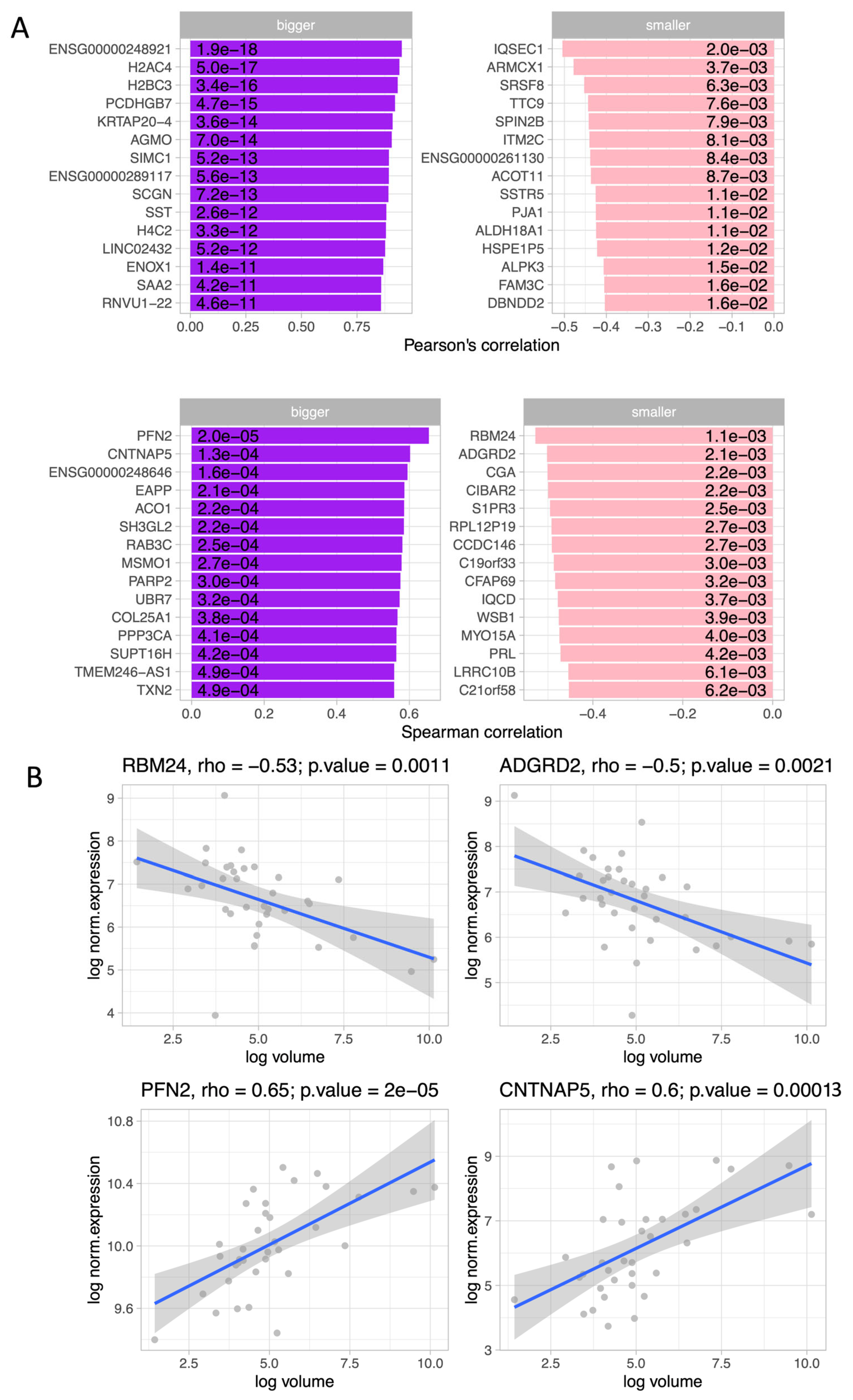
3.2. General Characteristics of Transcriptome Data
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CAGE | Cap Analysis Gene Expression |
| CD | Cushing’s disease |
| CPitNETs | corticotroph pituitary neuroendocrine tumors |
| DE | differentially expressed |
| FDR | false discovery rate |
| GLM | generalized linear model |
| GO | Gene Ontology |
| IHC | immunohistochemistry |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| TSS | transcription start sites |
| WikiPathways | Wikipedia pathways |
References
- Chandler, W.F.; Barkan, A.L.; Hollon, T.; Sakharova, A.; Sack, J.; Brahma, B.; Schteingart, D.E. Outcome of transsphenoidal surgery for Cushing disease: a single-center experience over 32 years. Neurosurgery 2016, 78, 216–223. [Google Scholar] [CrossRef]
- Semple, P.L.; Vance, M.L.; Findling, J.; Laws, E.R., Jr. Transsphenoidal surgery for Cushing’s disease: outcome in patients with a normal magnetic resonance imaging scan. Neurosurgery 2000, 46, 553–559. [Google Scholar] [CrossRef]
- Sheehan, J.M.; Lopes, M.B.; Sheehan, J.P.; Ellegala, D.; Webb, K.M.; Laws, E.R., Jr. Results of transsphenoidal surgery for Cushing’s disease in patients with no histologically confirmed tumor. Neurosurgery 2000, 47, 33–39. [Google Scholar] [CrossRef]
- Grober, Y.; Grober, H.; Wintermark, M.; Jane, J.A.; Oldfield, E.H. Comparison of MRI techniques for detecting microadenomas in Cushing’s disease. J Neurosurg. 2018, 128, 1051–1057. [Google Scholar] [CrossRef] [PubMed]
- Stroud, A.; Dhaliwal, P.; Alvarado, R.; Winder, M.J.; Jonker, B.P.; Grayson, J.W.; Hamizan, A.; Harvey, R.J.; McCormack, A. Outcomes of pituitary surgery for Cushing’s disease: a systematic review and meta-analysis. Pituitary 2020, 23, 595–609. [Google Scholar] [CrossRef]
- Capatina, C.; Hinojosa-Amaya, J.M.; Poiana, C.; Fleseriu, M. Management of patients with persistent or recurrent Cushing’s disease after initial pituitary surgery. Expert Rev Endocrinol Metab 2020, 15, 321–339. [Google Scholar] [CrossRef]
- Fleseriu, M.; Auchus, R.; Bancos, I.; Ben-Shlomo, A.; Bertherat, J.; Biermasz, N.R.; Boguszewski, C.L.; Bronstein, M.D.; Buchfelder, M.; Carmichael, J.D.; et al. Consensus on Diagnosis and Management of Cushing’s Disease: A Guideline Update. Lancet Diabetes Endocrinol 2021, 9, 847–875. [Google Scholar] [CrossRef]
- Casar-Borota, O.; Boldt, H.B.; Engström, B.E.; Andersen, M.S.; Baussart, B.; Bengtsson, D.; Berinder, K.; Ekman, B.; Feldt-Rasmussen, U.; Höybye, C.; et al. Corticotroph Aggressive Pituitary Tumors and Carcinomas Frequently Harbor ATRX Mutations. J Clin Endocrinol Metab 2021, 106. 4, 1183–1194. [Google Scholar] [CrossRef] [PubMed]
- Trouillas, J.; Roy, P.; Sturm, N.; Dantony, E.; Cortet-Rudelli, C.; Viennet, G.; Bonneville, J.F.; Assaker, R.; Auger, C.; Brue, T.; Cornelius, A.; et al. A new prognostic clinicopathological classification of pituitary adenomas: a multicentric case–control study of 410 patients with 8 years post-operative follow-up. Acta Neuropathol 2013, 126, 123–135. [Google Scholar] [CrossRef]
- Simon, J.; Theodoropoulou, M. Genetics of Cushing’s disease. Journal of Neuroendocrinology 2022, 13148. [Google Scholar] [CrossRef] [PubMed]
- Peculis, R.; Niedra, H.; Rovite, V. Large scale molecular studies of pituitary neuroendocrine tumors: novel markers, mechanisms and translational perspectives. Cancers 2021, 13, 1395. [Google Scholar] [CrossRef]
- Neou, M.; Villa, C.; Armignacco, R.; Jouinot, A.; Raffin-Sanson, M.L.; Septier, A.; Letourneur, F.; Diry, S.; Diedisheim, M.; Izac, B.; et al. Pangenomic classification of pituitary neuroendocrine tumors. Cancer cell 2020. 37, 1, 123-134.e5. [CrossRef]
- Salomon, M.P.; Wang, X.; Marzese, D.M.; Hsu, S.C.; Nelson, N.; Zhang, X.; Matsuba, C.; Takasumi, Y.; Ballesteros-Merino, C.; Fox, B.A.; et al. The epigenomic landscape of pituitary adenomas reveals specific alterations and differentiates among acromegaly, Cushing’s disease and endocrine-inactive subtypes. Clinical Cancer Research 2018, 24, 17, 4126–4136. [Google Scholar] [CrossRef] [PubMed]
- Kawaji, H.; Lizio, M.; Itoh, M.; Kanamori-Katayama, M.; Kaiho, A.; Nishiyori-Sueki, H.; Shin, J.W.; Kojima-Ishiyama, M.; Kawano, M.; Murata, M.; et al. Comparison of CAGE and RNA-seq transcriptome profiling using clonally amplified and single-molecule next-generation sequencing. Genome Res. 2014, 24, 4, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Murata, M.; Nishiyori-Sueki, H.; Kojima-Ishiyama, M.; Carninci, P.; Hayashizaki, Y.; Itoh, M. Detecting expressed genes using CAGE. Methods in molecular biology 2014, 1164, 67–85. [Google Scholar] [CrossRef]
- The Gene Ontology Resource. Available online: https://geneontology.org/ (accessed on 10 May 2024).
- Kyoto Encyclopedia of Genes and Genomes. Available online: https://www.genome.jp/kegg/ (accessed on 18 May 2024).
- WikiPathways 2024: next generation pathway database. Available online: https://www.wikipathways.org/ (accessed on 10 June 2024).
- Neumann, L.C.; Weinhäusel, A.; Thomas, S.; Horsthemke, B.; Lohmann, D.R.; Zeschnigk, M. EFS shows biallelic methylation in uveal melanoma with poor prognosis as well as tissue-specific methylation. BMC Cancer 2011, 11, 380. [Google Scholar] [CrossRef]
- Miura, K.; Akashi, T.; Namiki, T.; Hishima, T.; Bae, Y.; Sakurai, U.; Murano, K.; Shiraishi, J.; Warabi, M.; Tanizawa, T.; et al. Engrailed Homeobox 1 and Cytokeratin 19 are independent Diagnostic markers of eccrine porocarcinoma and distinguish it from squamous cell carcinoma. Am J Clin Pathol 2020. 154:499-509. [CrossRef]
- Zhang, X.; Cui, Y.; He, M. Lipocalin-1 expression as a prognosticator marker of survival in breast cancer patients. Breast Care 2020, 15, 272–280. [Google Scholar] [CrossRef]
- Davis, H.; Raja, E.; Miyazono, K.; Tsubakihara, Y.; Moustakas, A. Mechanisms of Action of Bone Morphogenetic Proteins in Cancer. Cytokine Growth Factor Rev 2016, 27, 81–92. [Google Scholar] [CrossRef]
- Meng, W.; Xiao, H.; Zhao, R.; Li, D.; Li, K.; Meng, Y.; Chen, J.; Wang, Y.; Liao, Y. The Prognostic Value of Bone Morphogenetic Proteins and Their Receptors in Lung Adenocarcinoma. Front. Oncol 2021, 11, 608239. [Google Scholar] [CrossRef]
- Shi, D.L. RBM24 in the post-transcriptional regulation of cancer progression: anti-tumor or pro-tumor activity? Cancers 2022, 14, 1843. [Google Scholar] [CrossRef]
- Qin, H.; Ni, H.; Liu, Y.; Yuan, Y.; Tao, X.; Li, X.; Zheng, L. RNA-binding proteins in tumor progression. J. Hematol. Oncol. 2020, 13, 90. [Google Scholar] [CrossRef]
- He, B.; Wu, C.; Sun, W.; Qiu, Y.; Li, J.; Liu, Z.; Jing, T.; Wang, H.; Liao, Y. Mir-383 increases the cisplatin sensitivity of lung adenocarcinoma cells through inhibition of the RBM24-mediated NF-kappaB signaling pathway. Int. J. Oncol. 2021, 59, 1–16. [Google Scholar] [CrossRef]
- Seltzer, J.; Ashton, C.E.; Scotton, T.C.; Pangal, D.; Carmichael, J.D.; Zada, G. Gene and protein expression in pituitary corticotroph adenomas: a systematic review of the literature. Neurosurg Focus 2015, 38, 2, E17. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.A.; Zhong, H.G.; Qin, Y.Z.; Wei, W.; Li, Z.; Huang, M.; Luo, X. ARMCX family gene expression analysis and potential prognostic biomarkers for prediction of clinical outcome in patients with gastric carcinoma. BioMed Research International Volume 2020. [Google Scholar] [CrossRef] [PubMed]
- Lv, C.; Yang, D.; Zhang, D.; Shen, J.; Wang, Z.; He, S.; Meng, L.; Song, J.; Zhao, J. Expression of ARMCX1 in gastric cancer contributes to prognosis and influences chemotherapy. Journal of Immunology Research 2023, 262, 3317. [Google Scholar] [CrossRef] [PubMed]
- Shenglan, C.; Jayasree, K.I.V. Identification of tetratricopeptide repeat domain 9, a hormonally regulated protein. Biochemical and Biophysical Research Communications 2006, 345, 1, 310–317. [Google Scholar] [CrossRef]
- Dhahri, H.; Saintilnord, W.N.; Chandler, D.; Fondufe-Mittendorf, Y.N. Beyond the usual suspects: examining the role of understudied histone variants in breast cancer. Int. J. Mol. Sci. 2024, 25, 6788. [Google Scholar] [CrossRef]
- Shang, B.; Gao, A.; Pan, Y.; Zhang, G.; Tu, J.; Zhou, Y.; Yang, P.; Cao, Z.; Wei, Q.; Ding, Y.; et al. CT45A1 acts as a new proto-oncogene to trigger tumorigenesis and cancer metastasis. Cell Death and Disease 2014, 5, e1285. [Google Scholar] [CrossRef]
- Cooley, L.S.; Rudewicz, J.; Souleyreau, W.; Emanuelli, A.; Alvarez-Arenas, A.; Clarke, K.; Falciani, F.; Dufies, M.; Lambrechts, D.; Modave, E.; et al. Experimental and computational modeling for signature and biomarker discovery of renal cell carcinoma progression. Mol Cancer 2021, 20, 136. [Google Scholar] [CrossRef]
- Marquard, F.E.; Jücker, M. PI3K/AKT/mTOR Signaling as a Molecular Target in Head and Neck Cancer. Biochem. Pharm. 2020, 172, 113729. [Google Scholar] [CrossRef]
- Miricescu, D.; Totan, A.; Stanescu-Spinu, I.I.; Badoiu, S.C.; Stefani, C.; Greabu, M. PI3K/AKT/mTOR signaling pathway in breast cancer: from molecular landscape to clinical aspects. Int. J. Mol. Sci. 2020, 22, 173. [Google Scholar] [CrossRef]
- Derwich, A.; Sykutera, M.; Bromińska, B.; Rubiś, B.; Ruchała, M.; Sawicka-Gutaj, N. The role of activation of PI3K/AKT/mTOR and RAF/MEK/ERK pathways in aggressive pituitary adenomas—new potential therapeutic approach—a systematic review. Int. J. Mol. Sci. 2023, 24, 10952. [Google Scholar] [CrossRef]
- Treppiedi, D.; Barbieri, A.M.; Di Muro, G.; Marra, G.; Mangili, F.; Catalano, R.; Esposito, E.; Ferrante, E.; Serban, A.L.; Locatelli, M.; et al. Genetic profiling of a cohort of italian patients with ACTH-secreting pituitary tumors and characterization of a novel Usp8 gene variant. Cancers 2021, 13, 4022. [Google Scholar]
- Sajjad, E.A.; Zieliński, G.; Maksymowicz, M.; Hutnik, L.; Bednarczuk, T.; Włodarski, P. mTOR is frequently active in GH-secreting pituitary adenomas without influencing their morphopathological features. Endocr. Pathol. 2013, 24, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Zhang, H.; Yang, J.; Zheng, Z.; Liu, K. Expression mode and prognostic value of FXYD family members in colon cancer. Aging 2021, 13, 18404–18422. [Google Scholar] [CrossRef]
- Yee, K.X.; Lee, Y.C.; Nguyen, H.D. Uncovering the role of FXYD3 as a potential oncogene and early biomarker in pancreatic cancer. Am J Cancer Res 2024, 14, 9, 4353. [Google Scholar] [CrossRef] [PubMed]
- Sbiera, S.; Perez-Rivas, L.G.; Taranets, L.; Weigand, I.; Flitsch, J.; Graf, E.; Monoranu, C.M.; Saeger, W.; Hagel, C.; Honegger, J.; Assie, G.; Hermus, A.R.; Stalla, G.K.; et al. Driver mutations in USP8 wild-type Cushing’s disease. Neuro Oncology 2019, 21, 10, 1273–1283. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ma, L.; Zhao, Y.; Liu, M.; Ye, W.; Li, X. Research progress on the role of the Wnt signaling pathway in pituitary adenoma. Front. Endocrinol. 2023, 14, 1216817. [Google Scholar] [CrossRef]
- Zhu, D.; Wang, Z.; Tian, T.; Wu, X.; He, D.; Zhu, Y.; Liu, D.; Wang, H. Prevalence and clinical characteristics of Crooke’s cell adenomas in 101 patients with T-PIT-positive pituitary adenomas: case series and literature review. Front Endocrinol 2022, 13, 947085. [Google Scholar] [CrossRef]
- Garbicz, F.; Mehlich, D.; Rak, B. Increased expression of the microRNA 106b~25 cluster and its host gene MCM7 in corticotroph pituitary adenomas is associated with tumor invasion and Crooke’s cell morphology. Pituitary 2017, 20, 450–463. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome Regulation by Long Noncoding RNAs. Annu Rev Biochem 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Adelman, K.; Egan, E. Non-Coding RNA: More Uses for Genomic Junk. Nature 2017, 543, 183–185. [Google Scholar] [CrossRef]
- Butz, H. Circulating Noncoding RNAs in Pituitary Neuroendocrine Tumors-Two Sides of the Same Coin. Int J Mol Sci 2022, 23, 5122. [Google Scholar] [CrossRef]
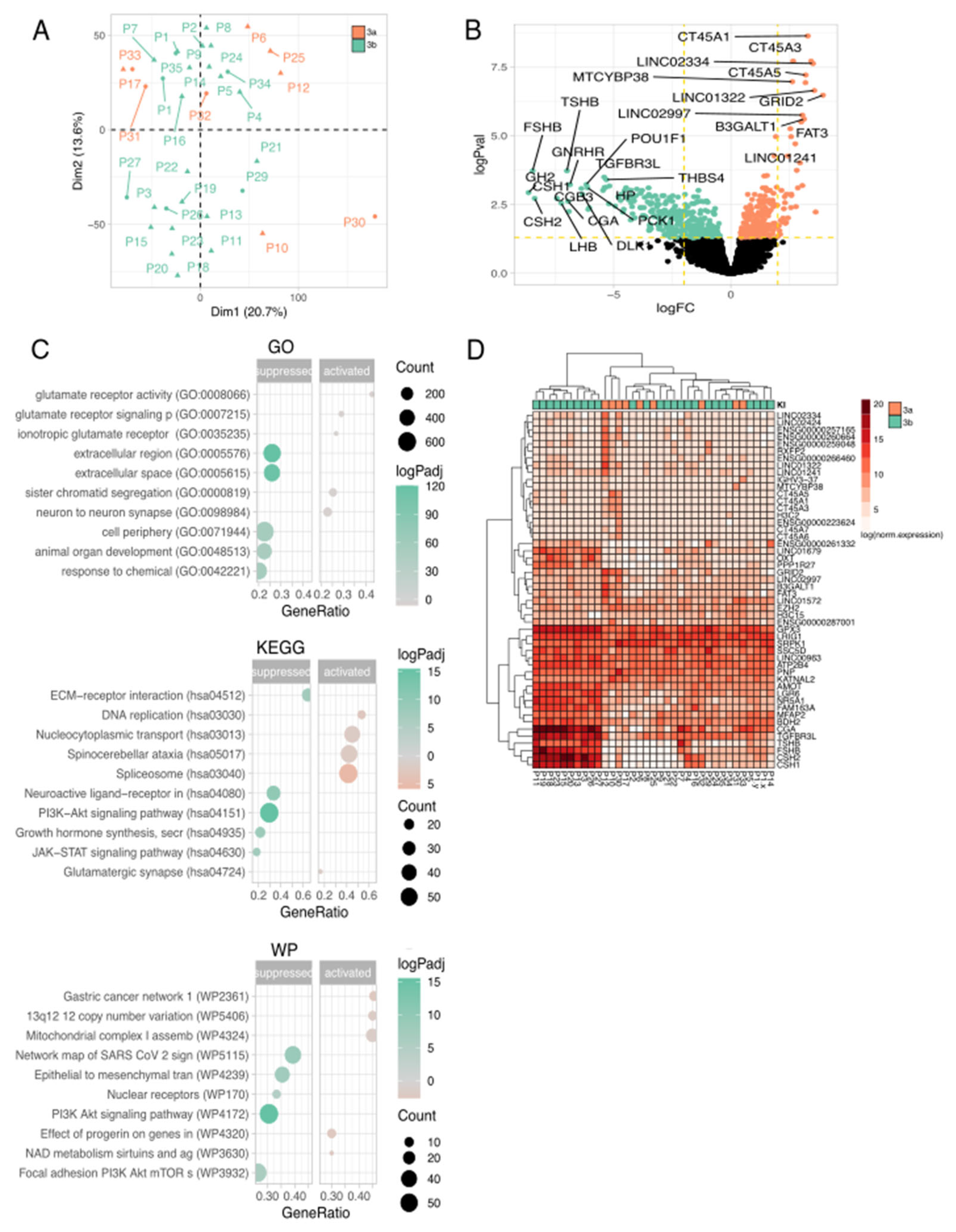
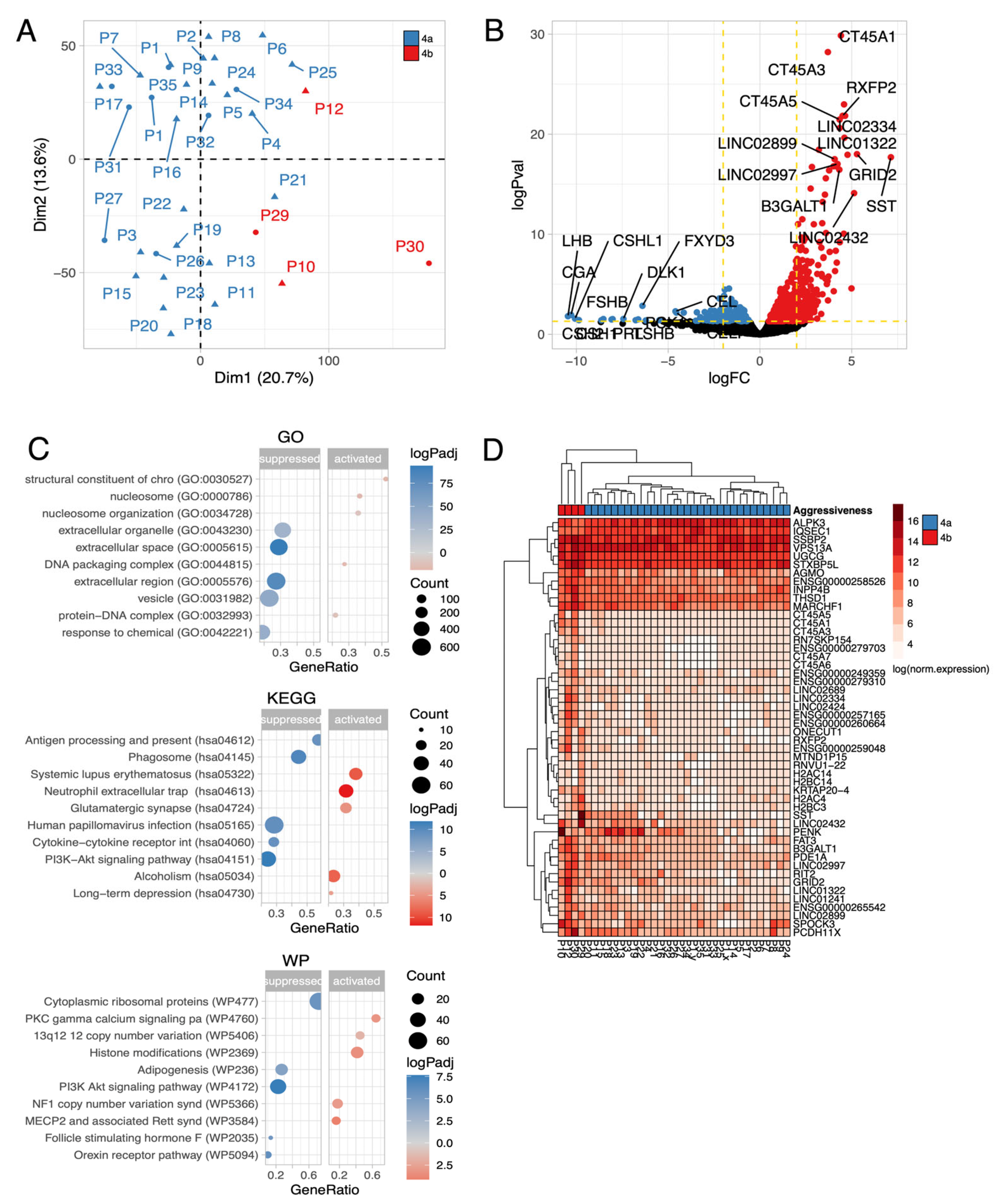
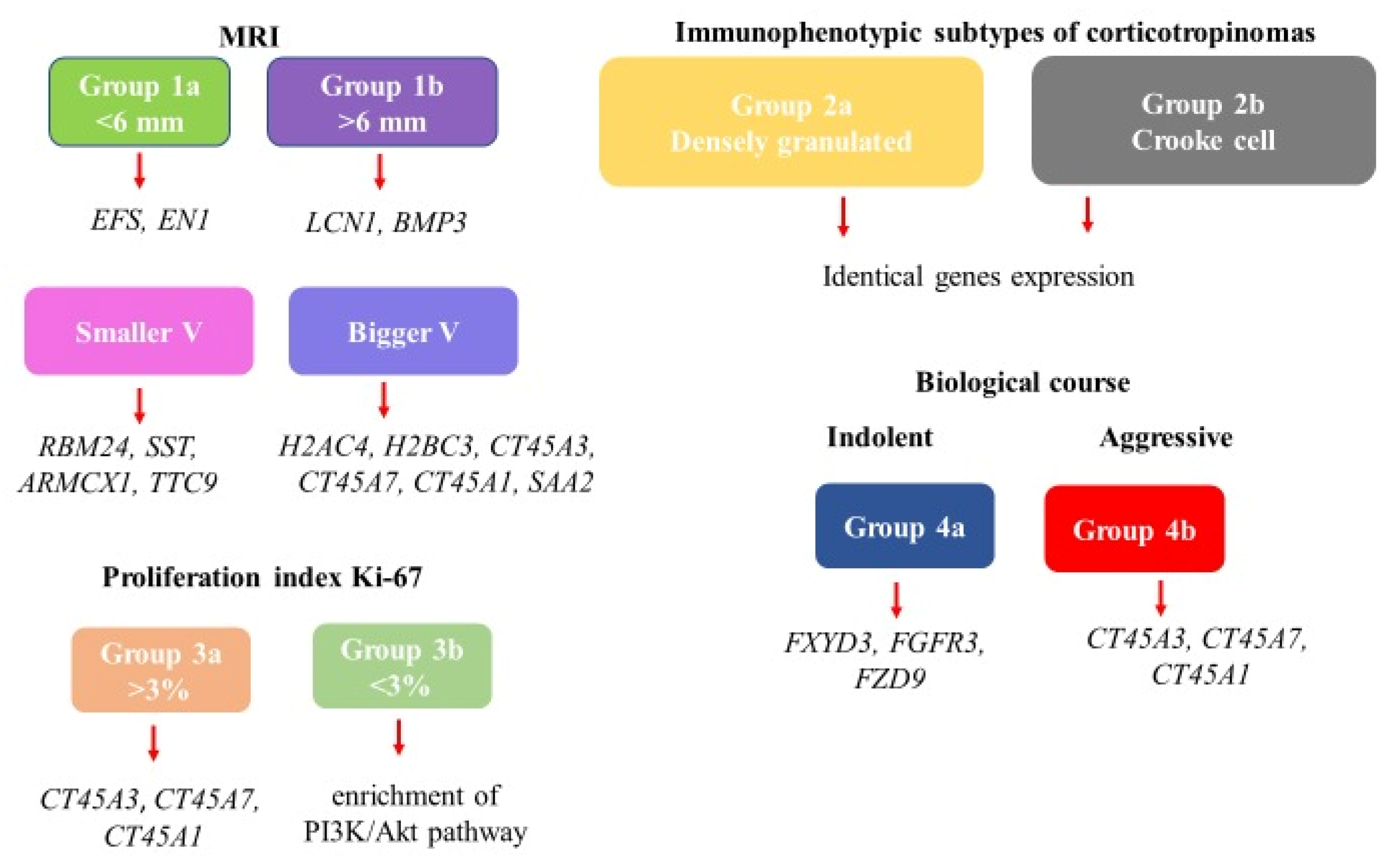
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




