1. Introduction
This study illustrates the case of a previously healthy, young adult who was hospitalised in the Netherlands for the treatment of a community-acquired necrotising
Staphylococcus aureus (MSSA) pneumonia with an underlying influenza A infection. The course of disease was fulminant, and the patient deceased on the second day of hospitalisation on the intensive care. Molecular evaluation revealed that the isolate belongs to Multi Locus Sequencing Type (ST) ST152 and was positive for an extensive repertoire of virulence genes including genes encoding for the cytotoxin Panton-Valentine leukocidine (PVL), exotoxin
edinB, and the recently identified exfoliative toxin E sequencing variant
etE2 from a case of severe necrotising fasciitis [
1].
This communication should raise awareness for the emergence of the MSSA ST152 lineage, which is associated with severe necrotising pneumonia accompanied by bacteraemia, and deep-seated soft tissue infections as frequent presentations. In all isolates of this lineage, the previously unrecognised exfolative toxin etE2 is present. The global dissemination of this high-risk PVL-positive community lineage, which harbors the exotoxin genes edinB and etE2 implicated in the destruction of tissue integrity, is a potential threat for public health and warrants monitoring.
2. Case Description
A 25-year-old previously healthy male presented with tachypnea and hypoxemia to the emergency department of a regional hospital in the Netherlands in 2024. The patient had been experiencing malaise for one week and had been prescribed amoxicillin one day before the admission. Upon admission; he was diagnosed with pneumosepsis and bilateral pneumonia; necessitating high-flow nasal cannula oxygen therapy in the intensive care unit. A nasopharyngeal swab and PCR tested positive for influenza A. Blood tests revealed no leucocytosis but showed an elevated C-reactive protein level of 108 mg/L (normal range: <5 mg/L) and an arterial lactate of 6.4 mmol/L (normal range: <2 mmol/L). Given the suspicion of bacterial superinfection, empirical antibiotic therapy with cefuroxime and ciprofloxacin was initiated after obtaining blood and sputum cultures
Despite treatment, the patient’s condition deteriorated, requiring endotracheal intubation and mechanical ventilation in the prone position. Hemodynamic support with escalating doses of norepinephrine and vasopressin were started, along with continuous hydrocortisone due to the severity of septic shock.
The following day, the patient was transferred to our tertiary intensive care unit. Shortly after arrival, he suffered cardiac arrest, prompting cardiopulmonary resuscitation for approximately 20 minutes. During resuscitation, large amounts of pink, watery sputum were expelled from the lungs. After achieving return of spontaneous circulation, venovenous extracorporeal membrane oxygenation and continuous renal replacement therapy were initiated. The antibiotic regimen was escalated to cefotaxime, levofloxacin, and clindamycin. Despite these interventions, the patient’s condition continued to deteriorate, with the development of liver failure, coagulopathy, refractory hyperlactatemia, heart failure, pericardial effusion, and loss of brain function, with dilated non-reactive pupils. Bronchoscopy revealed diffusely swollen mucosa and significant watery secretions. Extensive transfusions led to worsening pulmonary oedema.
Given the lack of clinical stabilization, the medical team concluded that recovery was unlikely. Best supportive care was initiated, and the patient passed away later that day. Post mortem, the blood and sputum cultures identified S. aureus.
2.1. Isolate Characterisation
The
S. aureus isolate UMCG-ST152-2024-1 grew from a blood culture, and in pure culture from sputum samples. Analysis of short-read sequencing (Miseq, Ilumina) as described previously [
2], revealed that the isolate belongs to MLST ST152. The isolate was susceptible to all antibiotics tested by Vitek2 except for trimethoprim and penicillin, interpreted by EUCAST v.13.1. The resistance was confirmed by agar diffusion disk testing with a zone of 6 mm to trimethoprim, and 10 mm to penicillin, and the associated resistance genes
blaZ and
dfrG were detected by sequencing. In
Table 1, we present the repertoire of virulence genes identified by SeqSphere [
3]. The list of virulence factors is extensive; however, we emphasisethe presence of PVL encoding genes,
edinB and
etE2 since they are epidemiologically important molecular markers.
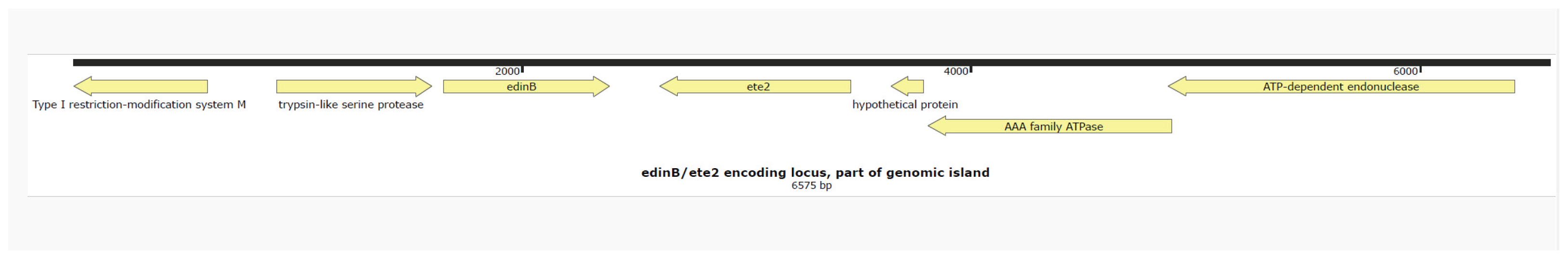
Figure 1 shows that the toxin genes
etE2 and
edinB were detected in co-located position on a previously described genomic island [
1]. The
lukF-
PV and
lukS-PV genes encoding for the PVL-cytotoxin were located on ΦSa2 phage. In
Figure 2, the phylogenetic relation of the isolate is presented using public sequencing data of isolates from clonal complex (CC) 152 [
4]. The clinical isolate is in a clade that consists mostly of MSSA strains. The MSSA isolate ERR3861662 obtained from the Democratic Republic of the Congo is the most recent common ancestor of UMCG-ST152-2024-1, and it clusters with isolates that have been isolated from Nigeria n=3, Gabon n=2, Reunion (French) n=2 and one isolate from Denmark which is identified as MRSA. The co-located virulence genes
etE2 and
edinB were present in the same nucleotide composition in all isolates of the CC152 lineage.
Virulence genes were detected using SeqSphere+ v9.0.8. Exfolative toxin etE2 was manually detected and submitted to the Virulence Factor Database. ACME: arginine catabolic mobile element; MSCRAMM: Microbial Surface Components Recognizing Adhesive Matrix Molecules; PVL: Panton-Valentine leukocidin
The genetic map was visualised using Snapgene V 5.1.7.
The radial neighbor-joining tree of S. aureus tree was generated by SeqSphere+ v9.0.8 analyzing 140 genomes. In this analysis 1861 genes were compared using published schema. The red dot represents isolate UMCG-ST152-2024-1, the white dots are MSSA isolates, and the blue dots MRSA isolates.
3. Discussion
We present the case of a fatal community-acquired necrotising pneumoniae caused by S. aureus in a young adult with underlying influenza A infection. At the University Medical Center Groningen, severe necrotising S aureus infections had sporadically been seen before. When we encounter such infections, we sequence the isolates to determine the sequence type of the S aureus and explore if exotoxins are present that could have contributed to the severity of the infection. Although disease presentation and severity of infection generally depend on combinations of virulence and host-factors, some specific well-characterized virulence factors are associated with typical disease presentations.
Remarkably, when comparing the genomic data in the UMCG database, we found a closest match with a case of severe necrotising
S. aureus infection by MSSA ST152, presenting in 2022 [
1]. This patient had been admitted to the intensive care suffering from severe necrotising cellulitis. The isolate in that study showed a similar virulence factor profile to the present case. In that study, we reported the identification of a genomic island encoding the exotoxins
edinB, and
etE2 in the isolate’s genome. Both toxins encoded by these genes are directly associated with tissue destruction. The gene
etE2 is a novel exfolative toxin encoding gene variant of
etE-. The
etE gene has been detected in ovine
S. aureus strains, whereas we exclusively detect
etE2 in human
S. aureus isolates [
5]. Degradation experiments of epidermal cells have shown host-specificity of
etE, which may explain that the toxin variants have consistently been detected in different species. Exfolative toxins in general are very specific serine proteases, causing laesions in tissue by cutting desmosomes, which are cell-adherence molecules. The typical presentations of disease are bullous impetigo and staphylococcal scalded skin syndrome [
6].
Our study shows that
edinB and the co-located
etE2 are consistently present in the CC152 lineage. EDIN-B is a C3-like ADP-ribosyltransferase that catalyses the ribosylation of Rho GTPases. The inhibition of the Rho GTPases results in modification of the actin cytoskeleton of the host cells and devastate stress fibers, which are involved in cell contractility [
7]. Exposure of cells to EDIN-B results in the loss of tissue integrity by the formation of large transcellular tunnels by this exotoxin [
8]. In a mouse model of
S aureus pneumonia, the role of EDIN-B was identified as an important factor in the translocation of
S aureus to the bloodstream by comparing wildtype and knock-out strains [
9]. Also, clinical reports suggest that EDIN-B is a virulence factor associated with invasive infection. In diabetic ulcers,
edinB-positive
S aureus is prevalent at a much higher rate in deep-seated ulcer infections compared to low-grade infections. In addition, several studies have reported increased percentages of
edin-positive
S aureus in deep-seated or bloodstream infection [
10]. However, since
edinB is co-located on a genomic island with exfoliative exotoxins as we and others have shown [
11], it is unclear if EDIN acts independently as causative factor in deep-seated infections.
The PVL-positive MSSA ST152 is a hyperepidemic lineage, predominantly circulating in Africa and the Caribbean. Reports of MSSA ST152 in Europe are uncommon so far, in contrast to the MRSA clade of ST152, which is most prevalent in Europe [
4]. The ST152 lineage is commonly associated with wound infections [
12,
13].
Necrotising pneumonia is a disease entity that is characterized by a rapidly worsening of the symptoms, leukopenia, airway hemorrhages, severe respiratory failure, a high mortality rate and necrotic destruction of wide areas of the lung [
14]. This disease is often caused by a
S. aureus superinfection upon a viral airway infection. If the infection is by PVL positive strains, the course of disease is severe in young, otherwise healthy adults. In 2022, two cases of severe necrotising pneumonia PVL-positive MSSA ST152 requiring intensive care treatment have been reported in the Indian Ocean region [
15]. In both cases, an underlying SARS-CoV-2 respiratory infection had been detected, but no other immuno-compromising disease. Like our case, both the patients had concurrent bacteremia with
S. aureus. In 2021, in a case of severe necrotising pneumonia and bloodstream infection from the Faroe Islands, PVL-positive MSSA ST152 had been reported as causative pathogen. This patient had an underlying Influenza B infection, was 47 years old, and previously healthy [
16].
The pathological association between preceding influenza infection and PVL-positive
S. aureus causing necrotising pneumonia has already been proposed in 2014. The proposed theory for this association is that the lungs are infiltrated by inflammatory cells during viral infection. Subsequently, PVL produced by
S. aureus forms membrane pores causing massive destruction of leukocytes with subsequent release of neutrophile serine proteases that destroy human tissue [
17]. While experimental data supports this theory, there are other membrane-active toxins such as PSMs and pore forming toxins such as alpha-toxin that cause the lysis of leukocytes and red blood cells [
18]. PVL could be one factor in a multifactorial etiology of disease contributed by the numerous virulence factors that are produced by
S. aureus implicated in lysis of leukocytes, tissue destruction and immune evasion. Whereas the exact contribution to the severity of disease of each virulence factor separately is difficult to establish in humans, it is sure that the PVL-positive MSSA ST152 lineage is outstanding in the plentitude of virulence factors.
In conclusion, severe infections caused by MSSA ST152 positive for an extensive repertoire of virulence factors including PVL, edinB and etE2 appear to be an emerging issue in Europe and other regions. The presented cases, and recent other casuistic studies report that isolates of this lineage cause life-threatening necrotising pneumonia with concurrent bacteraemia in previously healthy persons suffering from underlying Influenza or SARS-CoV2 infections. In future surveillance projects, the typing of MSSA isolated from blood cultures could be considered for risk assessment of this lineage. On our request, the characterized exfoliative toxin gene etE, which thus far exclusively had been detected in animals, and its sequencing variant etE2 have been included in the Virulence Factor Database, so that they can be detected by NGS-based analysis tools.
Authors’ contributions
W.J.S. collected the data and drafted the manuscript, M.A.F. supervised the molecular research and analysis, E.M., K.F. and C.H.B. participated in writing and editing of the manuscript, B.S. identified the case, participated in the concept and editing of the manuscript, E.B. coordinated and edited the manuscript, all authors revised and approved the final version of the manuscript.
Institutional Review Board Statement
No ethical approval was necessary for this case study. The conduct and reporting of this study was in line with the Declaration of Helsinki, as revised in 2013.
Informed Consent Statement
Written informed consent was obtained from the parents of the patient.
Data availability
The sequences of the isolates are available from the European Nucleotide Archive (ENA) under project number: PRJEB88875.
Conflicts of Interest
None.
References
- Sabat AJ, Wouthuyzen-Bakker M, Rondags A, Hughes L, Akkerboom V, Koutsopetra O, et al. Case Report: Necrotizing fasciitis caused by Staphylococcus aureus positive for a new sequence variant of exfoliative toxin E. Front. Genet. 2002;13:964358. [CrossRef]
- Lisotto, P., Raangs, E. C., Couto, N., Rosema, S., Lokate, M., Zhou, X., Friedrich, et al. Long-read sequencing-based in silico phage typing of vancomycin-resistant Enterococcus faecium. BMC Genomics. 2021, 22(1), Article 758. [CrossRef]
- Strauß L, Ruffing U, Abdulla S, Alabi A, Akulenko R, Garrine M, et al. Detecting Staphylococcus aureus Virulence and Resistance Genes: a Comparison of Whole-Genome Sequencing and DNA Microarray Technology. J Clin Microbiol. 2016 Apr;54(4):1008-16. [CrossRef]
- Baig S, Rhod Larsen A, Martins Simões P, Laurent F, Johannesen TB, Lilje B, Tristan A, et al. Evolution and Population Dynamics of Clonal Complex 152 Community-Associated Methicillin-Resistant Staphylococcus aureus. mSphere. 2020 Jul 1;5(4):e00226-20. [CrossRef]
- Imanishi I, Nicolas A, Caetano AB, Castro TLP, Tartaglia NR, Mariutti R, et al. Exfoliative toxin E, a new Staphylococcus aureus virulence factor with host-specific activity. Sci Rep. 2019 Nov 8;9(1):16336.
- Bukowski M, Wladyka B, Dubin G. Exfoliative toxins of Staphylococcus aureus. Toxins (Basel). 2010 May;2(5):1148-65. [CrossRef]
- Wiegers W, Just I, Müller H, Hellwig A, Traub P, Aktories K. Alteration of the cytoskeleton of mammalian cells cultured in vitro by Clostridium botulinum C2 toxin and C3 ADP-ribosyltransferase. Eur J Cell Biol. 1991 Apr;54(2):237-45.
- Boyer L, Doye A, Rolando M, Flatau G, Munro P, Gounon P, et al. Induction of transient macroapertures in endothelial cells through RhoA inhibition by Staphylococcus aureus factors. J Cell Biol. 2006 Jun 5;173(5):809-19.
- Courjon J, Munro P, Benito Y, Visvikis O, Bouchiat C, Boyer L, et al. EDIN-B Promotes the Translocation of Staphylococcus aureus to the Bloodstream in the Course of Pneumonia. Toxins. 2015; 7(10):4131-4142. [CrossRef]
- Czech A, Yamaguchi T, Bader L, Linder S, Kaminski K, Sugai M, Aepfelbacher M. Prevalence of Rho-inactivating epidermal cell differentiation inhibitor toxins in clinical Staphylococcus aureus isolates. J Infect Dis. 2001 Sep 15;184(6):785-8. [CrossRef]
- Yamaguchi T, Nishifuji K, Sasaki M, Fudaba Y, Aepfelbacher M, Takata T, et al. Identification of the Staphylococcus aureus etd pathogenicity island which encodes a novel exfoliative toxin, ETD, and EDIN-B. Infect Immun. 2002 Oct;70(10):5835-45.
- Egyir B, Owusu-Nyantakyi C, Bortey A, Rabbi Amuasi G, Owusu FA, et al. . Whole genome sequencing revealed high proportions of ST152 MRSA among clinical Staphylococcus aureus isolates from ten hospitals in Ghana. mSphere 9:e00446-24. [CrossRef]
- Shittu AO, Okon K, Adesida S, Oyedara O, Witte W, Strommenger B, et al. Antibiotic resistance and molecular epidemiology of Staphylococcus aureus in Nigeria. BMC Microbiol. 2011 May 5;11:92. [CrossRef]
- Gillet Y, Issartel B, Vanhems P, Fournet JC, Lina Get al. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet. 2002 Mar 2;359(9308):753-9. [CrossRef]
- Allou N, Allyn J, Traversier N, Baron M, Blondé R, Dupieux C, et al. SARS-CoV-2 with Panton-Valentine leukocidin-producing Staphylococcus aureus healthcare-associated pneumonia in the Indian Ocean. Heliyon. 2022 Sep;8(9):e10422. [CrossRef]
- Larsen SAH, Kyhl K, Baig S, Petersen A, Av Steinum MR, Clemmensen S, et al. Life-Threatening Necrotizing Pneumonia with Panton-Valentine Leukocidin-Producing, Methicillin-Sensitive Staphylococcus aureus in a Healthy Male Co-Infected with Influenza B. Infect Dis Rep. 2021 Dec 26;14(1):12-19.
- Löffler B, Niemann S, Ehrhardt C, Horn D, Lanckohr C, Lina G, et al. Pathogenesis of Staphylococcus aureus necrotizing pneumonia: the role of PVL and an influenza coinfection. Expert Rev Anti Infect Ther. 2013 Oct;11(10):1041-51.
- Otto, M. Staphylococcus aureus toxins. Current Opinion in Microbiology. 2014, 17:32–37.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).