Submitted:
04 June 2025
Posted:
05 June 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Lipid Metabolism
2.1. Role of Lipid Rafts in Viral Entry
2.2. Fatty Acid Synthesis, Lipid Droplets and β-Oxidation Dysregulation
2.3. Disrupted Cholesterol Homeostasis and Innate Immune Evasion
3. Energy Metabolism and Mitochondrial Dynamics
3.1. Glycolysis and the Warburg-like Effect in Viral Infections
3.2. Mitochondrial Dysfunction and Bioenergetic Failure
3.3. Cellular Metabolic Sensors and Viral Manipulation
4. Metabolic Alterations in Amino Acid and Nucleotide Pathways
4.1. Amino Acid Metabolism
4.2. Nucleotide Metabolism
5. Oxidative Stress and the Inflammatory Reaction
5.1. Intracellular Factors Linking Oxidative Stress and Inflammation
5.2. Alterations in Endogenous Antioxidant Systems
6. Analysis of Peripheral Circulating Metabolites and Search for Metabolic Biomarkers
6.1. Omics Disciplines and the Rationale for Metabolic Biomarkers in Viral Infections
6.2. Lipid Signatures and the Lipidome
6.3. Altered Carbohydrate Flux, Energy Imbalance, and Perturbations in Amino Acid and Nucleotide Metabolism
6.4. Redox Imbalance and the Antioxidant Response
7. Identification of Potential Therapeutic Targets
7.1. Inhibiting Viral Entry
7.2. Targeting Lipid Metabolism
7.3. Inhibiting Glycolysis
7.4. Modulating Mitochondrial Dynamics and Bioenergetics
7.5. Antioxidant System Modulation
7.6. Modulation of the Kynurenine Pathway
7.7. Multi-Omics Guided Therapies and Personalized Interventions
8. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACE2 | Angiotensin-converting enzyme 2 |
| AMPK | AMP-activated protein kinase |
| ATP | Adenosine triphosphate |
| AUC | Area under the curve |
| AhR | Aryl hydrocarbon receptor |
| BCAA | Branched-chain amino acids |
| CAR 8:0 | O-octanoyl-L-carnitine |
| CAT | Catalase |
| COVID-19 | Coronavirus disease 2019 |
| CPT | Carnitine palmitoyltransferases |
| CCL2 | Chemokine (C-C motif) ligand 2 |
| DGAT | Diacylglycerol O-acyltransferase |
| Drp1 | Dynamin-related protein 1 |
| FASN | Fatty acid synthase |
| GC | Gas chromatography |
| GLUT | Glucose transporter |
| GSH | Glutathione |
| HDL | High-density lipoproteins |
| HETE | Hydroxyeicosatetraenoic acid |
| HIF-1α | Hypoxia-inducible factor 1-α |
| HODE | Hydroxyoctadecadienoic acid |
| IDO1 | Indoleamine 2,3-dioxygenase 1 |
| IFN | Interferon |
| IL | Interleukin |
| Kyn/Trp | Kynurenine/tryptophan |
| LC | Liquid chromatography |
| LD | Lipid droplets |
| LDL | Low-density lipoproteins |
| LPC | Lysophosphatidylcholine |
| LPE | Lysophosphatidylethanolamine |
| MAVS | Mitochondrial antiviral-signaling protein |
| MCP-1 | Monocyte chemoattractant protein-1 |
| Mdivi-1 | Mitochondrial division inhibitor 1 |
| MS | Mass spectrometry |
| mTOR | Mechanistic target of rapamycin |
| MTP | Mitochondrial trifunctional protein |
| NAC | N-acetylcysteine |
| NADH | Reduced nicotinamide adenine dinucleotide |
| NADPH | Reduced nicotinamide adenine dinucleotide phosphate |
| NF-κB | Nuclear factor kappa B |
| NLRP3 | NOD-, LRR- and pyrin domain-containing protein 3 |
| NMR | Nuclear magnetic resonance |
| NOX | NADPH oxidase |
| NRF2 | Nuclear factor erythroid 2–related factor 2 |
| OXPHOS | Oxidative phosphorylation |
| PGC-1α | Peroxisome proliferator-activated receptor-γ coactivator 1-α |
| PC | Phosphatidylcholine |
| PFKFB3 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| PI | Phosphatidylinositol |
| PI3K | Phosphoinositide 3-kinase |
| 3PO | 3-(3-pyridinyl)-1-(4-pyridinyl)-2-propen-1-one |
| PON1 | Paraoxonase-1 |
| ROC | Receiver operating characteristic |
| ROS | Reactive oxygen species |
| RLR | Retinoic acid-inducible gene-I-like receptors, |
| RSV | Respiratory syncytial virus |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| SOD | Superoxide dismutase |
| SREBP | Sterol regulatory element-binding proteins |
| SREBP2-LDLR | Sterol regulatory element-binding protein 2 and low-density lipoprotein receptor |
| TLR | Toll-like receptors |
| TMPRSS2 | Transmembrane protease, serine 2 |
| TNF-α | Tumor necrosis factor-α |
| VLDL | Very low-density lipoproteins |
References
- Safiri, S.; Mahmoodpoor, A.; Kolahi, A.A.; Nejadghaderi, S.A.; Sullman, M.J.M.; Mansournia, M.A.; Ansarin, K.; Collins, G.S.; Kaufman, J.S.; Abdollahi, M. Global burden of lower respiratory infections during the last three decades. Front. Public Health. 2023, 10, 1028525. [Google Scholar] [CrossRef] [PubMed]
- Gravenstein, S. Foreword: Prevention of COVID-19, influenza, and respiratory syncytial virus in at-risk populations. Infect. Dis. Ther. 2025, 14 (Suppl 1), 1–3. [Google Scholar] [CrossRef] [PubMed]
- Hanage, W.P.; Schaffner, W. Burden of acute respiratory infections caused by influenza virus, respiratory syncytial virus, and SARS-CoV-2 with consideration of older adults: A narrative review. Infect. Dis. Ther. 2025, 14 (Suppl 1), 5–37. [Google Scholar] [CrossRef] [PubMed]
- Kleinehr, J.; Wilden, J.J.; Boergeling, Y.; Ludwig, S.; Hrincius, E.R. Metabolic modifications by common respiratory viruses and their potential as new antiviral targets. Viruses 2021, 13, 2068. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.N.S.; Arjona, S.P.; Santerre, M.; Sawaya, B.E. Hallmarks of metabolic reprogramming and their role in viral pathogenesis. Viruses 2022, 14, 602. [Google Scholar] [CrossRef]
- Shahpar, A.; Sofiani, V.H.; Nezhad, N.Z.; Charostad, M.; Ghaderi, R.; Farsiu, N.; Kiskani, A.K.; Pezeshki, S.; Nakhaie, M. A narrative review: exploring viral-induced malignancies through the lens of dysregulated cellular metabolism and glucose transporters. BMC Cancer 2024, 24, 1329. [Google Scholar] [CrossRef]
- Konaklieva, M.I.; Plotkin, B.J. Targeting host-specific metabolic pathways-opportunities and challenges for anti-infective therapy. Front. Mol. Biosci. 2024, 11, 1338567. [Google Scholar] [CrossRef]
- Zumla, A.; Rao, M.; Wallis, R.S.; Kaufmann, S.H.; Rustomjee, R.; Mwaba, P.; Vilaplana, C.; Yeboah-Manu, D.; Chakaya, J.; Ippolito, G.; Azhar, E.; Hoelscher, M.; Maeurer, M; Host-Directed Therapies Network consortium. Host-directed therapies for infectious diseases: current status, recent progress, and future prospects. Lancet Infect. Dis. 2016, 16, e47–e63. [CrossRef]
- Palmer, C.S. Innate metabolic responses against viral infections. Nat. Metab. 2022, 4, 1245–1259. [Google Scholar] [CrossRef]
- Mazzon, M.; Mercer, J. Lipid interactions during virus entry and infection. Cell. Microbiol. 2014, 16, 1493–1502. [Google Scholar] [CrossRef]
- Waheed, A.A.; Freed, E.O. The role of lipids in retrovirus replication. Viruses 2010, 2, 1146–1180. [Google Scholar] [CrossRef]
- Simons, K.; Sampaio, J.L. Membrane organization and lipid rafts. Cold Spring Harb. Perspect. Biol. 2011, 3, a004697. [Google Scholar] [CrossRef] [PubMed]
- Pike, L.J. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res. 2006, 47, 1597–1598. [Google Scholar] [CrossRef] [PubMed]
- Ripa, I., Andreu, S.; López-Guerrero, J.A.; Bello-Morales, R. Membrane rafts: Portals for viral entry. Front. Microbiol. 2021, 12, 631274. [CrossRef]
- Lorizate, M.; Kräusslich, H.G. Role of lipids in virus replication. Cold Spring Harb. Perspect. Biol. 2011, 3, a004820. [Google Scholar] [CrossRef]
- Marsh, M.; Helenius, A. Virus entry: open sesame. Cell 2006, 124, 729–740. [Google Scholar] [CrossRef]
- Glende, J.; Schwegmann-Wessels, C.; Al-Falah, M.; Pfefferle, S.; Qu, X.; Deng, H.; Drosten, C.; Naim, H.Y.; Herrler, G. Importance of cholesterol-rich membrane microdomains in the interaction of the S protein of SARS-coronavirus with the cellular receptor angiotensin-converting enzyme 2. Virology 2008, 381, 215–221. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, D.X.; Tam, J.P. Lipid rafts are involved in SARS-CoV entry into Vero E6 cells. Biochem. Biophys. Res. Commun. 2008, 369, 344–349. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; Müller, M.A.; Drosten, C.; Pöhlmann, S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e8. [CrossRef]
- Chakraborty, S.; Veettil, M.V.; Bottero, V.; Chandran, B. Kaposi’s sarcoma-associated herpesvirus interacts with EphrinA2 receptor to amplify signaling essential for productive infection. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 7146–7147. [Google Scholar] [CrossRef]
- Jiang, Y.; Liu, S.; Shen, S.; Guo, H.; Huang, H.; Wei, W. Methyl-β-cyclodextrin inhibits EV-D68 virus entry by perturbing the accumulation of virus particles and ICAM-5 in lipid rafts. Antiviral Res. 2020, 176, 104752. [CrossRef]
- Roncato, R.; Angelini, J.; Pani, A.; Talotta, R. Lipid rafts as viral entry routes and immune platforms: A double-edged sword in SARS-CoV-2 infection? Biochim. Biophys. Acta Mol. Cell. Biol. Lipids. 2022, 1867, 159140. [Google Scholar] [CrossRef]
- Bukrinsky, M.I.; Mukhamedova, N.; Sviridov, D. Lipid rafts and pathogens: the art of deception and exploitation. J. Lipid Res. 2020, 61, 601–610. [Google Scholar] [CrossRef]
- Song, M.S.; Lee, D.K.; Lee, C.Y.; Park, S.C.; Yang, J. Host subcellular organelles: Targets of viral manipulation. Int. J. Mol. Sci. 2024, 25, 1638. [Google Scholar] [CrossRef]
- Rawat, S.S.; Viard, M.; Gallo, S.A.; Rein, A.; Blumenthal, R.; Puri, A. Modulation of entry of enveloped viruses by cholesterol and sphingolipids. Mol. Membr. Biol. 2003, 20, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Harrison, S.C. Viral membrane fusion. Virology 2015, 479–480, 498–507. [CrossRef]
- Barrett, C.T.; Dutch, R.E. Viral membrane fusion and the transmembrane domain. Viruses 2020, 12, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Basturea, G. Endocytosis Mater. Methods 2019, 9, 2752. [CrossRef]
- Kaksonen, M.; Roux, A. Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2018, 19, 313–326. [Google Scholar] [CrossRef]
- Mayor, S.; Parton, R.G.; Donaldson, J.G. Clathrin-independent pathways of endocytosis. Cold Spring Harb. Perspect. Biol. 2014, 6, a016758. [Google Scholar] [CrossRef]
- Sandvig, K.; Kavaliauskiene, S.; Skotland, T. Clathrin-independent endocytosis: an increasing degree of complexity. Histochem. Cell Biol. 2018, 150, 107–118. [Google Scholar] [CrossRef]
- Shafaq-Zadah, M.; Dransart, E.; Johannes, L. Clathrin-independent endocytosis, retrograde trafficking, and cell polarity. Curr. Opin. Cell Biol. 2020, 65, 112–121. [Google Scholar] [CrossRef]
- Ruzzi, F.; Cappello, C.; Semprini, M.S.; Scalambra, L.; Angelicola, S.; Pittino, O.M.; Landuzzi, L.; Palladini, A.; Nanni, P.; Lollini, P.L. Lipid rafts, caveolae, and epidermal growth factor receptor family: friends or foes? Cell. Commun. Signal. 2024, 22, 489. [Google Scholar] [CrossRef]
- McMahon, K.A.; Zajicek, H.; Li, W.P.; Peyton, M.J.; Minna, J.D.; Hernandez, V.J.; Luby-Phelps, K.; Anderson, R.G. SRBC/cavin-3 is a caveolin adapter protein that regulates caveolae function. EMBO J. 2009, 28, 1001–1015. [Google Scholar] [CrossRef]
- Kovtun, O.; Tillu, V.A.; Ariotti, N.; Parton, R.G.; Collins, B.M. Cavin family proteins and the assembly of caveolae. J. Cell Sci. 2015, 128, 1269–1278. [CrossRef]
- Vicinanza, M.; D’angelo, G.; Di Campli, A.; De Matteis, M.A. Function and dysfunction of the PI system in membrane trafficking. EMBO J. 2008, 27, 2457–2470. [Google Scholar] [CrossRef]
- Vanhaesebroeck, B.; Stephens, L.; Hawkins, P. PI3K signalling: the path to discovery and understanding. Nat. Rev. Mol. Cell. Biol. 2012, 13, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Chakraborty, S.; Basu, A. Critical role of lipid rafts in virus entry and activation of phosphoinositide 3′ kinase/Akt signaling during early stages of Japanese encephalitis virus infection in neural stem/progenitor cells. J. Neurochem. 2010, 115, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Bohdanowicz, M.; Grinstein, S. Role of phospholipids in endocytosis, phagocytosis, and macropinocytosis. Physiol. Rev. 2013, 93, 69–106. [Google Scholar] [CrossRef] [PubMed]
- Marjuki, H.; Gornitzky, A.; Marathe, B.M.; Ilyushina, N.A.; Aldridge, J.R.; Desai, G.; Webby, R.J.; Webster, R.G. Influenza A virus-induced early activation of ERK and PI3K mediates V-ATPase-dependent intracellular pH change required for fusion. Cell. Microbiol. 2011, 13, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, Y.; Tsuda, M.; Hattori, T.; Sasaki, J.; Sasaki, T.; Miyazaki, T.; Ohba, Y. The Ras-PI3K signaling pathway is involved in clathrin-independent endocytosis and the internalization of influenza viruses. PLoS ONE 2011, 6, e16324. [Google Scholar] [CrossRef]
- Palacios-Rápalo, S.N.; De Jesús-González, L.A.; Cordero-Rivera, C.D.; Farfan-Morales, C.N.; Osuna-Ramos, J.F.; Martínez-Mier, G.; Quistián-Galván, J.; Muñoz-Pérez, A.; Bernal-Dolores, V.; Del Ángel, R.M.; Reyes-Ruiz, J.M. Cholesterol-rich lipid rafts asplatforms for SARS-CoV-2 entry. Front. Immunol. 2021, 12, 796855. [Google Scholar] [CrossRef]
- El Khoury, M.; Naim, H.Y. Lipid rafts disruption by statins negatively impacts the interaction between SARS-CoV-2 S1 subunit and ACE2 in intestinal epithelial cells. Front. Microbiol. 2024, 14, 1335458. [Google Scholar] [CrossRef]
- Zhang, Y.; Whittaker, G.R. Influenza entry pathways in polarized MDCK cells. Biochem. Biophys. Res. Commun. 2014, 450, 234–239. [Google Scholar] [CrossRef]
- Bolland, W.; Marechal, I.; Petiot, C.; Porrot, F.; Guivel-Benhassine, F.; Brelot, A.; Casartelli, N.; Schwartz, O.; Buchrieser, J. SARS-CoV-2 entry and fusion are independent of ACE2 localization to lipid rafts. J. Virol. 2025, 99, e0182324. [Google Scholar] [CrossRef]
- Rossman, J.S.; Leser, G.P.; Lamb, R.A. Filamentous influenza virus enters cells via macropinocytosis. J. Virol. 2012, 86, 10950–10960. [Google Scholar] [CrossRef]
- de Vries, E.; Tscherne, D.M.; Wienholts, M.J.; Cobos-Jiménez, V.; Scholte, F.; García-Sastre, A.; Rottier, P.J.; de Haan, C.A. Dissection of the influenza A virus endocytic routes reveals macropinocytosis as an alternative entry pathway. PLoS Pathog. 2011, 7, e1001329. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.K.; Gupta, D.; Lal, S.K. Host lipid rafts play a major role in binding and endocytosis of Influenza A virus. Viruses 2018, 10, 650. [Google Scholar] [CrossRef]
- Fleming, E.H.; Kolokoltsov, A.A.; Davey, R.A.; Nichols, J.E.; Roberts, N.J. Jr. Respiratory syncytial virus F envelope protein associates with lipid rafts without a requirement for other virus proteins. J. Virol. 2006, 80, 12160–12170. [Google Scholar] [CrossRef]
- Shaikh, F.Y.; Crowe, J.E. Jr. Molecular mechanisms driving respiratory syncytial virus assembly. Future Microbiol. 2013, 8, 123–131. [Google Scholar] [CrossRef]
- Chandel, N.S. Lipid metabolism. Cold Spring Harb. Perspect. Biol. 2021, 13, a040576. [Google Scholar] [CrossRef]
- Chu, J.; Xing, C.; Du, Y.; Duan, T.; Liu, S.; Zhang, P.; Cheng, C.; Henley, J.; Liu, X.; Qian, C.; Yin, B.; Wang, H.Y.; Wang, RF. Pharmacological inhibition of fatty acid synthesis blocks SARS-CoV-2 replication. Nat. Metab. 2021, 3, 1466–1475. [Google Scholar] [CrossRef]
- Williams, C.G.; Jureka, A.S.; Silvas, J.A.; Nicolini, A.M.; Chvatal, S.A.; Carlson-Stevermer, J.; Oki, J.; Holden, K.; Basler, C.F. Inhibitors of VPS34 and fatty-acid metabolism suppress SARS-CoV-2 replication. Cell. Rep. 2021, 36, 109479. [Google Scholar] [CrossRef]
- Limsuwat, N.; Boonarkart, C.; Phakaratsakul, S.; Suptawiwat, O.; Auewarakul, P. Influence of cellular lipid content on influenza A virus replication. Arch. Virol. 2020, 165, 1151–1161. [Google Scholar] [CrossRef]
- Ohol, Y.M.; Wang, Z.; Kemble, G.; Duke, G. Direct inhibition of cellular fatty acid synthase impairs replication of respiratory syncytial virus and other respiratory viruses. PLoS One 2015, 10, e0144648. [Google Scholar] [CrossRef]
- Aliyari, S.R.; Ghaffari, A.A.; Pernet, O.; Parvatiyar, K.; Wang, Y.; Gerami, H.; Tong, A.J.; Vergnes, L.; Takallou, A.; Zhang, A.; Wei, X.; Chilin, L.D.; Wu, Y.; Semenkovich, C.F.; Reue, K.; Smale, S.T.; Lee, B.; Cheng, G. Suppressing fatty acid synthase by type I interferon and chemical inhibitors as a broad spectrum anti-viral strategy against SARS-CoV-2. Acta Pharm. Sin. B. 2022, 12, 1624–1635. [Google Scholar] [CrossRef]
- Wölk, M.; Fedorova, M. The lipid droplet lipidome. FEBS Lett. 2024, 598, 1215–1225. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Mak, H.Y.; Lukmantara, I.; Li, Y.E.; Hoehn, K.L.; Huang, X.; Du, X.; Yang, H. CDP-DAG synthase 1 and 2 regulate lipid droplet growth through distinct mechanisms. J. Biol. Chem. 2019, 294, 16740–16755. [Google Scholar] [CrossRef] [PubMed]
- Monks, J.; Orlicky, D.J.; Libby, A.E.; Dzieciatkowska, M.; Ladinsky, M.S.; McManaman, J.L. Perilipin-2 promotes lipid droplet-plasma membrane interactions that facilitate apocrine lipid secretion in secretory epithelial cells of the mouse mammary gland. Front. Cell. Dev. Biol. 2022, 10, 958566. [Google Scholar] [CrossRef] [PubMed]
- Dias, S.S.G.; Soares, V.C.; Ferreira, A.C.; Sacramento, C.Q.; Fintelman-Rodrigues, N.; Temerozo, J.R.; Teixeira, L.; Nunes da Silva, M.A.; Barreto, E.; Mattos, M.; de Freitas, C.S.; Azevedo-Quintanilha, I.G.; Manso, P.P.A.; Miranda, M.D.; Siqueira, M.M.; Hottz, E.D.; Pão, C.R.R.; Bou-Habib, D.C.; Barreto-Vieira, D.F.; Bozza, F.A.; Souza, T.M.L.; Bozza, P.T. Lipid droplets fuel SARS-CoV-2 replication and production of inflammatory mediators. PLoS Pathog. 2020, 16, e1009127. [Google Scholar] [CrossRef]
- Chawla, K.; Subramanian, G.; Rahman, T.; Fan, S.; Chakravarty, S.; Gujja, S.; Demchak, H.; Chakravarti, R.; Chattopadhyay, S. Autophagy in virus infection: A race between host immune response and viral antagonism. Immuno. 2022, 2, 153–169. [Google Scholar] [CrossRef]
- Herrera-Moro Huitron, L.; De Jesús-González, L.A.; Martínez-Castillo, M.; Ulloa-Aguilar, J.M.; Cabello-Gutierrez, C.; Helguera-Repetto, C.; Garcia-Cordero, J.; León Juárez, M. Multifaceted nature of lipid droplets in viral interactions and pathogenesis. Microorganisms 2023, 11, 1851. [Google Scholar] [CrossRef]
- Kuss-Duerkop, S.K.; Wang, J.; Mena, I.; White, K.; Metreveli, G.; Sakthivel, R.; Mata, M.A.; Muñoz-Moreno, R.; Chen, X.; Krammer, F.; Diamond, M.S.; Chen, Z.J.; García-Sastre, A.; Fontoura, B.M.A. Influenza virus differentially activates mTORC1 and mTORC2 signaling to maximize late stage replication. PLoS Pathog. 2017, 13, e1006635. [Google Scholar] [CrossRef]
- Dai, P.; Tang, Z.; Qi, M.; Liu, D.; Bajinka, O.; Tan, Y. Dispersion and utilization of lipid droplets mediates respiratory syncytial virus-induced airway hyperresponsiveness. Pediatr. Allergy Immunol. 2022, 33, e13651. [Google Scholar] [CrossRef]
- Cheung, W.; Gill, M.; Esposito, A.; Kaminski, C.F.; Courousse, N.; Chwetzoff, S.; Trugnan, G.; Keshavan, N.; Lever, A.; Desselberger, U. Rotaviruses associate with cellular lipid droplet components to replicate in viroplasms, and compounds disrupting or blocking lipid droplets inhibit viroplasm formation and viral replication. J. Virol. 2010, 84, 6782–6798. [Google Scholar] [CrossRef]
- Longo, N.; Frigeni, M.; Pasquali, M. Carnitine transport and fatty acid oxidation. Biochim. Biophys. Acta 2016, 1863, 2422–2435. [Google Scholar] [CrossRef]
- Talley, J.T.; Mohiuddin, SS. Biochemistry, Fatty Acid Oxidation; StatPearls [Internet]. StatPearls Publishing: Treasure Island (FL), USA, 2025. [PubMed]
- Tanner, L.B.; Chng, C.; Guan, X.L.; Lei, Z.; Rozen, S.G.; Wenk, M.R. Lipidomics identifies a requirement for peroxisomal function during influenza virus replication. J. Lipid Res. 2014, 55, 1357–1365. [Google Scholar] [CrossRef] [PubMed]
- Andrade Silva, M.; da Silva, A.R.P.A.; do Amaral, M.A.; Fragas, M.G.; Câmara, N.O.S. Metabolic alterations in SARS-CoV-2 infection and its implication in kidney dysfunction. Front. Physiol. 2021, 12, 624698. [Google Scholar] [CrossRef] [PubMed]
- Keshavarz, M.; Solaymani-Mohammadi, F.; Namdari, H.; Arjeini, Y.; Mousavi, M.J.; Rezaei, F. Metabolic host response and therapeutic approaches to influenza infection. Cell. Mol. Biol. Lett. 2020, 25, 15. [Google Scholar] [CrossRef]
- Pérez, S.E.; Gooz, M.; Maldonado, E.N. Mitochondrial dysfunction and metabolic disturbances induced by viral infections. Cells 2024, 13, 1789. [Google Scholar] [CrossRef]
- Feingold, K.R. Introduction to Lipids and Lipoproteins. In Endotext [Internet]; Feingold, K.R., Anawalt, B., Blackman, M.R., Boyce, A., Chrousos, G., Corpas, E., de Herder, W.W., Dhatariya, K., Dungan, K., Hofland, J., Kalra, S., Kaltsas, G., Kapoor, N., Koch, C., Kopp, P., Korbonits, M., Kovacs, C.S., Kuohung, W., Laferrère, B., Levy, M., McGee, E.A., McLachlan, R., New, M., Purnell, J., Sahay, R., Shah, A.S., Singer, F., Sperling, M.A., Stratakis, C.A., Trence, D.L., Wilson, D. Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2024. Bookshelf ID: NBK305896. [PubMed]
- Navab, M.; Reddy, S.T.; Van Lenten, B.J.; Fogelman, A.M. HDL and cardiovascular disease: atherogenic and atheroprotective mechanisms. Nat. Rev. Cardiol. 2011, 8, 222–232. [Google Scholar] [CrossRef]
- Catapano, A.L.; Pirillo, A.; Bonacina, F.; Norata, G.D. HDL in innate and adaptive immunity. Cardiovasc. Res. 2014, 103, 372–383. [Google Scholar] [CrossRef]
- Camps, J.; Iftimie, S.; García-Heredia, A.; Castro, A.; Joven, J. Paraoxonases and infectious diseases. Clin. Biochem. 2017, 50, 804–811. [Google Scholar] [CrossRef]
- G, H.B.; Rao, V.S.; Kakkar, V.V. Friend turns foe: Transformation of anti-inflammatory HDL to proinflammatory HDL during acute-phase response. Cholesterol 2011, 2011, 274629. [Google Scholar] [CrossRef]
- Camps, J.; Castañé, H.; Rodríguez-Tomàs, E.; Baiges-Gaya, G.; Hernández-Aguilera, A.; Arenas, M.; Iftimie, S.; Joven, J. On the role of paraoxonase-1 and chemokine ligand 2 (C-C motif) in metabolic alterations linked to inflammation and disease. A 2021 update. Biomolecules 2021, 11, 971. [Google Scholar] [CrossRef]
- Camps, J.; Iftimie, S.; Arenas, M.; Castañé, H.; Jiménez-Franco, A.; Castro, A.; Joven, J. Paraoxonase-1: How a xenobiotic detoxifying enzyme has become an actor in the pathophysiology of infectious diseases and cancer. Chem. Biol. Interact. 2023, 380, 110553. [Google Scholar] [CrossRef]
- Mallol, R.; Amigó, N.; Rodríguez, M.A.; Heras, M.; Vinaixa, M.; Plana, N.; Rock, E.; Ribalta, J.; Yanes, O.; Masana, L.; Correig, X. Liposcale: a novel advanced lipoprotein test based on 2D diffusion-ordered 1H NMR spectroscopy. J. Lipid Res. 2015, 56, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Ballout, R.A.; Kong, H.; Sampson, M.; Otvos, J.D.; Cox, A.L.; Agbor-Enoh, S.; Remaley, AT. The NIH lipo-COVID study: A pilot NMR investigation of lipoprotein subfractions and other metabolites in patients with severe COVID-19. Biomedicines 2021, 9, 1090. [Google Scholar] [CrossRef] [PubMed]
- Lodge, S.; Nitschke, P.; Kimhofer, T.; Coudert, J.D.; Begum, S.; Bong, S.H.; Richards, T.; Edgar, D.; Raby, E.; Spraul, M.; Schaefer, H.; Lindon, J.C.; Loo, R.L.; Holmes, E.; Nicholson, J.K. NMR spectroscopic windows on the systemic effects of SARS-CoV-2 infection on plasma lipoproteins and metabolites in relation to circulating cytokines. J. Proteome Res. 2021, 20, 1382–1396. [Google Scholar] [CrossRef] [PubMed]
- Schmelter, F.; Föh, B.; Mallagaray, A.; Rahmöller, J.; Ehlers, M.; Lehrian, S.; von Kopylow, V.; Künsting, I.; Lixenfeld, A.S.; Martin, E.; Ragab, M.; Meyer-Saraei, R.; Kreutzmann, F.; Eitel, I.; Taube, S.; Käding, N.; Jantzen, E.; Graf, T.; Sina, C.; Günther, U.L. Metabolic and lipidomic markers differentiate COVID-19 from non-hospitalized and other intensive care patients. Front. Mol. Biosci. 2021, 8, 737039. [Google Scholar] [CrossRef]
- Rössler, T.; Berezhnoy, G.; Singh, Y.; Cannet, C.; Reinsperger, T.; Schäfer, H.; Spraul, M.; Kneilling, M.; Merle, U.; Trautwein, C. Quantitative serum NMR spectroscopy stratifies COVID-19 patients and sheds light on interfaces of host metabolism and the immune response with cytokines and clinical parameters. Metabolites 2022, 12, 1277. [Google Scholar] [CrossRef]
- Iftimie, S.; Amigó, N.; Martínez-Micaelo, N.; López-Azcona, A.F.; Martínez-Navidad, C.; Castañé, H.; Jiménez-Franco, A.; Ribalta, J.; Parra, S.; Castro, A.; Camps, J.; Joven, J. Differential analysis of lipoprotein and glycoprotein profiles in bacterial infections and COVID-19 using proton nuclear magnetic resonance and machine learning. Heliyon 2024, 10, e37115. [Google Scholar] [CrossRef]
- Van Lenten, B.J.; Wagner, A.C.; Anantharamaiah, G.M.; Garber, D.W.; Fishbein, M.C.; Adhikary, L.; Nayak, D.P.; Hama, S.; Navab, M.; Fogelman, A.M. Influenza infection promotes macrophage traffic into arteries of mice that is prevented by D-4F, an apolipoprotein A-I mimetic peptide. Circulation 2002, 106, 1127–1132. [Google Scholar] [CrossRef]
- Heinzl, M.W.; Freudenthaler, M.; Fellinger, P.; Kolenchery, L.; Resl, M.; Klammer, C.; Obendorf, F.; Schinagl, L.; Berger, T.; Egger, M.; Dieplinger, B.; Clodi, M. High-density lipoprotein predicts intrahospital mortality in influenza. J. Clin. Med. 2024, 13, 7242. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, J.; Xu, W.; Chen, J.; Tang, Y.; Xiong, S.; Li, Y.; Zhang, H.; Li, M.; Liu, Z. Cholesterol-rich lysosomes induced by respiratory syncytial virus promote viral replication by blocking autophagy flux. Nat. Commun. 2024, 15, 6311. [Google Scholar] [CrossRef]
- Doyle, A.; Goodson, B.A.; Kolaczkowski, O.M.; Liu, R.; Jia, J.; Wang, H.; Han, X.; Ye, C.; Bradfute, S.B.; Kell, A.M.; Lemus, M.R.; Pu. J. Manipulation of host cholesterol by SARS-CoV-2. bioRxiv [Preprint]. 2024, 2024.11.13.623299. [CrossRef]
- Li, Y.J.; Chen, C.Y.; Yang, J.H.; Chiu, Y.F. Modulating cholesterol-rich lipid rafts to disrupt influenza A virus infection. Front. Immunol. 2022, 13, 982264. [Google Scholar] [CrossRef]
- Carter, T.; Iqbal, M. The influenza A virus replication cycle: A comprehensive review. Viruses 2024, 16, 316. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, S.; Wang, J.; Guo, X.; Song, Y.; Fu, K.; Gao, Z.; Liu, D.; He, W.; Yang, L.L. Energy metabolism in health and diseases. Signal Transduct. Target. Ther. 2025, 10, 69. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.P.; Amar, S.; Gehlot, P.; Patra, S.K.; Kanwar, N.; Kanwal, A. Mitochondrial modulations, autophagy pathways shifts in viral infections: Consequences of COVID-19. Int. J. Mol. Sci. 2021, 22, 8180. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, E.L.; Lagunoff, M. Viral activation of cellular metabolism. Virology 2015, 479–480, 609–618. [CrossRef]
- Singh, S.; Singh, P.K.; Suhail, H.; Arumugaswami, V.; Pellett, P.E.; Giri, S.; Kumar, A. AMP-activated protein kinase restricts Zika virus replication in endothelial cells by potentiating innate antiviral responses and inhibiting glycolysis. J. Immunol. Baltim. Md. 2020, 1950, 1810–1824. [Google Scholar] [CrossRef]
- Pouysségur, J.; Marchiq, I.; Parks, S.K.; Durivault, J.; Ždralević, M.; Vucetic, M. 'Warburg effect' controls tumor growth, bacterial, viral infections and immunity-Genetic deconstruction and therapeutic perspectives. Semin. Cancer Biol. 2022, 86, 334–346. [Google Scholar] [CrossRef]
- Codo, A.C.; Davanzo, G.G.; Monteiro, L.B.; de Souza, G.F.; Muraro, S.P.; Virgilio-da-Silva, J.V.; Prodonoff, J.S.; Carregari, V.C.; de Biagi Junior, C.A.O.; Crunfli, F.; Jimenez Restrepo, J.L.; Vendramini, P.H.; Reis-de-Oliveira, G.; Bispo Dos Santos, K.; Toledo-Teixeira, D.A.; Parise, P.L.; Martini, M.C.; Marques, R.E.; Carmo, H.R.; Borin, A.; Coimbra, L.D.; Boldrini, V.O.; Brunetti, N.S.; Vieira, A.S.; Mansour, E.; Ulaf, R.G.; Bernardes, A.F.; Nunes, T.A.; Ribeiro, L.C.; Palma, A.C.; Agrela, M.V.; Moretti, M.L.; Sposito, A.C.; Pereira, F.B.; Velloso, L.A.; Vinolo, M.A.R.; Damasio, A.; Proença-Módena, J.L.; Carvalho, R.F.; Mori, M.A.; Martins-de-Souza, D.; Nakaya, H.I.; Farias, A.S.; Moraes-Vieira, P.M. Elevated glucose levels favor SARS-CoV-2 infection and monocyte response through a HIF-1α/glycolysis-dependent Axis. Cell. Metab. 2020, 32, 437-446.e5. [CrossRef]
- Santos, A.F.; Póvoa, P.; Paixão, P.; Mendonça, A.; Taborda-Barata, L. Changes in glycolytic pathway in SARS-COV 2 infection and their importance in understanding the severity of COVID-19. Front. Chem. 2021, 9, 685196. [Google Scholar] [CrossRef]
- Guarnieri, J.W.; Lie, T.; Albrecht, Y.E.S.; Hewin, P.; Jurado, K.A.; Widjaja, G.A.; Zhu, Y.; McManus, M.J.; Kilbaugh, T.J.; Keith, K.; Potluri, P.; Taylor, D.; Angelin, A.; Murdock, D.G.; Wallace, D.C. Mitochondrial antioxidants abate SARS-COV-2 pathology in mice. Proc. Natl. Acad. Sci. U. S. A. 2024, 121, e2321972121. [Google Scholar] [CrossRef]
- Guo, X.; Zhu, Z.; Zhang, W.; Meng, X.; Zhu, Y.; Han, P.; Zhou, X.; Hu, Y.; Wang, R. Nuclear translocation of HIF-1α induced by influenza A (H1N1) infection is critical to the production of proinflammatory cytokines. Emerg. Microbes Infect. 2017, 6, e39. [Google Scholar] [CrossRef]
- Meng, X.; Zhu, Y.; Yang, W.; Zhang, J.; Jin, W.; Tian, R.; Yang, Z.; Wang, R. HIF-1α promotes virus replication and cytokine storm in H1N1 virus-induced severe pneumonia through cellular metabolic reprogramming. Virol. Sin. 2024, 39, 81–96. [Google Scholar] [CrossRef]
- Zhao, C.; Chen, J.; Cheng, L.; Xu, K.; Yang, Y.; Su, X. Deficiency of HIF-1α enhances influenza A virus replication by promoting autophagy in alveolar type II epithelial cells. Emerg. Microbes Infect. 2020, 9, 691–706. [Google Scholar] [CrossRef]
- Reyes, A.; Duarte, L.F.; Farías, M.A.; Tognarelli, E.; Kalergis, A.M.; Bueno, S.M.; González, P.A. Impact of hypoxia over human viral infections and key cellular processes. Int. J. Mol. Sci. 2021, 22, 7954. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhu, Y.; Ren, C:; Yang, S.; Tian, S.; Chen, H.; Jin, M.; Zhou, H. Influenza A virus protein PB1-F2 impairs innate immunity by inducing mitophagy. Autophagy 2021, 17, 496–511. [CrossRef]
- Chen, L.F.; Cai, J.X.; Zhang, J.J.; Tang. Y.J.; Chen, J.Y.; Xiong, S.; Li, Y.L.; Zhang, H.; Liu, Z.; Li, M.M. Respiratory syncytial virus co-opts hypoxia-inducible factor-1α-mediated glycolysis to favor the production of infectious virus. mBio. 2023, 14, e0211023. [CrossRef]
- Kahan, S.M.; Wherry, E.J.; Zajac, A.J. T cell exhaustion during persistent viral infections. Virology 2015, 479-480, 180–193. [CrossRef]
- Wu, L.; Yan, Z.; Jiang, Y.; Chen, Y.; Du, J.; Guo, L.; Xu, J.; Luo, Z.; Liu, Y. Metabolic regulation of dendritic cell activation and immune function during inflammation. Front. Immunol. 2023, 14, 1140749. [Google Scholar] [CrossRef] [PubMed]
- Stefano, G.B.; Weissenberger, S.; Ptacek, R.; Anders, M.; Raboch, J.; Büttiker, P. Viruses and mitochondrial dysfunction in neurodegeneration and cognition: An evolutionary perspective. Cell. Mol. Neurobiol. 2024, 44, 68. [Google Scholar] [CrossRef] [PubMed]
- Gay, L.; Desquiret-Dumas, V.; Nagot, N.; Rapenne, C.; Van de Perre, P.; Reynier, P.; Molès, J.P. Long-term persistence of mitochondrial dysfunctions after viral infections and antiviral therapies: A review of mechanisms involved. J. Med. Virol. 2024, 96, e29886. [Google Scholar] [CrossRef] [PubMed]
- Elesela, S.; Lukacs, N.W. Role of mitochondria in viral infections. Life (Basel) 2021, 11, 232. [Google Scholar] [CrossRef]
- Purandare, N.; Ghosalkar, E.; Grossman, L.I.; Aras, S. Mitochondrial oxidative phosphorylation in viral infections. Viruses 2023, 15, 2380. [Google Scholar] [CrossRef]
- Guarnieri, J.W.; Dybas, J.M.; Fazelinia, H.; Kim, M.S.; Frere, J.; Zhang, Y.; Soto Albrecht, Y.; Murdock, D.G.; Angelin, A.; Singh, L.N.; Weiss, S.L.; Best, S.M.; Lott, M.T.; Zhang, S.; Cope, H.; Zaksas, V.; Saravia-Butler, A.; Meydan, C.; Foox, J.; Mozsary, C.; Bram, Y.; Kidane, Y.; Priebe, W.; Emmett, M.R.; Meller, R.; Demharter, S.; Stentoft-Hansen, V.; Salvatore, M.; Galeano, D.; Enguita, F.J.; Grabham, P.; Trovao, N.S.; Singh, U.; Haltom, J.; Heise, M.T.; Moorman, N.J.; Baxter, V.K.; Madden, E.A.; Taft-Benz, S.A.; Anderson, E.J.; Sanders, W.A.; Dickmander, R.J.; Baylin, S.B.; Wurtele, E.S.; Moraes-Vieir, P.M.; Taylor, D.; Mason, C.E.; Schisler, J.C.; Schwartz, R.; Beheshti, A.; Wallace, D.C. Core mitochondrial genes are down-regulated during SARS-CoV-2 infection of rodent and human hosts. Sci. Transl. Med. 2023, 15, eabq1533. [Google Scholar] [CrossRef]
- Miller, B.; Silverstein, A.; Flores, M.; Cao, K.; Kumagai, H.; Mehta, H.H.; Yen, K.; Kim, S.J.; Cohen, P. Host mitochondrial transcriptome response to SARS-CoV-2 in multiple cell models and clinical samples. Sci. Rep. 2021, 11, 3. [Google Scholar] [CrossRef]
- Westermann, B. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell. Biol. 2010, 11, 872–884. [Google Scholar] [CrossRef]
- Mehmood, T.; Nasir, Q.; Younis, I.; Muanprasat, C. Inhibition of mitochondrial dynamics by mitochondrial division inhibitor-1 suppresses cell migration and metastatic markers in colorectal cancer HCT116 cells. J. Exp. Pharmacol. 2025, 17, 143–157. [Google Scholar] [CrossRef]
- Barbier, V.; Lang, D.; Valois, S.; Rothman, A.L.; Medin, C.L. Dengue virus induces mitochondrial elongation through impairment of Drp1-triggered mitochondrial fission. Virology 2017, 500, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Archer, S.L.; Dasgupta, A.; Chen, K.H.; Wu, D.; Baid, K.; Mamatis, J.E.; Gonzalez, V.; Read, A.; Bentley, R.E.; Martin, A.Y.; Mewburn, J.D.; Dunham-Snary, K.J.; Evans, G.A.; Levy, G.; Jones, O.; Al-Qazazi, R.; Ring, B.; Alizadeh, E.; Hindmarch, C.C.; Rossi, J.; Lima, P.D.; Falzarano, D.; Banerjee, A.; Colpitts, C.C. SARS-CoV-2 mitochondriopathy in COVID-19 pneumonia exacerbates hypoxemia. Redox Biol. 2022, 58, 102508. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Delgado, I.; López-Pastor, A.R.; González-Jiménez, A.; Ramos-Acosta, C.; Hernández-Garate, Y.; Martínez-Micaelo, N.; Amigó, N.; Espino-Paisán, L.; Anguita, E.; Urcelay, E. Long-term mitochondrial and metabolic impairment in lymphocytes of subjects who recovered after severe COVID-19. Cell. Biol. Toxicol. 2025, 41, 27. [Google Scholar] [CrossRef]
- Tábara, L.C.; Morris, J.L.; Prudent, J. The complex dance of organelles during mitochondrial division. Trends Cell. Biol. 2021, 31, 241–253. [Google Scholar] [CrossRef]
- Duan, X.; Liu, R.; Lan, W.; Liu, S. The essential role of mitochondrial dynamics in viral infections. Int. J. Mol. Sci. 2025, 26, 1955. [Google Scholar] [CrossRef]
- Vazquez, C.; Horner, S.M. MAVS coordination of antiviral innate immunity. J. Virol. 2015, 89, 6974–6977. [Google Scholar] [CrossRef]
- Qi, Y.; Yin, J.; Xia, W.; Yang, S. Exploring the role of mitochondrial antiviral signaling protein in cardiac diseases. Front. Immunol. 2025, 16, 1540774. [Google Scholar] [CrossRef]
- Li, X.; Hou, P.; Ma, W.; Wang, X.; Wang, H.; Yu, Z.; Chang, H.; Wang, T.; Jin, S.; Wang, X.; Wang, W.; Zhao, Y.; Zhao, Y.; Xu, C.; Ma, X.; Gao, Y.; He, H. SARS-CoV-2 ORF10 suppresses the antiviral innate immune response by degrading MAVS through mitophagy. Cell. Mol. Immunol. 2022, 19, 67–78. [Google Scholar] [CrossRef]
- Swanson, K.V.; Deng, M.; Ting, J.P. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 2019, 19, 477–489. [Google Scholar] [CrossRef]
- Hsieh, L.L.; Looney, M.; Figueroa, A.; Massaccesi, G.; Stavrakis, G.; Anaya, E.U.; D'Alessio, F.R.; Ordonez, A.A.; Pekosz, A.S.; DeFilippis, V.R.; Karakousis, P.C.; Karaba, A.H.; Cox, AL. Bystander monocytic cells drive infection-independent NLRP3 inflammasome response to SARS-CoV-2. mBio. 2024, 15, e0081024. [Google Scholar] [CrossRef]
- Pandey, K.P.; Zhou, Y. Influenza A virus infection activates NLRP3 inflammasome through trans-Golgi network dispersion. Viruses 2022, 14, 88. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Shin, O.S. Zika virus modulates mitochondrial dynamics, mitophagy, and mitochondria-derived vesicles to facilitate viral replication in trophoblast cells. Front. Immunol. 2023, 14, 1203645. [Google Scholar] [CrossRef] [PubMed]
- Shang, C.; Liu, Z.; Zhu, Y.; Lu, J.; Ge, C.; Zhang, C.; Li, N.; Jin, N.; Li, Y.; Tian, M.; Li, X. SARS-CoV-2 causes mitochondrial dysfunction and mitophagy impairment. Front. Microbiol. 2022, 12, 780768. [Google Scholar] [CrossRef] [PubMed]
- Bhutta, M.S.; Gallo, E.S.; Borenstein, R. Multifaceted role of AMPK in viral infections. Cells 2021, 10, 1118. [Google Scholar] [CrossRef]
- Camps, J.; Rodríguez-Gallego, E.; García-Heredia, A.; Triguero, I.; Riera-Borrull, M.; Hernández-Aguilera, A.; Luciano-Mateo, F.; Fernández-Arroyo, S.; Joven, J. Paraoxonases and chemokine (C-C motif) ligand-2 in noncommunicable diseases. Adv. Clin. Chem. 2014, 63, 247–308. [Google Scholar] [CrossRef]
- Hardie, D.G.; Ross, F.A.; Hawley, S.A. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell. Biol. 2012, 13, 251–262. [Google Scholar] [CrossRef]
- Mankouri, J.; Tedbury, P.R.; Gretton, S.; Hughes, M.E.; Griffin, S.D.; Dallas, M.L.; Green, K.A.; Hardie, D.G.; Peers, C.; Harris, M. Enhanced hepatitis C virus genome replication and lipid accumulation mediated by inhibition of AMP-activated protein kinase. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 11549–11554. [Google Scholar] [CrossRef]
- Deretic, V. Autophagy in inflammation, infection, and immunometabolism. Immunity 2021, 54, 437–453. [Google Scholar] [CrossRef]
- Cheng, J.; Wang, Y.; Yin, L.; Liang, W.; Zhang, J.; Ma, C.; Zhang, Y.; Liu, B.; Wang, J.; Zhao, W.; Li, M.; Wei, L. The nonstructural protein 1 of respiratory syncytial virus hijacks host mitophagy as a novel mitophagy receptor to evade the type I IFN response in HEp-2 cells. mBio. 2023, 14, e0148023. [Google Scholar] [CrossRef]
- Le Sage, V.; Cinti, A.; Amorim, R.; Mouland, A.J. Adapting the stress response: Viral subversion of the mTOR signaling pathway. Viruses 2016, 8, 152. [Google Scholar] [CrossRef]
- Vincent, H.A.; Ziehr, B.; Moorman, N.J. Human cytomegalovirus strategies to maintain and promote mRNA translation. Viruses 2016, 8, 97. [Google Scholar] [CrossRef] [PubMed]
- Majeed, S.T.; Batool, A.; Majeed, R.; Bhat, N.N.; Zargar, M.A.; Andrabi, K.I. mTORC1 induces eukaryotic translation initiation factor 4E interaction with TOS-S6 kinase 1 and its activation. Cell Cycle 2021, 20, 839–854. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Lei, X.; Zhao, G.; Guo, R.; Cui, N. mTOR in programmed cell death and its therapeutic implications. Cytokine Growth Factor Rev. 2023, 71–72, 66–81. [CrossRef]
- Twu, W.I.; Lee, J.Y.; Kim, H.; Prasad, V.; Cerikan, B.; Haselmann, U.; Tabata, K.; Bartenschlager, R. Contribution of autophagy machinery factors to HCV and SARS-CoV-2 replication organelle formation. Cell. Rep. 2021, 37, 110049. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. mTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef]
- Dunn, E.F.; Connor, J.H. HijAkt: The PI3K/Akt pathway in virus replication and pathogenesis. Prog. Mol. Biol. Transl. Sci. 2012, 106, 223–250. [Google Scholar] [CrossRef]
- Greene, K. S:; Choi, A.; Yang, N.; Chen, M.; Li, R.; Qiu, Y.; Ezzatpour, S.; Rojas, K.S.; Shen, J.; Wilson, K.F.; Katt, W.P.; Aguilar, H.C.; Lukey, M.J.; Whittaker, G.R.; Cerione, R.A. Glutamine metabolism is essential for coronavirus replication in host cells and in mice. J. Biol. Chem. 2025, 301, 108063. [Google Scholar] [CrossRef]
- Smallwood, H.S.; Duan, S.; Morfouace, M.; Rezinciuc, S.; Shulkin, B.L.; Shelat, A.; Zink, E.E.; Milasta, S.; Bajracharya, R.; Oluwaseum, A.J.; Roussel, M.F.; Green, D.R.; Pasa-Tolic, L.; Thomas, P.G. Targeting metabolic reprogramming by influenza infection for therapeutic intervention. Cell. Rep. 2017, 19, 1640–1653. [Google Scholar] [CrossRef]
- Lu, Y.; Xu, S.; Sun, H.; Shan, J.; Shen, C.; Ji, J.; Lin, L.; Xu, J.; Peng, L.; Dai, C.; Xie, T. Analysis of temporal metabolic rewiring for human respiratory syncytial virus infection by integrating metabolomics and proteomics. Metabolomics 2023, 19, 30. [Google Scholar] [CrossRef]
- Yoo, H.C.; Yu, Y.C.; Sung, Y.; Han, J.M. Glutamine reliance in cell metabolism. Exp. Mol. Med. 2020, 52, 1496–1516. [Google Scholar] [CrossRef]
- Cruzat, V.; Macedo Rogero, M.; Noel Keane, K.; Curi, R.; Newsholme, P. Glutamine: Metabolism and immune function, supplementation and clinical translation. Nutrients 2018, 10, 1564. [Google Scholar] [CrossRef]
- Wu, G.; Meininger, C.J.; McNeal, C.J.; Bazer, F.W.; Rhoads, J.M. Role of L-arginine in nitric oxide synthesis and health in humans. Adv. Exp. Med. Biol. 2021, 1332, 167–187. [Google Scholar] [CrossRef] [PubMed]
- Moraes, T.J. Arginase and respiratory viral infections. The Open Nitric Oxide Journal, 2010, 2, 64–68. [CrossRef]
- van den Berg, M.P.; Meurs, H.; Gosens, R. Targeting arginase and nitric oxide metabolism in chronic airway diseases and their co-morbidities. Curr. Opin. Pharmacol. 2018, 40, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Dean, M.J.; Ochoa, J.B.; Sanchez-Pino, M.D.; Zabaleta, J.; Garai, J.; Del Valle, L.; Wyczechowska, D.; Baiamonte, L.B.; Philbrook, P.; Majumder, R.; Vander Heide, R.S.; Dunkenberger, L.; Thylur, R.P.; Nossaman, B.; Roberts, W.M.; Chapple, A.G.; Wu, J.; Hicks, C.; Collins, J.; Luke, B.; Johnson, R.; Koul, H.K.; Rees, C.A.; Morris, C.R.; Garcia-Diaz, J.; Ochoa, A.C. Severe COVID-19 is characterized by an impaired type I interferon response and elevated levels of arginase producing granulocytic myeloid derived suppressor cells. Front. Immunol. 2021, 12, 695972. [Google Scholar] [CrossRef] [PubMed]
- Churiso, G.; Husen, G.; Bulbula, D.; Abebe, L. Immunity cell responses to RSV and the role of antiviral inhibitors: A systematic review. Infect. Drug Resist. 2022, 15, 7413–7430. [Google Scholar] [CrossRef]
- West, E.E.; Merle, N.S.; Kamiński, M.M.; Palacios, G.; Kumar, D.; Wang, L.; Bibby. J.A.; Overdahl, K.; Jarmusch, A.K.; Freeley, S.; Lee, D.Y.; Thompson, J.W.; Yu, Z.X.; Taylor, N.; Sitbon, M.; Green, D.R.; Bohrer, A.; Mayer-Barber, K.D.; Afzali, B.; Kazemian, M.; Scholl-Buergi, S.; Karall, D.; Huemer, M.; Kemper, C. Loss of CD4+ T cell-intrinsic arginase 1 accelerates Th1 response kinetics and reduces lung pathology during influenza infection. Immunity 2023, 56, 2036–2053.e12. [CrossRef]
- Mounce, B.C.; Olsen, M.E.; Vignuzzi, M.; Connor, J.H. Polyamines and their role in virus infection. Microbiol. Mol. Biol. Rev. 2017, 81, e00029–17. [Google Scholar] [CrossRef]
- Firpo, M.R.; Mastrodomenico, V.; Hawkins, G.M.; Prot, M.; Levillayer, L.; Gallagher, T.; Simon-Loriere, E.; Mounce, B.C. Targeting polyamines inhibits coronavirus infection by reducing cellular attachment and entry. ACS Infect. Dis. 2021, 7, 1423–1432. [Google Scholar] [CrossRef]
- Cruz-Pulido, Y.E.; Mounce, B.C. Good cop, bad cop: Polyamines play both sides in host immunity and viral replication. Semin. Cell. Dev. Biol. 2023, 146, 70–79. [Google Scholar] [CrossRef]
- Lionetto, L.; Ulivieri, M.; Capi, M.; De Bernardini, D.; Fazio, F.; Petrucca, A.; Pomes, L.M.; De Luca, O.; Gentile, G.; Casolla, B.; Curto, M.; Salerno, G.; Schillizzi, S.; Torre, M.S.; Santino, I.; Rocco, M.; Marchetti, P.; Aceti, A.; Ricci, A.; Bonfini, R.; Nicoletti, F.; Simmaco, M.; Borro, M. Increased kynurenine-to-tryptophan ratio in the serum of patients infected with SARS-CoV2: An observational cohort study. Biochim. Biophys. Acta Mol. Basis. Dis. 2021, 1867, 166042. [Google Scholar] [CrossRef]
- Thomas, T.; Stefanoni, D.; Reisz, J.A.; Nemkov, T.; Bertolone, L.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hansen, K.C.; Hod, E.A.; Spitalnik, S.L.; D'Alessandro, A. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight 2020, 5, e140327. [Google Scholar] [CrossRef]
- Dehhaghi, M.; Heydari, M.; Panahi, H.K.S.; Lewin, S.R.; Heng, B.; Brew, B.J.; Guillemin, G.J. The roles of the kynurenine pathway in COVID-19 neuropathogenesis. Infection 2024, 52, 2043–2059. [Google Scholar] [CrossRef]
- Karimi, Z.; Chenari, M.; Rezaie, F.; Karimi, S.; Parhizgari, N.; Mokhtari-Azad, T. Proposed pathway linking respiratory infections with depression. Clin. Psychopharmacol. Neurosci. 2022, 20, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Stone, T.W.; Williams, R.O. Modulation of T cells by tryptophan metabolites in the kynurenine pathway. Trends Pharmacol. Sci. 2023, 44, 442–456. [Google Scholar] [CrossRef] [PubMed]
- Shih, A.Y.; Erb, H.; Sun, X.; Toda, S.; Kalivas, P.W.; Murphy, T.H. Cystine/glutamate exchange modulates glutathione supply for neuroprotection from oxidative stress and cell proliferation. J. Neurosci. 2006, 26, 10514–10523. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Haas de Mello, A.; Morris, D.R.; Jones-Hall, Y.L.; Ivanciuc, T.; Sattler, R.A.; Paessler, S.; Menachery, V.D.; Garofalo, R.P.; Casola, A. SARS-CoV-2 inhibits NRF2-mediated antioxidant responses in airway epithelial cells and in the lung of a murine model of infection. Microbiol. Spectr. 2023, 11, e0037823. [Google Scholar] [CrossRef]
- Fernandes, I.G.; de Brito, C.A.; Dos Reis, V.M.S.; Sato, M.N.; Pereira, N.Z. SARS-CoV-2 and other respiratory viruses: What does oxidative stress have to do with it? Oxid. Med. Cell. Longev. 2020, 2020, 8844280. [Google Scholar] [CrossRef]
- Yang, X.; Liu, X.; Nie, Y.; Zhan, F.; Zhu, B. Oxidative stress and ROS-mediated cellular events in RSV infection: potential protective roles of antioxidants. Virol. J. 2023, 20, 224. [Google Scholar] [CrossRef]
- Jones, J.T.; Qian, X.; van der Velden, J.L.; Chia, S.B.; McMillan, D.H.; Flemer, S.; Hoffman, S.M.; Lahue, K.G.; Schneider, R.W.; Nolin, J.D.; Anathy, V.; van der Vliet, A.; Townsend, D.M.; Tew, K.D.; Janssen-Heininger, Y.M. Glutathione S-transferase pi modulates NF-κB activation and pro-inflammatory responses in lung epithelial cells. Redox Biol. 2016, 8, 375–382. [Google Scholar] [CrossRef]
- Mazzarino, R.C. Targeting future pandemics, a case for de novo purine synthesis and basic research. Front. Immunol. 2021, 12, 694300. [Google Scholar] [CrossRef]
- Qin, C.; Rao, Y.; Yuan, H.; Wang, T.Y.; Zhao, J.; Espinosa, B.; Liu, Y.; Zhang, S.; Savas, A.C.; Liu, Q.; Zarinfar, M.; Rice, S.; Henley, J.; Comai, L.; Graham, N.A.; Chen, C.; Zhang, C.; Feng, P. SARS-CoV-2 couples evasion of inflammatory response to activated nucleotide synthesis. Proc. Natl. Acad. Sci. U. S. A. 2022, 119, e2122897119. [Google Scholar] [CrossRef]
- Diehl, F.F.; Miettinen, T.P.; Elbashir, R.; Nabel, C.S.; Darnell, A.M.; Do, B.T.; Manalis, S.R.; Lewis, C.A.; Vander Heiden, M.G. Nucleotide imbalance decouples cell growth from cell proliferation. Nat. Cell. Biol. 2022, 24, 1252–1264. [Google Scholar] [CrossRef]
- Sahan, A.Z.; Hazra, T.K.; Das, S. The pivotal role of DNA repair in infection mediated-inflammation and cancer. Front. Microbiol. 2018, 9, 663. [Google Scholar] [CrossRef] [PubMed]
- Okesli, A.; Khosla, C.; Bassik, M.C. Human pyrimidine nucleotide biosynthesis as a target for antiviral chemotherapy. Curr. Opin. Biotechnol. 2017, 48, 127–134. [Google Scholar] [CrossRef]
- Luganini, A.; Boschi, D.; Lolli, M.L.; Gribaudo, G. DHODH inhibitors: What will it take to get them into the clinic as antivirals? Antiviral Res. 2025, 236, 106099. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, D.; Rubini, M.; Burns, J.S. The potential of purinergic signaling to thwart viruses including SARS-CoV-2. Front. Immunol. 2022, 13, 904419. [Google Scholar] [CrossRef] [PubMed]
- Pacheco-Hernández, L.M.; Ramírez-Noyola, J.A.; Gómez-García, I.A.; Ignacio-Cortés, S.; Zúñiga, J.; Choreño-Parra, J.A. Comparing the cytokine storms of COVID-19 and pandemic influenza. J. Interferon Cytokine Res. 2022, 42, 369–392. [Google Scholar] [CrossRef]
- Ali, E.S.; Ben-Sahra, I. Regulation of nucleotide metabolism in cancers and immune disorders. Trends Cell. Biol. 2023, 33, 950–966. [Google Scholar] [CrossRef]
- Khomich, O.A.; Kochetkov, S.N.; Bartosch, B.; Ivanov, A.V. Redox biology of respiratory viral infections. Viruses 2018, 10, 392. [Google Scholar] [CrossRef]
- Forrester, S.J.; Kikuchi, D.S.; Hernandes, M.S.; Xu, Q.; Griendling, K.K. Reactive oxygen species in metabolic and inflammatory signaling. Circ. Res. 2018, 122, 877–902. [Google Scholar] [CrossRef]
- Amatore, D.; Sgarbanti, R.; Aquilano, K.; Baldelli, S.; Limongi, D.; Civitelli, L.; Nencioni, L.; Garaci, E.; Ciriolo, M.R.; Palamara, A.T. Influenza virus replication in lung epithelial cells depends on redox-sensitive pathways activated by NOX4-derived ROS. Cell. Microbiol. 2015, 17, 131–145. [Google Scholar] [CrossRef]
- Yang, W.S.; Yi, Y.S.; Kim, D.; Kim, M.H.; Park, J.G.; Kim, E.; Lee, S.Y.; Yoon, K.; Kim, J.H.; Park, J.; Cho, J.Y. Nuclear factor kappa-B- and activator protein-1-mediated immunostimulatory activity of compound K in monocytes and macrophages. J. Ginseng Res. 2017, 41, 298–306. [Google Scholar] [CrossRef]
- Hong, Y.; Boiti, A.; Vallone, D.; Foulkes, N.S. Reactive oxygen species signaling and oxidative stress: Transcriptional regulation and evolution. Antioxidants (Basel) 2024, 13, 312. [Google Scholar] [CrossRef] [PubMed]
- Bhol, N.K.; Bhanjadeo, M.M.; Singh, A.K.; Dash, U.C.; Ojha, R.R.; Majhi, S.; Duttaroy, A.K.; Jena, A.B. The interplay between cytokines, inflammation, and antioxidants: mechanistic insights and therapeutic potentials of various antioxidants and anti-cytokine compounds. Biomed. Pharmacother. 2024, 178, 117177. [Google Scholar] [CrossRef] [PubMed]
- Naiditch, H.; Betts, M.R.; Larman, H.B.; Levi, M.; Rosenberg, A.Z. Immunologic and inflammatory consequences of SARS-CoV-2 infection and its implications in renal disease. Front. Immunol. 2025, 15, 1376654. [Google Scholar] [CrossRef] [PubMed]
- Blevins, H.M.; Xu, Y.; Biby, S.; Zhang, S. The NLRP3 inflammasome pathway: A review of mechanisms and inhibitors for the treatment of inflammatory diseases. Front. Aging Neurosci. 2022, 14, 879021. [Google Scholar] [CrossRef]
- Ziehr, B.K.; MacDonald, J.A. Regulation of NLRPs by reactive oxygen species: A story of crosstalk. Biochim. Biophys. Acta Mol. Cell. Res. 2024, 1871, 119823. [Google Scholar] [CrossRef]
- Kolattukudy, P.E.; Niu, J. Inflammation, endoplasmic reticulum stress, autophagy, and the monocyte chemoattractant protein-1/CCR2 pathway. Circ Res. 2012, 110, 174–189. [Google Scholar] [CrossRef]
- Pandey, E.; Nour, A.S.; Harris, E.N. Prominent receptors of liver sinusoidal endothelial cells in liver homeostasis and disease. Front. Physiol. 2020, 11, 873. [Google Scholar] [CrossRef]
- Andersson, U.; Ottestad, W.; Tracey, K.J. Extracellular HMGB1: a therapeutic target in severe pulmonary inflammation including COVID-19? Mol. Med. 2020, 26, 42. [Google Scholar] [CrossRef]
- Shamilov, R.; Ackley, T.W.; Aneskievich, B.J. Enhanced wound healing -and inflammasome- associated gene expression in TNFAIP3-interacting protein 1-(TNIP1-) deficient HaCaT keratinocytes parallels reduced reepithelialization. Mediators Inflamm. 2020, 2020, 5919150. [Google Scholar] [CrossRef]
- Relja, B.; Land, W.G. Damage-associated molecular patterns in trauma. Eur. J. Trauma. Emerg. Surg. 2020, 46, 751–775. [Google Scholar] [CrossRef]
- Afrose, S.S.; Junaid, M.; Akter, Y.; Tania, M.; Zheng, M.; Khan, M.A. Targeting kinases with thymoquinone: a molecular approach to cancer therapeutics. Drug Discov. Today 2020, 25, 2294–2306. [Google Scholar] [CrossRef] [PubMed]
- Dantonio, P.M.; Klein, M.O.; Freire, M.R.V.B.; Araujo, C.N.; Chiacetti, A.C.; Correa, R.G. Exploring major signaling cascades in melanomagenesis: a rationale route for targetted skin cancer therapy. Biosci. Rep. 2018, 38, BSR20180511. [Google Scholar] [CrossRef] [PubMed]
- Slaine, P.D.; Kleer, M.; Duguay, B.A.; Pringle, E.S.; Kadijk, E.; Ying, S. , Balgi, A.; Roberge, M.; McCormick, C.; Khaperskyy, D.A. Thiopurines activate an antiviral unfolded protein response that blocks influenza A virus glycoprotein accumulation. J. Virol. 2021, 95, e00453–21. [Google Scholar] [CrossRef] [PubMed]
- Féral, K.; Jaud, M.; Philippe, C.; Di Bella, D.; Pyronnet, S.; Rouault-Pierre, K.; Mazzolini, L.; Touriol, C. ER stress and unfolded protein response in leukemia: Friend, Foe, or Both? Biomolecules 2021, 11, 199. [Google Scholar] [CrossRef]
- Robinson, C.M.; Talty, A.; Logue, S.E.; Mnich, K.; Gorman, A.M.; Samali, A. An emerging role for the unfolded protein response in pancreatic cancer. Cancers (Basel) 2021, 13, 261. [Google Scholar] [CrossRef]
- Rashid, H.O.; Yadav, R.K.; Kim, H.R.; Chae, H.J. ER stress: Autophagy induction, inhibition and selection. Autophagy 2015, 11, 1956–1977. [Google Scholar] [CrossRef]
- Dymkowska, D. The involvement of autophagy in the maintenance of endothelial homeostasis: The role of mitochondria. Mitochondrion 2021, 57, 131–147. [Google Scholar] [CrossRef]
- Polonikov, A. Endogenous deficiency of glutathione as the most likely cause of serious manifestations and death in COVID-19 patients. ACS Infect. Dis. 2020, 6, 1558–1562. [Google Scholar] [CrossRef]
- Silvagno, F.; Vernone, A.; Pescarmona, G.P. The role of glutathione in protecting against the severe inflammatory response triggered by COVID-19. Antioxidants (Basel) 2020, 9, 624. [Google Scholar] [CrossRef]
- De Angelis, M.; Amatore, D.; Checconi, P.; Zevini, A.; Fraternale, A.; Magnani, M.; Hiscott, J.; De Chiara, G.; Palamara, A.T.; Nencioni, L. Influenza virus down-modulates G6PD expression and activity to induce oxidative stress and promote its replication. Front. Cell. Infect. Microbiol. 2022, 11, 804976. [Google Scholar] [CrossRef]
- Hosakote, Y.M.; Liu, T.; Castro, S.M.; Garofalo, R.P.; Casola, A. Respiratory syncytial virus induces oxidative stress by modulating antioxidant enzymes. Am. J. Respir. Cell. Mol. Biol. 2009, 41, 348–357. [Google Scholar] [CrossRef] [PubMed]
- Fukai, T.; Ushio-Fukai, M. Superoxide dismutases: role in redox signaling, vascular function, and diseases. Antioxid. Redox Signal. 2011, 15, 1583–1606. [Google Scholar] [CrossRef] [PubMed]
- Chu, J.; Hua, L.; Liu, X.; Xiong, H.; Jiang, F.; Zhou, W.; Wang, L.; Xue, G. Superoxide dismutase alterations in COVID-19: implications for disease severity and mortality prediction in the context of omicron variant infection. Front. Immunol. 2024, 15, 1362102. [Google Scholar] [CrossRef] [PubMed]
- Tavassolifar, M.J.; Aghdaei, H.A.; Sadatpour, O.; Maleknia, S.; Fayazzadeh, S.; Mohebbi, S.R.; Montazer, F.; Rabbani, A.; Zali, M.R.; Izad, M.; Meyfour, A. New insights into extracellular and intracellular redox status in COVID-19 patients. Redox Biol. 2023, 59, 102563. [Google Scholar] [CrossRef]
- Choi, A.M.; Knobil, K.; Otterbein, S.L.; Eastman, D.A.; Jacoby, D.B. Oxidant stress responses in influenza virus pneumonia: gene expression and transcription factor activation. Am. J. Physiol. 1996, 271, L383–L391. [Google Scholar] [CrossRef]
- Chen, F.; Chen, L.; Liang, J.; Chen, Z.; Zhang, C.; Zhang, Z.; Yang, J. Potential role of superoxide dismutase 3 (SOD3) in resistance to Influenza A virus infection. Antioxidants (Basel) 2023, 12, 354. [Google Scholar] [CrossRef]
- Hasan Anber, Z.N.; Oied Saleh, B.; Hassan Majed, R. Assessment of oxidative stress parameters in Iraqi male patients with Covid-19; A case control study. Rep. Biochem. Mol. Biol. 2024, 13, 167–173. [Google Scholar] [CrossRef]
- Rodríguez-Tomàs, E.; Iftimie, S.; Castañé, H.; Baiges-Gaya, G.; Hernández-Aguilera, A.; González-Viñas, M.; Castro, A.; Camps, J.; Joven, J. Clinical performance of paraoxonase-1-related variables and novel markers of inflammation in coronavirus disease-19. A machine learning approach. Antioxidants (Basel) 2021, 10, 991. [Google Scholar] [CrossRef]
- Gabaldó, X.; Juanpere, M.; Castañé, H.; Rodríguez-Tomàs, E.; López-Azcona, A.F.; Baiges-Gaya, G.; Castro, L.; Valverde-Díaz, E.; Muñoz-Blázquez, A.; Giménez-Cuenca, L.; Felipo-Balada, L.; Ballester, F.; Pujol, I.; Simó, J.M.; Castro, A.; Iftimie, S.; Camps, J.; Joven, J. Usefulness of the measurement of serum paraoxonase-1 arylesterase activity in the diagnoses of COVID-19. Biomolecules 2022, 12, 879. [Google Scholar] [CrossRef]
- Cho, K.H.; Kim, J.R.; Lee, I.C.; Kwon, H.J. Native high-density lipoproteins (HDL) with higher paraoxonase exerts a potent antiviral effect against SARS-CoV-2 (COVID-19), while glycated HDL lost the antiviral activity. Antioxidants (Basel) 2021, 10, 209. [Google Scholar] [CrossRef]
- Thaker, S.K.; Ch'ng, J.; Christofk, H.R. Viral hijacking of cellular metabolism. BMC Biol. 2019, 17, 59. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Chen, J.; Liang, Q.; Zheng, H.; Ye, Y.; Nan, W.; Zhang, X.; Gao, H.; Li, Y. Metabolomics profile in acute respiratory distress syndrome by nuclear magnetic resonance spectroscopy in patients with community-acquired pneumonia. Respir. Res. 2022, 23, 172. [Google Scholar] [CrossRef] [PubMed]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Camps, J.; Jiménez-Franco, A.; García-Pablo, R.; Joven, J.; Arenas, M. Artificial intelligence-driven integration of multi-omics and radiomics: A new hope for precision cancer diagnosis and prognosis. Biochim. Biophys. Acta Mol. Basis Dis. 2025, 1871, 167841. [Google Scholar] [CrossRef]
- Hu, L.; Liu, J.; Zhang, W.; Wang, T.; Zhang, N.; Lee, Y.H.; Lu, H. Functional metabolomics decipher biochemical functions and associated mechanisms underlie small-molecule metabolism. Mass Spectrom. Rev. 2020, 39, 417––433. [Google Scholar] [CrossRef]
- Fuertes-Martín, R.; Taverner, D.; Vallvé, J.; Paredes, S.; Masana, L.; Correig Blanchar, X. Characterization of 1H NMR plasma glycoproteins as a new strategy to identify inflammatory patterns in rheumatoid arthritis. J. Proteome Res. 2018, 17, 3730–3739. [Google Scholar] [CrossRef]
- Fuertes-Martin, R.; Moncayo, S.; Insenser, M.; Martínez-García, M.A.; Luque-Ramírez, M.; Grau, N.A.; Blanchar, X.C.; Escobar-Morreale, H.F. Glycoprotein A and B height-to-width ratios as obesity-independent novel biomarkers of low-grade chronic inflammation in women with polycystic ovary syndrome (PCOS) J. Proteome Res. 2019, 18, 4038–4045. [Google Scholar] [CrossRef]
- Riera-Borrull, M.; Rodríguez-Gallego, E.; Hernández-Aguilera, A.; Luciano, F.; Ras, R.; Cuyàs, E.; Camps, J.; Segura-Carretero, A.; Menendez, J.A.; Joven, J.; Fernández-Arroyo, S. Exploring the process of energy generation in pathophysiology by targeted metabolomics: Performance of a simple and quantitative method. J. Am. Soc. Mass Spectrom. 2016, 27, 168–177. [Google Scholar] [CrossRef]
- Cuyàs, E.; Fernández-Arroyo, S.; Verdura, S.; García, R.Á.; Stursa, J.; Werner, L.; Blanco-González, E.; Montes-Bayón, M.; Joven, J.; Viollet, B.; Neuzil, J.; Menendez, J.A. Metformin regulates global DNA methylation via mitochondrial one-carbon metabolism. Oncogene 2018, 37, 963–970. [Google Scholar] [CrossRef]
- Nikolskiy, I.; Siuzdak, G.; Patti, G.J. Discriminating precursors of common fragments for large-scale metabolite profiling by triple quadrupole mass spectrometry. Bioinformatics 2015, 31, 2017–2023. [Google Scholar] [CrossRef]
- Patti, G.J. Separation strategies for untargeted metabolomics. J. Sep. Sci. 2011, 34, 3460–3469. [Google Scholar] [CrossRef] [PubMed]
- Ivanisevic, J.; Want, E.J. From samples to insights into metabolism: uncovering biologically relevant information in LC-HRMS metabolomics data. Meta 2019, 9, 308. [Google Scholar] [CrossRef] [PubMed]
- Strimbu K, Tavel JA. What are biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463–466. [CrossRef]
- FDA-NIH Biomarker Working Group. BEST (Biomarkers, EndpointS, and other Tools) Resource [Internet]; Silver Spring: Maryland, USA, 2016; Bookshelf ID: NBK326791. [PubMed]
- Pepe, M.S.; Janes, H.; Longton, G.; Leisenring, W.; Newcomb, P. Limitations of the odds ratio in gauging the performance of a diagnostic, prognostic, or screening marker. Am. J. Epidemiol. 2004, 159, 882–890. [Google Scholar] [CrossRef] [PubMed]
- Castañé, H.; Iftimie, S.; Baiges-Gaya, G.; Rodríguez-Tomàs, E.; Jiménez-Franco, A.; López-Azcona, A.F.; Garrido, P.; Castro, A.; Camps, J.; Joven, J. Machine learning and semi-targeted lipidomics identify distinct serum lipid signatures in hospitalized COVID-19-positive and COVID-19-negative patients. Metabolism 2022, 131, 155197. [Google Scholar] [CrossRef]
- Mai, M.; Tönjes, A.; Kovacs, P.; Stumvoll, M.; Fiedler, G.M.; Leichtle, A.B. Serum levels of acylcarnitines are altered in prediabetic conditions. PLoS One 2013, 8, e82459. [Google Scholar] [CrossRef]
- Ayres, J.S. A metabolic handbook for the COVID-19 pandemic. Nat. Metab. 2020, 2, 572–585. [Google Scholar] [CrossRef]
- Otsubo, C.; Bharathi, S.; Uppala, R.; Ilkayeva, O.R.; Wang, D.; McHugh, K.; Zou, Y.; Wang, J.; Alcorn, J.F.; Zuo, Y.Y.; Hirschey, M.D.; Goetzman, E.S. Long-chain acylcarnitines reduce lung function by inhibiting pulmonary surfactant. J. Biol. Chem. 2015, 290, 23897–23904. [Google Scholar] [CrossRef]
- Wu, D.; Shu, T.; Yang, X.; Song, J.X.; Zhang, M.; Yao, C.; Liu, W.; Huang, M.; Yu, Y.; Yang, Q.; Zhu, T.; Xu, J.; Mu, J.; Wang, Y.; Wang, H.; Tang, T.; Ren, Y.; Wu, Y.; Lin, S.H.; Qiu, Y.; Zhang, D.Y.; Shang, Y.; Zhou, X. Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl. Sci. Rev. 2020, 7, 1157–1168. [Google Scholar] [CrossRef]
- Song, J.W.; Lam, S.M.; Fan, X.; Cao, W.J.; Wang, S.Y.; Tian, H.; Chua, G.H.; Zhang, C.; Meng, F.P.; Xu, Z.; Fu, J.L.; Huang, L.; Xia, P.; Yang, T.; Zhang, S.; Li, B.; Jiang, T.J.; Wang, R.; Wang, Z.; Shi, M.; Zhang, J.Y.; Wang, F.S.; Shui, G. Omics-driven systems interrogation of metabolic dysregulation in COVID-19 pathogenesis. Cell. Metab. 2020, 32, 188–202. [Google Scholar] [CrossRef]
- Barberis, E.; Timo, S.; Amede, E.; Vanella, V.V.; Puricelli, C.; Cappellano, G.; Raineri, D.; Cittone, M.G.; Rizzi, E.; Pedrinelli, A.R.; Vassia, V.; Casciaro, F.G.; Priora, S.; Nerici, I.; Galbiati, A.; Hayden, E.; Falasca, M.; Vaschetto, R.; Sainaghi, P.P.; Dianzani, U.; Rolla, R.; Chiocchetti, A.; Baldanzi, G.; Marengo, E.; Manfredi, M. Large-scale plasma analysis revealed new mechanisms and molecules associated with the host response to SARS-CoV-2. Int. J. Mol. Sci. 2020, 21, 8623. [Google Scholar] [CrossRef]
- Fraser, D.D.; Slessarev, M.; Martin, C.M.; Daley, M.; Patel, M.A.; Miller, M.R.; Patterson, E.K.; O'Gorman, D.B.; Gill, S.E.; Wishart, D.S.; Mandal, R.; Cepinskas, G. Metabolomics profiling of critically ill coronavirus disease 2019 patients: identification of diagnostic and prognostic biomarkers. Crit. Care Explor. 2020, 2, e0272. [Google Scholar] [CrossRef] [PubMed]
- Delafiori, J.; Navarro, L.C.; Siciliano, R.F.; de Melo, G.C.; Busanello, E.N.B.; Nicolau, J.C.; Sales, G.M.; de Oliveira, A.N.; Val, F.F.A.; de Oliveira, D.N.; Eguti, A.; Dos Santos, L.A.; Dalçóquio, T.F.; Bertolin, A.J.; Abreu-Netto, R.L.; Salsoso, R.; Baía-da-Silva, D.; Marcondes-Braga, F.G.; Sampaio, V.S.; Judice, C.C.; Costa, F.T.M.; Durán, N.; Perroud, M.W.; Sabino, E.C.; Lacerda, M.V.G.; Reis, L.O.; Fávaro, W.J.; Monteiro, W.M.; Rocha, A.R.; Catharino, R.R. Covid-19 automated diagnosis and risk assessment through metabolomics and machine learning. Anal. Chem. 2021, 93, 2471–2479. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Zhang, Z.; Feng, G.; Chen, M.; Wan, Q.; Lin, J.; Wu, L.; Nie, W.; Chen, S. Distinct lipid metabolic dysregulation in asymptomatic COVID-19. iScience 2021, 24, 102974. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.M.; Zhang, C.; Wang, Z.; Ni, Z.; Zhang, S.; Yang, S.; Huang, X.; Mo, L.; Li, J.; Lee, B.; Mei, M.; Huang, L.; Shi, M.; Xu, Z.; Meng, F.P.; Cao, W.J.; Zhou, M.J.; Shi, L.; Chua, G.H.; Li, B.; Cao, J.; Wang, J.; Bao, S.; Wang, Y.; Song, J.W.; Zhang, F.; Wang, F.S.; Shui, G. A multi-omics investigation of the composition and function of extracellular vesicles along the temporal trajectory of COVID-19. Nat. Metab. 2021, 3, 909–922. [Google Scholar] [CrossRef] [PubMed]
- Kyle, J.E.; Burnum-Johnson, K.E.; Wendler, J.P.; Eisfeld, A.J.; Halfmann, P.J.; Watanabe, T.; Sahr, F.; Smith, R.D.; Kawaoka, Y.; Waters, K.M.; Metz, T.O. Plasma lipidome reveals critical illness and recovery from human Ebola virus disease. Proc Natl Acad Sci U S A. 2019, 116, 3919–3928. [Google Scholar] [CrossRef]
- Nguyen, M.; Bourredjem, A.; Piroth, L.; Bouhemad, B.; Jalil, A.; Pallot, G.; Le Guern, N.; Thomas, C.; Pilot, T.; Bergas, V.; Choubley, H.; Quenot, J.P.; Charles, P.E.; Lagrost, L.; Deckert, V.; de Barros, J.P.; Guinot, P.G.; Masson, D.; Binquet, C.; Gautier, T.; Blot, M; Lymphonie study group. High plasma concentration of non-esterified polyunsaturated fatty acids is a specific feature of severe COVID-19 pneumonia. Sci. Rep. 2021, 11, 10824. [CrossRef]
- Bizkarguenaga, M.; Bruzzone, C.; Gil-Redondo, R.; SanJuan, I.; Martin-Ruiz, I.; Barriales, D.; Palacios, A.; Pasco, S.T.; González-Valle, B.; Laín, A.; Herrera, L.; Azkarate, A.; Vesga, M.A.; Eguizabal, C.; Anguita, J.; Embade, N.; Mato, J.M.; Millet, O. Uneven metabolic and lipidomic profiles in recovered COVID-19 patients as investigated by plasma NMR metabolomics. NMR Biomed. 2022, 35, e4637. [Google Scholar] [CrossRef]
- Bruzzone, C.; Bizkarguenaga, M.; Gil-Redondo, R.; Diercks, T.; Arana, E.; García de Vicuña, A.; Seco, M.; Bosch, A.; Palazón, A.; San Juan, I.; Laín, A.; Gil-Martínez, J.; Bernardo-Seisdedos, G.; Fernández-Ramos, D.; Lopitz-Otsoa, F.; Embade, N.; Lu, S.; Mato, J.M.; Millet, O. SARS-CoV-2 infection dysregulates the metabolomic and lipidomic profiles of serum. iScience 2020, 23, 101645. [Google Scholar] [CrossRef]
- Iftimie, S.; Gabaldó-Barrios, X., Penadés-Nadal, J.; Canela-Capdevila, M.; Piñana, R.; Jiménez-Franco, A.; López-Azcona, A.F.; Castañé, H.; Cárcel, M.; Camps, J.; Castro, A.; Joven, J. Serum levels of arachidonic acid, interleukin-6, and C-reactive protein as potential indicators of pulmonary viral infections: Comparative analysis of influenza A, respiratory syncytial virus infection, and COVID-19. Viruses 2024, 16, 1065. [CrossRef]
- Yan, B.; Chu, H.; Yang, D.; Sze, K.H.; Lai, P.M.; Yuan, S.; Shuai, H.; Wang, Y.; Kao, R.Y.; Chan, J.F.; Yuen, K.Y. Characterization of the lipidomic profile of human coronavirus-infected cells: Implications for lipid metabolism remodeling upon coronavirus replication. Viruses 2019, 11, 73. [Google Scholar] [CrossRef]
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang. C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; Ge, W.; Liu, W.; Liang, S.; Chen, H.; Zhang, Y.; Li, J.; Xu, J.; He, Z.; Chen, B.; Wang, J.; Yan, H.; Zheng, Y.; Wang, D.; Zhu, J.; Kong, Z.; Kang, Z.; Liang, X.; Ding, X.; Ruan, G.; Xiang, N.; Cai, X.; Gao, H.; Li, L.; Li, S.; Xiao, Q.; Lu, T.; Zhu, Y.; Liu, H.; Chen, H.; Guo, T. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell 2020, 182, 59–72. [CrossRef]
- Torrente-Rodríguez, R.M., Ruiz-Valdepeñas Montiel, V.; Iftimie, S.; Montero-Calle, A.; Pingarrón, J.M.; Castro, A.; Camps, J.; Barderas, R.; Campuzano, S.; Joven, J. Contributing to the management of viral infections through simple immunosensing of the arachidonic acid serum level. Mikrochim. Acta 2024, 191, 369. [CrossRef]
- Amigó, N.; Martínez-Micaelo, N.; Velasco, M.; Casas, M.L.; Correig, X.; Guijarro, C. Identification of a molecular signature associated with covid-19 severity using a comprehensive 1H-NMR serum metabolomics profiling strategy. Atherosclerosis 2024, 395 (Suppl.1), 117627. [Google Scholar] [CrossRef]
- López-Hernández, Y.; Oropeza-Valdez, J.J.; García Lopez, D.A.; Borrego, J.C.; Murgu, M.; Valdez, J.; López, J.A.; Monárrez-Espino, J. Untargeted analysis in post-COVID-19 patients reveals dysregulated lipid pathways two years after recovery. Front. Mol. Biosci. 2023, 10, 1100486. [Google Scholar] [CrossRef]
- Washirasaksiri, C.; Sayabovorn, N.; Ariyakunaphan, P.; Kositamongkol, C.; Chaisathaphol, T.; Sitasuwan, T.; Tinmanee, R.; Auesomwang, C.; Nimitpunya, P.; Woradetsittichai, D.; Chayakulkeeree, M.; Phoompoung, P.; Mayurasakorn, K.; Sookrung, N.; Tungtrongchitr, A.; Wanitphakdeedecha, R.; Muangman, S.; Senawong, S.; Tangjittipokin, W.; Sanpawitayakul, G.; Nopmaneejumruslers, C.; Vamvanij, V.; Phisalprapa, P.; Srivanichakorn, W. Long-term multiple metabolic abnormalities among healthy and high-risk people following nonsevere COVID-19. Sci. Rep. 2023, 13, 14336. [Google Scholar] [CrossRef] [PubMed]
- Kyo, M.; Zhu, Z.; Shibata, R.; Fujiogi, M.; Mansbach, J.M.; Camargo, C.A.; Hasegawa, K. Respiratory virus-specific nasopharyngeal lipidome signatures and severity in infants with bronchiolitis: A prospective multicenter study. J. Infect. Dis. 2023, 228, 1410–1420. [Google Scholar] [CrossRef] [PubMed]
- Loo, R.L.; Lodge, S.; Kimhofer, T.; Bong, S.H.; Begum, S.; Whiley, L.; Gray, N.; Lindon, J.C.; Nitschke, P.; Lawler, N.G.; Schäfer, H.; Spraul, M.; Richards, T.; Nicholson, J.K.; Holmes, E. Quantitative in-vitro diagnostic NMR spectroscopy for lipoprotein and metabolite measurements in plasma and serum: Recommendations for analytical artifact minimization with special reference to COVID-19/SARS-CoV-2 samples. J. Proteome Res. 2020, 19, 4428–4441. [Google Scholar] [CrossRef] [PubMed]
- Corn, G.; Lund, M.; Andersson, N.W.; Dohlmann, T.L.; Hlatky, M.A.; Wohlfahrt, J.; Melbye, M. Low-density lipoprotein cholesterol response to statins according to comorbidities and co-medications: A population-based study. Am. Heart J. 2024, 274, 102–112. [Google Scholar] [CrossRef]
- Zimodro, J.M.; Mucha, M.; Berthold, H.K.; Gouni-Berthold, I. Lipoprotein metabolism, dyslipidemia, and lipid-lowering therapy in women: A comprehensive review. Pharmaceuticals (Basel) 2024, 17, 913. [Google Scholar] [CrossRef]
- Berta, E.; Zsíros, N.; Bodor, M.; Balogh, I.; Lőrincz, H.; Paragh, G.; Harangi, M. Clinical aspects of genetic and non-genetic cardiovascular risk factors in familial hypercholesterolemia. Genes (Basel) 2022, 13, 1158. [Google Scholar] [CrossRef]
- Baiges-Gaya, G.; Iftimie, S.; Castañé, H.; Rodríguez-Tomàs, E.; Jiménez-Franco, A.; López-Azcona, A.F.; Castro, A.; Camps, J.; Joven, J. Combining semi-targeted metabolomics and machine learning to identify metabolic alterations in the serum and urine of hospitalized patients with COVID-19. Biomolecules 2023, 13, 163. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, A.H.; Song, Q.; Fang, H.; Liu, X.Y.; Su, J.; Yang, L.; Yu, M.D.; Wang, X.J. Functional metabolomics discover pentose and glucuronate interconversion pathways as promising targets for Yang Huang syndrome treatment with Yinchenhao Tang. RSC Adv. 2018, 8, 36831–36839. [Google Scholar] [CrossRef]
- Chen, S.; Niu, C.; Lv, W. Multi-omics insights reveal the remodeling of gut mycobiome with P. gingivalis. Front. Cell. Infect. Microbiol. 2022, 12, 937725. [Google Scholar] [CrossRef]
- Lu, X.; Liu, T.; Zhou, J.; Liu, J.; Yuan, Z.; Guo, L. Subgingival microbiome in periodontitis and type 2 diabetes mellitus: An exploratory study using metagenomic sequencing. J. Periodontal Implant. Sci. 2022, 52, 282–297. [Google Scholar] [CrossRef]
- Xiong, H.; Li, N.; Zhao, L.; Li, Z.; Yu, Y.; Cui, X.; Liu, Q.; Zhao, C. Integrated serum pharmacochemistry, metabolomics, and network pharmacology to reveal the material basis and mechanism of Danggui Shaoyao San in the treatment of primary dysmenorrhea. Front. Pharmacol. 2022, 13, 942955. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, K.; Zeng, M.; Qiao, B.; Zhou, B. Serum metabolomics analysis of the anti-inflammatory effects of gallic acid on rats with acute inflammation. Front. Pharmacol. 2022, 13, 830439. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, D.; Gao, X.; Wang, X.; Zhang, L. 2’- and 3’-ribose modifications of nucleotide analogues establish the structural basis to inhibit the viral replication of SARS-CoV-2. J. Phys. Chem. Lett. 2022, 13, 4111–4118. [Google Scholar] [CrossRef]
- Guo, X.; Wu, S.; Li, N.; Lin, Q.; Liu, L.; Liang, H.; Niu, Y.; Huang, Z.; Fu, X. Accelerated metabolite levels of aerobic glycolysis and the pentose phosphate pathway are required for efficient replication of infectious spleen and kidney necrosis virus in Chinese perch brain cells. Biomolecules 2019, 9, 440. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.; Kaminiski, R.; Deshmane, S.; Langford, D.; Khalili, K.; Amini, S.; Datta, P.K. Role of hexokinase-1 in the survival of HIV-1-infected macrophages. Cell Cycle 2015, 14, 980–989. [Google Scholar] [CrossRef]
- Stincone, A.; Prigione, A.; Cramer, T.; Wamelink, M.M.; Campbell, K.; Cheung, E.; Olin-Sandoval, V.; Grüning, N.M.; Krüger, A.; Tauqeer Alam, M.; Keller, M.A.; Breitenbach, M.; Brindle, K.M.; Rabinowitz, J.D.; Ralser, M. The return of metabolism: Biochemistry and physiology of the pentose phosphate pathway. Biol. Rev. Camb. Philos. Soc. 2015, 90, 927–963. [Google Scholar] [CrossRef]
- Chen, I.T.; Aoki, T.; Huang, Y.T.; Hirono, I.; Chen, T.C.; Huang, J.Y. White spot Syndrome virus induces metabolic changes resembling the Warburg effect in shrimp hemocytes in the early stage of infection. J. Virol. 2011, 85, 12919–12928. [Google Scholar] [CrossRef]
- Pérez-Torres, I.; Soto, M.E.; Guarner-Lans, V..; Manzano-Pech, L.; Soria-Castro, E. The possible role of glucose-6-phosphate dehydrogenase in the SARS-CoV-2 infection. Cells 2022, 11, 1982. [CrossRef]
- Bojkova, D.; Costa, R.; Reus, P.; Bechtel, M.; Jaboreck, M.C.; Olmer, R.; Martin, U.; Ciesek, S.; Michaelis, M.; Cinatl, J., Jr. Targeting the pentose phosphate pathway for SARS-CoV-2 therapy. Metabolites 2021, 11, 699. [Google Scholar] [CrossRef]
- Isaacs, C.E.; Kim, K.S.; Thormar, H. Inactivation of enveloped viruses in human bodily fluids by purified lipids. Ann. N. Y. Acad. Sci. 1994, 724, 457–464. [Google Scholar] [CrossRef]
- Nefedova, E.; Koptev, V.; Bobikova, A.S; Cherepushkina, V.; Mironova, T.; Afonyushkin, V.; Shkil, N.; Donchenko, N.; Kozlova, Y.; Sigareva, N.; Davidova, N.; Bogdanchikova, N.; Pestryakov, A.; Toledano-Magaña, Y. The infectious bronchitis coronavirus pneumonia model presenting a novel insight for the SARS-CoV-2 dissemination route. Vet. Sci. 2021, 8, 239. [Google Scholar] [CrossRef]
- Thormar, H.; Isaacs, C.E.; Brown, H.R.; Barshatzky, M.R.; Pessolano, T. Inactivation of enveloped viruses and killing of cells by fatty acids and monoglycerides. Antimicrob. Agents Chemother. 1987, 31, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Cheudjeu, A. Correlation of D-xylose with severity and morbidity-related factors of COVID-19 and possible therapeutic use of D-xylose and antibiotics for COVID-19. Life Sci. 2020, 260, 118335. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.S.; Ad Souza, M.; Raposo, N.R.B.; Ferreira, A.P.; Silva, S.S. Xylitol inhibits J774A.1 macrophage adhesion in vitro. Braz. Arch. Biol. Technol. 2011, 54, 1211–1216. [Google Scholar] [CrossRef]
- Xu, M.L.; Wi, G.; Kim, H.J.; Kim, H.J. Ameliorating effect of dietary xylitol on human respiratory syncytial virus (hRSV) infection. Biol. Pharm. Bull. 2016, 39, 540–546. [Google Scholar] [CrossRef]
- Yin, S.Y.; Kim, H.J.; Kim, H.J. Protective effect of dietary xylitol on influenza A virus infection. PLoS ONE 2014, 9, e84633. [Google Scholar] [CrossRef]
- Anand, S.; Mande, S.S. Diet, microbiota and gut-lung connection. Front. Microbiol. 2018, 9, 2147. [Google Scholar] [CrossRef]
- Shukla, S.D.; Budden, K.F.; Neal, R.; Hansbro, P.M. Microbiome effects on immunity, health and disease in the lung. Clin. Transl. Immunol. 2017, 6, e133. [Google Scholar] [CrossRef]
- Russell, S.L.; Gold, M.J.; Willing, B.P.; Thorson, L.; Mcnagny, K.M.; Finlay, B.B. Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microbes 2013, 4, 158–164. [Google Scholar] [CrossRef]
- Looft, T.; Allen, H.K. Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microbes 2012, 3, 463–467. [Google Scholar] [CrossRef]
- Xie, J.; Cho, H.; Lin, B.M.; Pillai, M.; Heimisdottir, L.H.; Bandyopadhyay, D.; Zou, F.; Roach, J.; Divaris, K.; Wu, D. Improved metabolite prediction using microbiome data-based elastic net models. Front. Cell. Infect. Microbiol. 2021, 11, 734416. [Google Scholar] [CrossRef]
- Wan, J.; Zhang, Y.; He, W.; Tian, Z.; Lin, J.; Liu, Z.; Li, Y.; Chen, M.; Han, S.; Liang, J.; Shi, Y.; Wang, X.; Zhou, L.; Cao, Y.; Liu, J.; Wu, K. Gut microbiota and metabolite changes in patients with ulcerative colitis and Clostridioides difficile infection. Front. Microbiol. 2022, 13, 802823. [Google Scholar] [CrossRef] [PubMed]
- Colonetti, K.; de Carvalho, E.L.; Rangel, D.L.; Pinto, P.M.; Roesch, L.F.W.; Pinheiro, F.C.; Schwartz, I.V.D. Are the bacteria and their metabolites contributing for gut inflammation on GSD-Ia patients? Metabolites 2022, 12, 873. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Ma, T.; Xu, N.; Jin, H.; Zhao, F.; Kwok, L.Y.; Zhang, H.; Zhang, S.; Sun, Z. Adjunctive probiotics alleviates asthmatic symptoms via modulating the gut microbiome and serum metabolome. Microbiol. Spectr. 2021, 9, e0085921. [Google Scholar] [CrossRef] [PubMed]
- Tong, W.; Hannou, S.A.; Wang, Y.; Astapova, I.; Sargsyan, A.; Monn, R.; Thiriveedi, V.; Li, D.; McCann, J.R.; Rawls, J.F.; Roper, J.; Zhang, G.F.; Herman, M.A. . The intestine is a major contributor to circulating succinate in mice. FASEB J. 2022, 36, e22546. [Google Scholar] [CrossRef]
- Nagata, N.; Takeuchi, T.; Masuoka, H.; Aoki, R.; Ishikane, M.; Iwamoto, N.; Sugiyama, M.; Suda, W.; Nakanishi, Y.; Terada-Hirashima, J.; Kimura, M.; Nishijima, T.; Inooka, H.; Miyoshi-Akiyama, T.; Kojima, Y.; Shimokawa, C.; Hisaeda, H.; Zhang, F.; Yeoh, Y.K.; Ng, S.C.; Uemura, N.; Itoi, T.; Mizokami, M.; Kawai, T.; Sugiyama, H.; Ohmagari, N.; Ohno, H. Human gut microbiota and its metabolites impact immune responses in COVID-19 and its complications. Gastroenterology 2023, 164, 272–288. [Google Scholar] [CrossRef]
- Liao, J.; Li, Q.; Lei, C.; Yu, W.; Deng, J.; Guo, J.; Han, Q.; Hu, L.; Li, Y.; Pan, J.; Zhang, H.; Chang, Y.F.; Tang, Z. Toxic effects of copper on the jejunum and colon of pigs: Mechanisms related to gut barrier dysfunction and inflammation influenced by the gut microbiota. Food Funct. 2021, 12, 9642–9657. [Google Scholar] [CrossRef]
- Yu, W.; Shang, J.; Guo, R.; Zhang, F.; Zhang, W.; Zhang, Y.; Wu, F.; Ren, H.; Liu, C.; Xiao, J.; Zhao, Z. The gut microbiome in differential diagnosis of diabetic kidney disease and membranous nephropathy. Ren. Fail. 2020, 42, 1100–1110. [Google Scholar] [CrossRef]
- Yin, J.; Li, Y.; Han, H.; Liu, Z.; Zeng, X.; Li, T.; Yin, Y. Long-term effects of lysine concentration on growth performance, intestinal microbiome, and metabolic profiles in a pig model. Food Funct. 2018, 9, 4153–4163. [Google Scholar] [CrossRef]
- Ren, L.; Zhang, W.; Zhang, J.; Zhang, J.; Zhang, H.; Zhu, Y.; Meng, X.; Yi, Z.; Wang, R. Influenza A virus (H1N1) infection induces glycolysis to facilitate viral replication. Virol. Sin. 2021, 36, 1532–1542. [Google Scholar] [CrossRef]
- Martín-Vicente, M.; González-Riaño, C.; Barbas, C.; Jiménez-Sousa, M.Á.; Brochado-Kith, O.; Resino, S.; Martínez, I. Metabolic changes during respiratory syncytial virus infection of epithelial cells. PLoS One 2020, 15, e0230844. [Google Scholar] [CrossRef]
- Fratta Pasini, A.M.; Stranieri, C.; Girelli, D.; Busti, F.; Cominacini, L. Is ferroptosis a key component of the process leading to multiorgan damage in COVID-19? Antioxidants (Basel) 2021, 10, 1677. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.; Bortolasci, C.C.; Puri, B.K.; Olive, L.; Marx, W.; O'Neil, A.; Athan, E.; Carvalho, A.F.; Maes, M.; Walder, K.; Berk, M. The pathophysiology of SARS-CoV-2: A suggested model and therapeutic approach. Life Sci. 2020, 258, 118166. [Google Scholar] [CrossRef] [PubMed]
- Cecchini, R.; Cecchini, A.L. SARS-CoV-2 infection pathogenesis is related to oxidative stress as a response to aggression. Med. Hypotheses. 2020, 143, 110102. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell. Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Ammer, E.; Nietzsche, S.; Rien, C.; Kühnl, A.; Mader, T.; Heller, R.; Sauerbrei, A.; Henke, A. The anti-obesity drug orlistat reveals anti-viral activity. Med. Microbiol. Immunol. 2015, 204, 635–645. [Google Scholar] [CrossRef]
- Kow, C.S.; Hasan, S.S. Meta-analysis of effect of statins in patients with COVID-19. Am. J. Cardiol. 2020, 134, 153–155. [Google Scholar] [CrossRef]
- Diaz-Arocutipa, C.; Melgar-Talavera, B.; Alvarado-Yarasca, Á.; Saravia-Bartra, M.M.; Cazorla, P.; Belzusarri, I.; Hernandez, A.V. Statins reduce mortality in patients with COVID-19: an updated meta-analysis of 147 824 patients. Int. J. Infect. Dis. 2021, 110, 374–381. [Google Scholar] [CrossRef]
- Onorato, D.; Pucci, M.; Carpene, G.; Henry, B.M.; Sanchis-Gomar, F.; Lippi, G. Protective effects of statins administration in European and North American patients infected with COVID-19: A meta-analysis. Semin. Thromb. Hemost. 2021, 47, 392–399. [Google Scholar] [CrossRef]
- Florêncio de Mesquita, C.; Rivera, A.; Araújo, B.; Durães, V.L.; Queiroz, I.; Carvalho, V.H.; Haque, T.; Bes, T.M. Adjunctive statin therapy in patients with Covid-19: A systematic review and meta-analysis of randomized controlled trials. Am. J. Med. 2024, 137, 966–973.e11. [CrossRef]
- Arnardottir, H.; Pawelzik, S.C.; Öhlund Wistbacka. U.; Artiach, G.; Hofmann, R.; Reinholdsson, I.; Braunschweig, F.; Tornvall, P.; Religa, D.; Bäck, M. Stimulating the resolution of inflammation through omega-3 polyunsaturated fatty acids in COVID-19: Rationale for the COVID-Omega-F trial. Front. Physiol. 2021, 11, 624657. [CrossRef]
- Huang, Z.; Chavda, V.P.; Vora, L.K.; Gajjar, N.; Apostolopoulos, V.; Shah, N.; Chen, Z.S. 2-deoxy-D-glucose and its derivatives for the COVID-19 treatment: An update. Front. Pharmacol. 2022, 13, 899633. [Google Scholar] [CrossRef]
- Bhatt, A.N.; Shenoy, S.; Munjal, S.; Chinnadurai, V.; Agarwal, A.; Vinoth Kumar, A.; Shanavas, A.; Kanwar, R.; Chandna, S. 2-deoxy-D-glucose as an adjunct to standard of care in the medical management of COVID-19: a proof-of-concept and dose-ranging randomised phase II clinical trial. BMC Infect. Dis. 2022, 22, 669. [Google Scholar] [CrossRef]
- Verma, A.; Adhikary, A.; Woloschak, G.; Dwarakanath, B.S.; Papineni, R.V.L. A combinatorial approach of a polypharmacological adjuvant 2-deoxy-D-glucose with low dose radiation therapy to quell the cytokine storm in COVID-19 management. Int. J. Radiat. Biol. 2020, 96, 1323–1328. [Google Scholar] [CrossRef] [PubMed]
- Sandepogu, T.S.; Dara, C.; Mallamgunta, S.; Jogi, S.; Sree Podila, K.; Chandrasekhar, J.; N, V.; Sivakumar, S. Role of 2-deoxy-D-glucose in enhancing the efficacy of standard of care for moderate to severe COVID-19: A comparative analysis of clinical outcomes. Cureus 2024, 16, e73993. [Google Scholar] [CrossRef] [PubMed]
- Ergashev, A.; Shi, F.; Liu, Z.; Pan, Z.; Xie, H.; Kong, L.; Wu, L.; Sun, H.; Jin, Y.; Kong, H.; Geng, D.; Ibrohimov, A.; Obeng, E.; Wang, Y.; Ma, F.; Chen, G.; Zhang, T. KAN0438757, a novel PFKFB3 inhibitor, prevent the progression of severe acute pancreatitis via the Nrf2/HO-1 pathway in infiltrated macrophage. Free Radic. Biol. Med. 2024, 10, 130–145. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Ye, Z.W.; Chu, N. Host PFKFB3-dependent glycolytic reprogramming as a broad-spectrum antiviral strategy. Open Forum Infect. Dis. 2023, 10 Suppl. 2, ofad500.976.
- Klarer, A.C.; O'Neal, J.; Imbert-Fernandez, Y.; Clem, A.; Ellis, S.R.; Clark, J.; Clem, B.; Chesney, J.; Telang, S. Inhibition of 6-phosphofructo-2-kinase (PFKFB3) induces autophagy as a survival mechanism. Cancer Metab. 2014, 2, 2. [Google Scholar] [CrossRef]
- Clem, B.F.; O'Neal, J.; Tapolsky, G.; Clem, A.L.; Imbert-Fernandez, Y.; Kerr, D.A. 2nd.; Klarer, A.C.; Redman, R.; Miller, D.M.; Trent, J.O.; Telang, S.; Chesney, J. Targeting 6-phosphofructo-2-kinase (PFKFB3) as a therapeutic strategy against cancer. Mol. Cancer Ther. 2013, 12, 1461–1470. [CrossRef]
- Boyd, S.; Brookfield, J.L.; Critchlow, S.E.; Cumming, I.A.; Curtis, N.J.; Debreczeni, J.; Degorce, S.L.; Donald, C.; Evans, N.J.; Groombridge, S.; Hopcroft, P.; Jones, N.P.; Kettle, J.G.; Lamont, S.; Lewis, H.J.; MacFaull, P.; McLoughlin, S.B.; Rigoreau, L.J.; Smith, J.M.; St-Gallay, S.; Stock, J.K.; Turnbull, A.P.; Wheatley, E.R.; Winter, J.; Wingfield, J. Structure-based design of potent and selective inhibitors of the metabolic kinase PFKFB3. J. Med. Chem. 2015, 58, 3611–3625. [Google Scholar] [CrossRef]
- Xu, X.; Zhao, Y.; Zhu, Z.; Wen, W.; Li, X. Mitofusin-mediated mitochondrial fusion inhibits Pseudorabies virus infection in porcine cells. Vet. Sci. 2025, 12, 368. [Google Scholar] [CrossRef]
- Gregorczyk-Zboroch, K.; Szulc-Dąbrowska, L.; Pruchniak, P.; Gieryńska, M.; Mielcarska, M.B.; Biernacka, Z.; Wyżewski, Z.; Lasocka, I.; Świtlik, W.; Szepietowska, A.; Kukier, P.; Kwiecień-Dębska, A.; Kłęk, J. Modifications of mitochondrial network morphology affect the MAVS-dependent immune response in L929 murine fibroblasts during Ectromelia virus infection. Pathogens 2024, 13, 717. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, X.; Ni, L.; Di, X.; Ma, B.; Niu, S.; Liu, C.; Reiter, R.J. COVID-19: Melatonin as a potential adjuvant treatment. Life Sci. 2020, 250, 117583. [Google Scholar] [CrossRef]
- Gutierrez-Mariscal, F.M.; Arenas-de Larriva, A.P.; Limia-Perez, L.; Romero-Cabrera, J.L.; Yubero-Serrano, E.M.; López-Miranda, J. Coenzyme Q10 supplementation for the reduction of oxidative stress: Clinical implications in the treatment of chronic diseases. Int. J. Mol. Sci. 2020, 21, 7870. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gutierrez-Mariscal, F.M.; Yubero-Serrano, E.M.; Villalba, J.M.; Lopez-Miranda, J. Coenzyme Q10: From bench to clinic in aging diseases, a translational review. Crit. Rev. Food Sci. Nutr. 2019, 59, 2240–2257. [Google Scholar] [CrossRef]
- Shikama, Y.; Otsuka, K.; Shikama, Y.; Furukawa, M.; Ishimaru, N.; Matsushita, K. Involvement of metformin and aging in salivary expression of ACE2 and TMPRSS2. Biofactors 2025, 51, e2154. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Shi, S.; Sun, H.; Zhou, L.; Wang, H.; Li, Y.; Gilson, E.; Lu, Y.; Hu, L.; Ye, J. Metformin alleviates inflammatory response and severity rate of COVID-19 infection in elderly individuals. Sci. Rep. 2025, 15, 11340. [Google Scholar] [CrossRef] [PubMed]
- Rocha, M.; Hernandez-Mijares, A.; Garcia-Malpartida, K.; Bañuls, C.; Bellod, L.; Victor, V.M. Mitochondria-targeted antioxidant peptides. Curr. Pharm. Des. 2010, 16, 3124–3131. [Google Scholar] [CrossRef]
- Gasanoff, E.S.; Yaguzhinsky, L. .; Garab, G. Cardiolipin, non-bilayer structures and mitochondrial bioenergetics: Relevance to cardiovascular disease. Cells 2021, 10, 1721. [Google Scholar] [CrossRef]
- Ding, X.W.; Robinson, M.; Li, R.; Aldhowayan, H.; Geetha, T.; Babu, J.R. Mitochondrial dysfunction and beneficial effects of mitochondria-targeted small peptide SS-31 in Diabetes Mellitus and Alzheimer's disease. Pharmacol. Res. 2021, 171, 105783. [Google Scholar] [CrossRef]
- Li, X.; Ma, L.; Fu, P. The mitochondrion-targeted antioxidants in kidney disease. Curr. Med. Chem. 2021, 28, 4190–4206. [Google Scholar] [CrossRef]
- Oliver, D.M.A.; Reddy, P.H. Small molecules as therapeutic drugs for Alzheimer's disease. Mol. Cell. Neurosci. 2019, 96, 47–62. [Google Scholar] [CrossRef]
- Jantz-Naeem, N.; Guvencli, N.; Böttcher-Loschinski, R.; Böttcher, M.; Mougiakakos, D.; Kahlfuss, S. Metabolic T-cell phenotypes: from bioenergetics to function. Am. J. Physiol. Cell Physiol. 2025, 328, C1062–C1075. [Google Scholar] [CrossRef]
- Official Study Title: NIRVANA: NIcotinamide Riboside in SARSCoV-2 pAtients for reNAl protection. Available online: https://cdn.clinicaltrials.gov/large-docs/16/NCT04818216/Prot_SAP_002.pdf (accessed on 15 May 2025).
- Naidu, A.S.; Wang, C.K.; Rao, P.; Mancini, F.; Clemens, R.A.; Wirakartakusumah, A.; Chiu, H.F.; Yen, C.H.; Porretta, S.; Mathai, I.; Naidu, S.A.G. Precision nutrition to reset virus-induced human metabolic reprogramming and dysregulation (HMRD) in long-COVID. NPJ Sci. Food. 2024, 8, 19. [Google Scholar] [CrossRef]
- Assimakopoulos, S.F.; Aretha, D.; Komninos, D.; Dimitropoulou, D.; Lagadinou, M.; Leonidou, L.; Oikonomou, I.; Mouzaki, A.; Marangos, M. N-acetyl-cysteine reduces the risk for mechanical ventilation and mortality in patients with COVID-19 pneumonia: a two-center retrospective cohort study. Infect. Dis. (Lond). 2021, 53, 847–854. [Google Scholar] [CrossRef]
- Ibrahim, H.; Perl, A.; Smith, D.; Lewis, T.; Kon, Z.; Goldenberg, R.; Yarta, K.; Staniloae, C.; Williams, M. Therapeutic blockade of inflammation in severe COVID-19 infection with intravenous N-acetylcysteine. Clin. Immunol. 2020, 219, 108544. [Google Scholar] [CrossRef]
- Geiler, J.; Michaelis, M.; Naczk, P.; Leutz, A.; Langer, K.; Doerr, H.W.; Cinatl, J. Jr. N-acetyl-L-cysteine (NAC) inhibits virus replication and expression of pro-inflammatory molecules in A549 cells infected with highly pathogenic H5N1 influenza A virus. Biochem. Pharmacol. 2010, 79, 413–420. [Google Scholar] [CrossRef] [PubMed]
- De Flora, S.; Grassi, C.; Carati, L. Attenuation of influenza-like symptomatology and improvement of cell-mediated immunity with long-term N-acetylcysteine treatment. Eur. Respir. J. 1997, 10, 1535–1541. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.Y.; Ye, J.J.; Zhang, D.W.; Hu, L.; Wu, H.M.; Fei, G.H. Melatonin rescues Influenza A virus-induced cellular energy exhaustion via OMA1-OPA1-S in acute exacerbation of COPD. J. Pineal Res. 2024, 76, e12991. [Google Scholar] [CrossRef]
- Xu, M.M.; Kang, J.Y.; Wang, Q.Y.; Zuo, X.; Tan, Y.Y.; Wei, Y.Y.; Zhang, D.W.; Zhang, L.; Wu, H.M.; Fei, G.H. Melatonin improves influenza virus infection-induced acute exacerbation of COPD by suppressing macrophage M1 polarization and apoptosis. Respir. Res. 2024, 25, 186. [Google Scholar] [CrossRef]
- Huang, Y.; Jiang, C.; Liu, X.; Tang, W.; Gui, H.; Sun, T.; Xu, D.; He, M.; Han, M.; Qiu, H.; Chen, M.; Huang, S. Melatonin suppresses TLR4-mediated RSV infection in the central nervous cells by inhibiting NLRP3 inflammasome formation and autophagy. J. Cell. Mol. Med. 2024, 28, e18338. [Google Scholar] [CrossRef]
- Qin, J.; Wang, G.; Han, D. Benefits of melatonin on mortality in severe-to-critical COVID-19 patients: A systematic review and meta-analysis of randomized controlled trials. Clinics (Sao Paulo) 2025, 80, 100638. [CrossRef]
- Kakad, U.U.; Khopkar-Kale, P.S.; Tripathy, S.P.; Bhawalkar, J.S. Potential of melatonin as a treatment option for long COVID: A call for research. Br. J. Clin. Pharmacol. 2025, 91, 493–494. [Google Scholar] [CrossRef]
- Tirkan, A.; Eskandari, D.; Roham, M.; Aloosh, O.; Ramim, T.; Afshar, H. Investigating the effectiveness of melatonin in the treatment of critically ill patients with COVID-19 hospitalized in the Intensive Care Unit: A double-blind randomized clinical trial. Med. J. Islam. Repub. Iran 2024, 38, 41. [Google Scholar] [CrossRef]
- Bahmyari, R.; Zare, M.; Sharma, R.; Agarwal, A.; Halvaei, I. The efficacy of antioxidants in sperm parameters and production of reactive oxygen species levels during the freeze-thaw process: A systematic review and meta-analysis. Andrologia 2020, 52, e13514. [Google Scholar] [CrossRef]
- Maio, N.; Cherry, S.; Schultz, D.C.; Hurst, B.L.; Linehan, W.M.; Rouault, T.A. TEMPOL inhibits SARS-CoV-2 replication and development of lung disease in the Syrian hamster model. iScience 2022, 25, 105074. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; Duan, Y.; Yu, J.; Wang, L.; Yang, K.; Liu, F.; Jiang, R.; Yang, X.; You, T.; Liu, X.; Yang, X.; Bai, F.; Liu, H.; Liu, X.; Guddat, L.W.; Xu, W.; Xiao, G.; Qin, C.; Shi, Z.; Jiang, H.; Rao, Z.; Yang, H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef]
- Gouédard, C.; Barouki, R.; Morel, Y. Dietary polyphenols increase paraoxonase 1 gene expression by an aryl hydrocarbon receptor-dependent mechanism. Mol. Cell. Biol. 2004, 24, 5209–5222. [Google Scholar] [CrossRef] [PubMed]
- Camps, J.; Marsillach, J.; Joven, J. Pharmacological and lifestyle factors modulating serum paraoxonase-1 activity. Mini Rev. Med. Chem. 2009, 9, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Mellor, A.L.; Munn, D.H. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat. Rev. Immunol. 2004, 4, 762–774. [Google Scholar] [CrossRef] [PubMed]
- Rothhammer, V.; Quintana, F.J. The aryl hydrocarbon receptor: an environmental sensor integrating immune responses in health and disease. Nat. Rev Immunol. 2019, 19, 184–197. [Google Scholar] [CrossRef] [PubMed]
- Cicin, I.; Plimack, E.R.; Gurney, H.; Leibowitz, R.; Alekseev, B.Y.; Parnis, F.X.; Peer, A.; Necchi, A.; Bellmunt, J.; Nishiyam, H.; Clark, J.; Munteanu, M.; Kataria, R.; Jia, C.; Powles, T.; Sternberg, C.N. Epacadostat plus pembrolizumab versus placebo plus pembrolizumab for advanced urothelial carcinoma: results from the randomized phase III ECHO-303/KEYNOTE-698 study. BMC Cancer 2024, 23 (Suppl 1), 1256. [Google Scholar] [CrossRef]
- Lara, P.N. Jr.; Villanueva, L.; Ibanez, C.; Erman, M.; Lee, J.L.; Heinrich, D.; Lipatov, O.N.; Gedye, C.; Gokmen, E.; Acevedo, A.; Semenov, A.; Park, S.H.; Gafanov, R.A.; Kose, F.; Jones, M.; Du, X.; Munteanu, M.; Perini, R.; Choueiri, T.K.; Motzer, R.J. A randomized, open-label, phase 3 trial of pembrolizumab plus epacadostat versus sunitinib or pazopanib as first-line treatment for metastatic renal cell carcinoma (KEYNOTE-679/ECHO-302). BMC Cancer 2024, 23 (Suppl 1), 1253. [Google Scholar] [CrossRef]
- Chen, F.; Zhao, D.; Huang, Y.; Wen, X.; Feng, S. Synergetic impact of combined navoximod with cisplatin mitigates chemo-immune resistance via blockading IDO1+ CAFs-secreted Kyn/AhR/IL-6 and pol ζ-prevented CIN in human oral squamous cell carcinoma. Life Sci. 2023, 335, 122239. [Google Scholar] [CrossRef]
- Guillemin, G.J.; Brew, B.J. Implications of the kynurenine pathway and quinolinic acid in Alzheimer's disease. Redox Rep. 2002, 7, 199–206. [Google Scholar] [CrossRef]
- Låg, M.; Skuland, T.; Ballangby, J.; Grytting, V.S.; Jørgensen, R.B.; Snilsberg, B.; Øvrevik, J.; Holme, J.A.; Refsnes, M. Mechanisms involved in pro-inflammatory responses to traffic-derived particulate matter (PM) in THP-1 macrophages compared to HBEC3-KT bronchial epithelial cells. Toxicology 2025, 516, 154174. [Google Scholar] [CrossRef]
- Bogan, K.L.; Brenner, C. Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu. Rev. Nutr. 2008, 28, 115–130. [Google Scholar] [CrossRef]
- Agus, A.; Planchais, J.; Sokol, H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef] [PubMed]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Overmyer, K.A.; Shishkova, E.; Miller, I.J.; Balnis, J.; Bernstein, M.N.; Peters-Clarke, T.M.; Meyer, J.G.; Quan, Q.; Muehlbauer, L.K.; Trujillo, E.A.; He, Y.; Chopra, A.; Chieng, H.C.; Tiwari, A.; Judson, M.A.; Paulson, B.; Brademan, D.R.; Zhu, Y.; Serrano, L.R.; Linke, V.; Drake, L.A.; Adam, A.P.; Schwartz, B.S.; Singer, H.A.; Swanson, S.; Mosher, D.F.; Stewart, R.; Coon, J.J.; Jaitovich, A. Large-scale multi-omic analysis of COVID-19 severity. Cell. Syst. 2021, 12, 23–40.e7. [CrossRef]
- Grimes, J.M.; Khan, S.; Badeaux, M.; Rao, R.M.; Rowlinson, S.W.; Carvajal, R.D. Arginine depletion as a therapeutic approach for patients with COVID-19. Int. J. Infect. Dis. 2021, 102, 566–570. [Google Scholar] [CrossRef] [PubMed]
- Greene, K.S.; Choi, A.; Chen, M.; Yang, N.; Li, R.; Qiu, Y.; Lukey, M.J.; Rojas, K.S.; Shen, J.; Wilson, K.F.; Katt, W.P.; Whittaker, G.R.; Cerione, R.A. Inhibiting glutamine metabolism blocks coronavirus replication in mammalian cells. bioRxiv [Preprint], 2023, 28, 2023.09.27.559756. [CrossRef]
- Rodríguez-Vera, D.; Salazar, J.R.; Soriano-Ursúa, M.A.; Guzmán-Pérez, J.; Vergara-Castañeda, A.; Muñoz-Durán, H.; Ramírez-Velez, G.L.; Vivar-Sierra, A.; Naranjo-Navarro, C.R.; Meza-Meneses, P.A.; Loza-Mejía, M.A.; Pinto-Almazán, R. Effectiveness of omega-3 fatty acid supplementation in improving the metabolic and inflammatory profiles of Mexican adults hospitalized with COVID-19. Diseases 2024, 12, 28. [Google Scholar] [CrossRef] [PubMed]
- Safaei Ardestani, S.Z.; Rahideh, S.T. The effect of omega-3 fatty acid supplementation on clinical and biochemical parameters of critically ill patients with COVID-19: a randomized clinical trial. J. Transl. Med. 2022, 20, 32. [Google Scholar] [CrossRef]
- Wang, K.; Khoramjoo, M.; Srinivasan, K.; Gordon, P.M.K.; Mandal, R.; Jackson, D.; Sligl, W.; Grant, M.B.; Penninger, J.M.; Borchers, C.H.; Wishart, D.S.; Prasad, V.; Oudit, G.Y. Sequential multi-omics analysis identifies clinical phenotypes and predictive biomarkers for long COVID. Cell. Rep. Med. 2023, 4, 101254. [Google Scholar] [CrossRef]
- Pinero, S.; Li, X.; Liu, L.; Li, J.; Lee, S.H.; Winter, M.; Nguyen, T.; Zhang, J.; Le, T.D. Integrative multi-omics framework for causal gene discovery in long COVID. medRxiv, 2025, 2025.02.09.25321751. [CrossRef]
- Centers for Disease Control and Prevention. Influenza severity assessment and estimated influenza illnesses, medical visits, hospitalizations, and deaths that occurred and those that were prevented by vaccination in the United States – 2023-2024 influenza season. https://www.cdc.gov/flu/whats-new/flu-summary-addendum-2023-2024.html (accessed 16 May 2025).
- Havers, F.P.; Whitaker, M.; Melgar, M.; Chatwani, B.; Chai, S.J.; Alden, N.B.; Meek, J.; Openo, K.P.; Ryan, P.A.; Kim, S.; Lynfield, R.; Shaw, Y.P.; Barney, G.; Tesini, B.L.; Sutton, M.; Talbot, H.K.; Olsen, K.P.; Patton, M.E.; RSV-NET Surveillance Team. Characteristics and outcomes among adults aged ≥60 years hospitalized with laboratory-confirmed respiratory syncytial virus - RSV-NET, 12 States, July 2022-June 2023. MMWR Morb. Mortal. Wkly. Rep. 2023, 72, 1075–1082. [CrossRef]
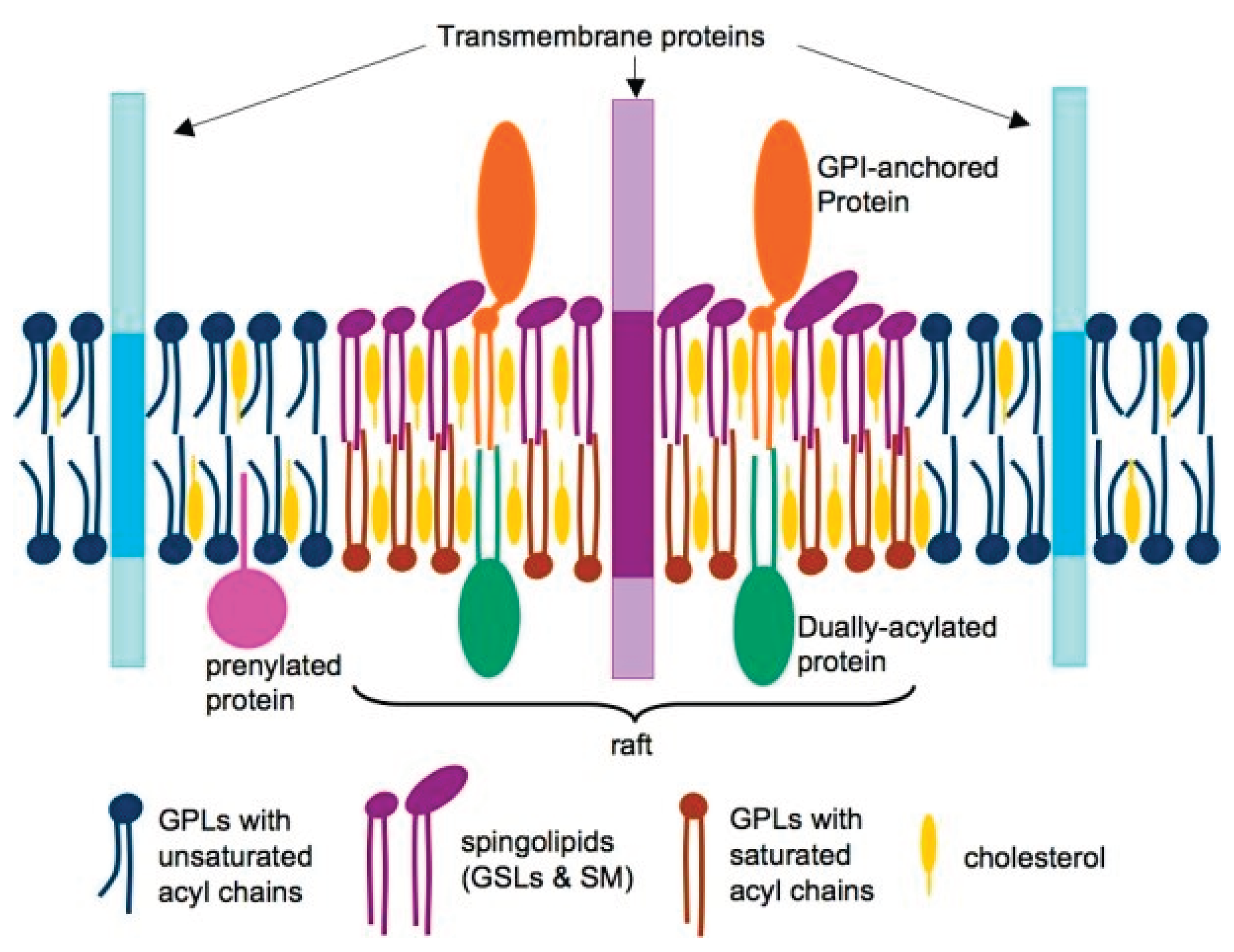
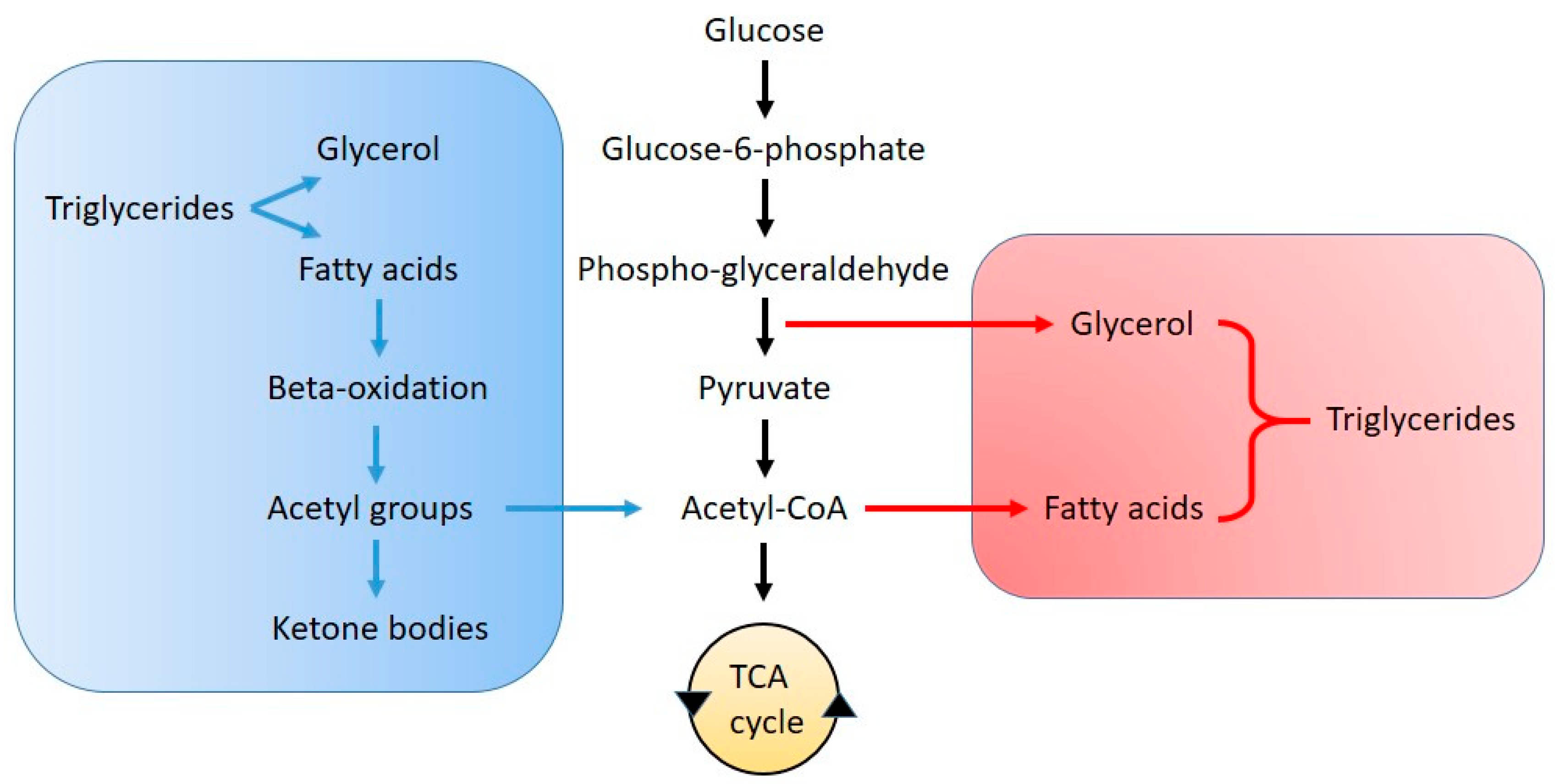
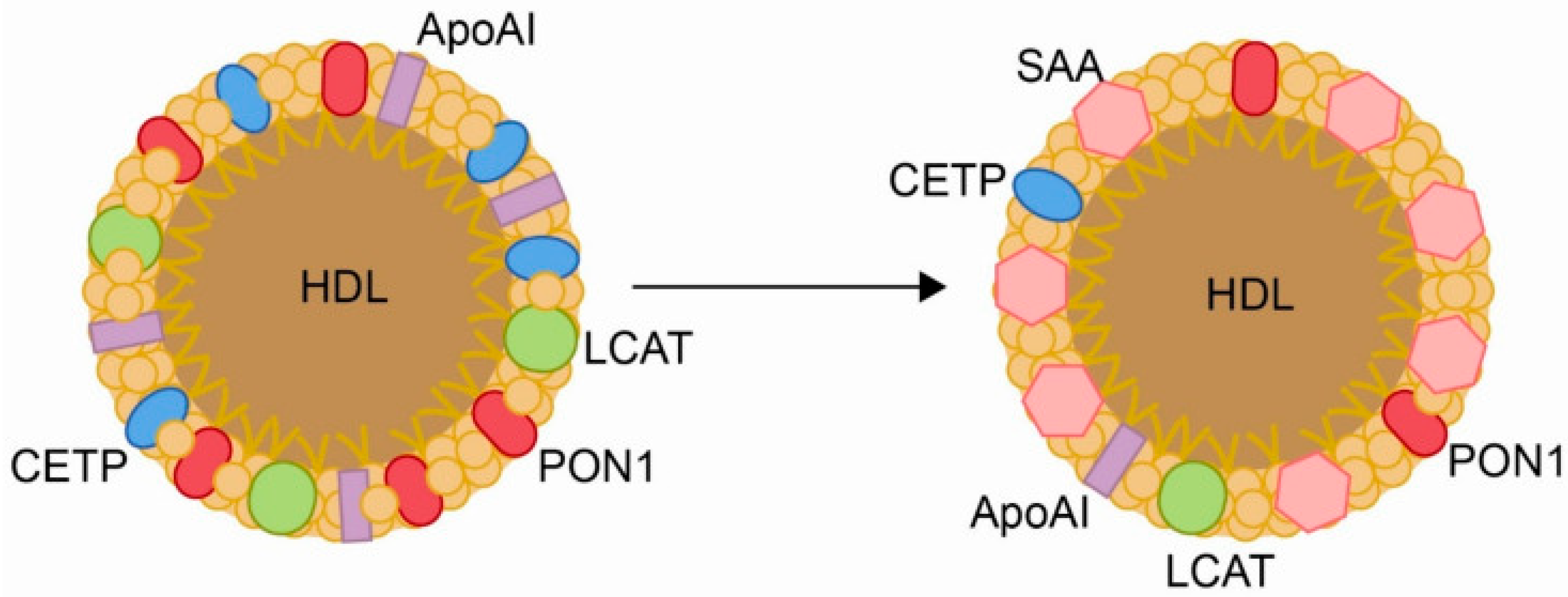
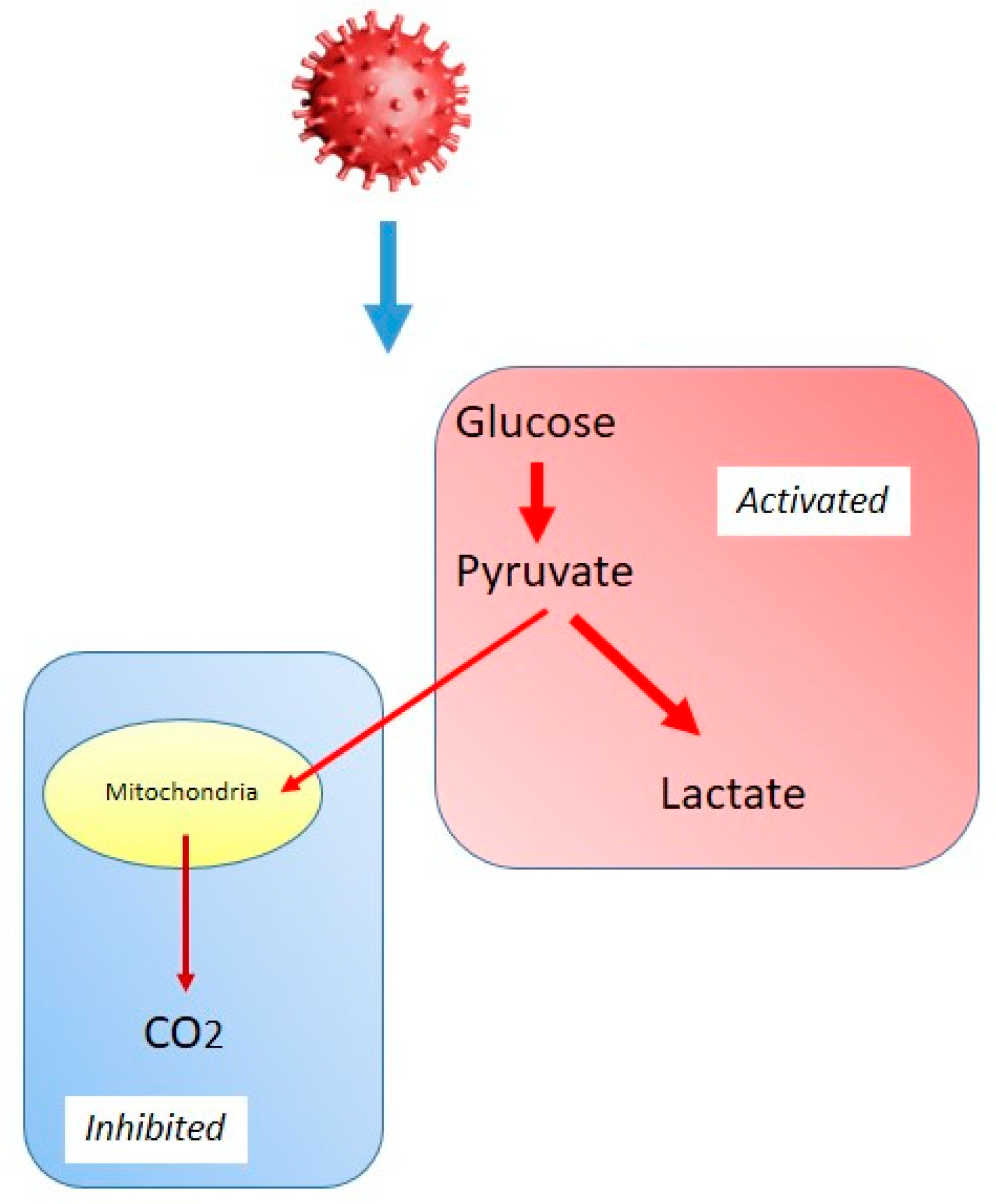
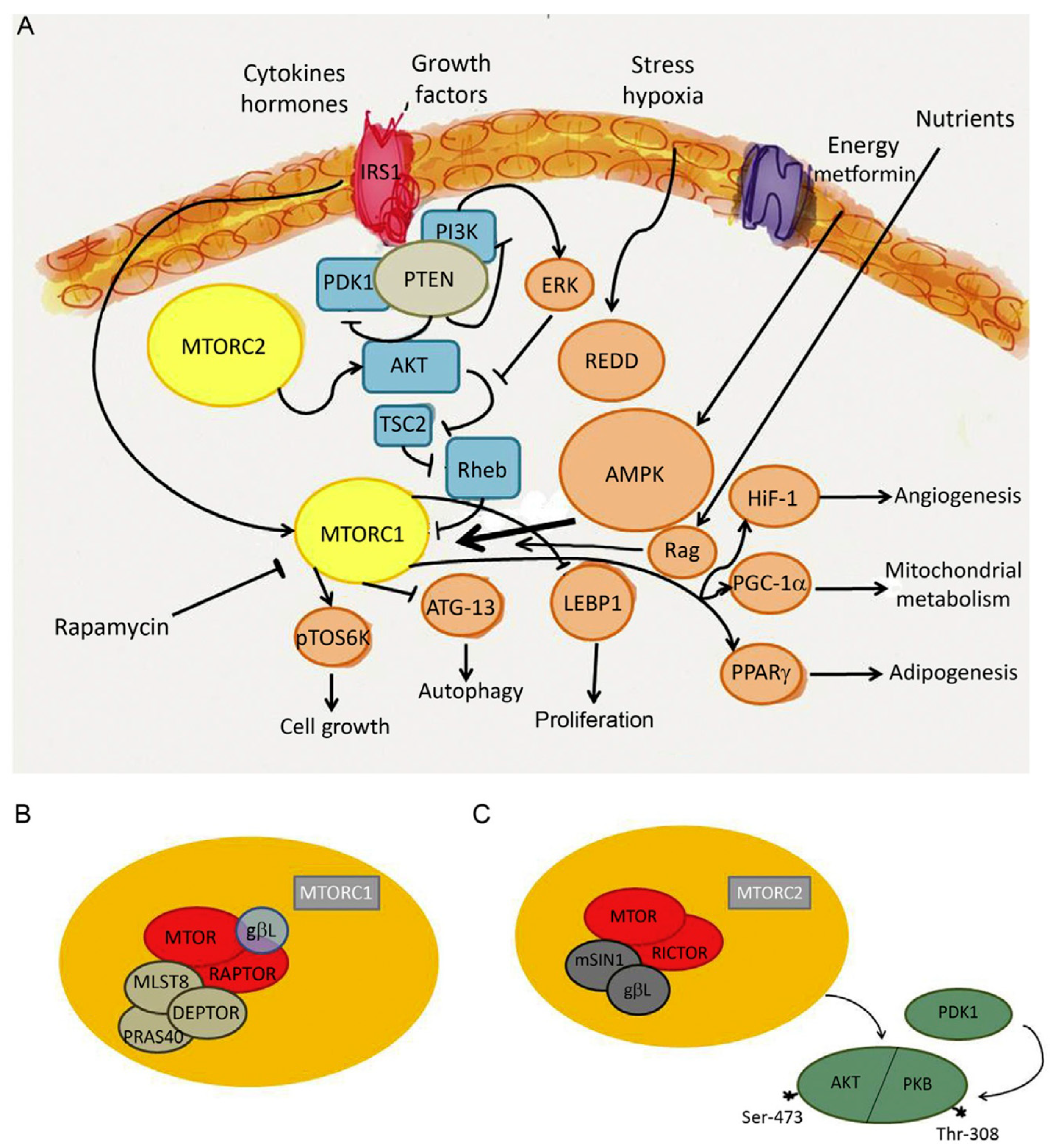
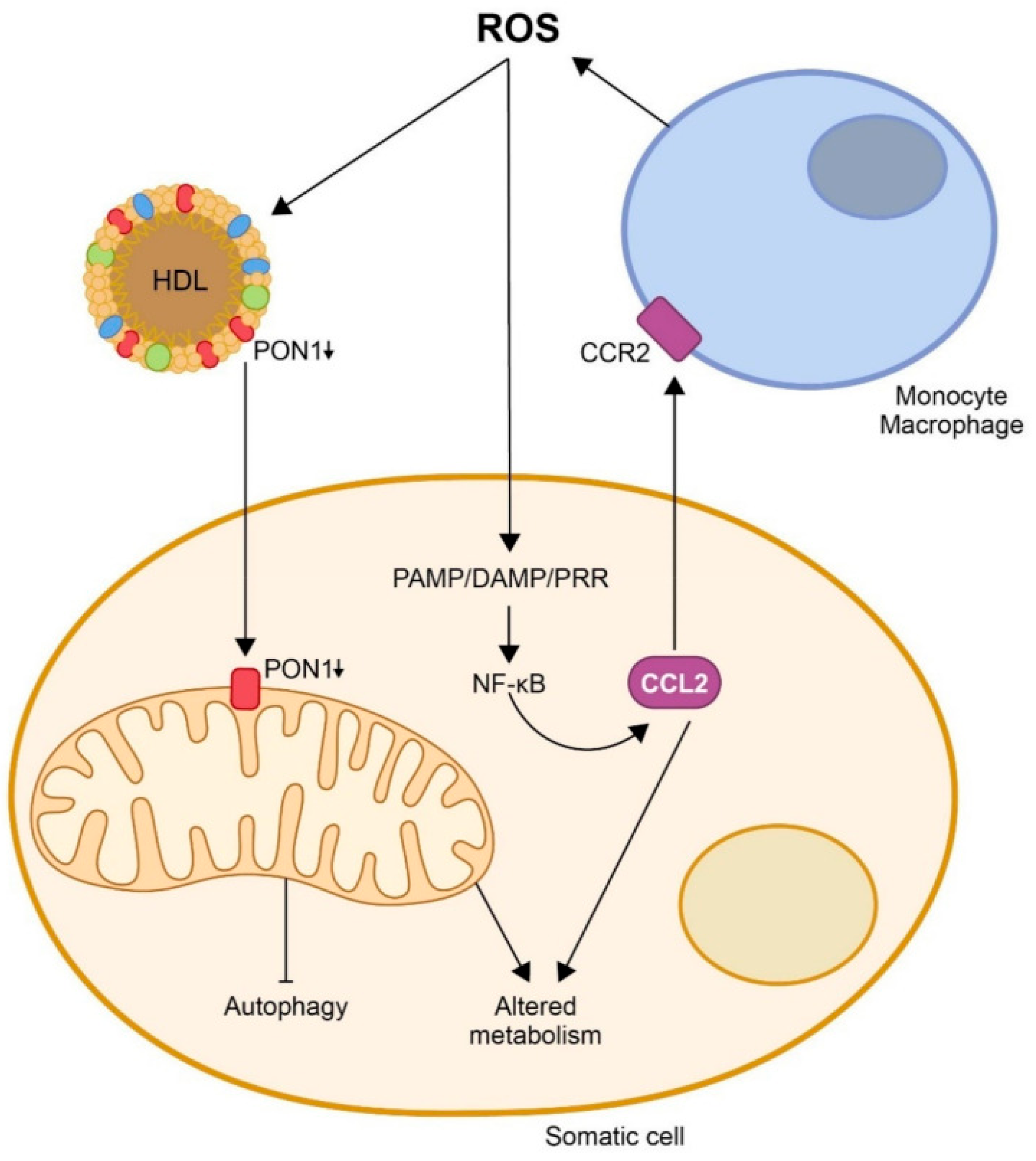
| Feature | SARS-CoV-2 | Influenza virus | RSV |
| Lipid rafts in viral entry | Spike protein binding to ACE2 and entry. | Mainly clathrin-dependent endocytosis involving lipid rafts. | May bind to heparan sulfate and use rafts for attachment and fusion. |
| Fatty acid synthesis | Increased FASN | Increased FASN | Increased FASN |
| LD involvement | Induces LD accumulation for immune evasion and viral assembly. | Depletes or degrades LD via mTOR activation to fuel viral replication; inhibits LD biogenesis. | Disperses and reduces LD content; mechanisms less well characterized, but likely interferes with LD-associated immune signaling. |
| β-oxidation alterations | Inhibits β-oxidation, causing lipid accumulation. | Suppresses β-oxidation; shifts toward lipogenesis. | Mitochondrial dysfunction affects β-oxidation; less studied. |
| Cholesterol dependency | Cholesterol metabolism dysregulation. | Cholesterol metabolism dysregulation. | Not well characterized. |
| Lipoprotein metabolism | Decreased HDL; increased VLDL and LDL. | Transient decreased HDL and increased LDL. | Not well characterized. |
| Feature | SARS-CoV-2 | Influenza virus | RSV |
| Glycolysis and OXPHOS | Increased glycolysis (Warburg-like shift); decreased OXPHOS. |
Increased glycolysis (Warburg-like shift); decreased OXPHOS. |
Increased glycolysis (Warburg-like shift); decreased OXPHOS. |
| Mitochondrial dysfunction |
Altered morphology, decreased function; increased ROS; inhibited MAVS; activated NLRP3. |
Altered morphology, decreased function; increased ROS; activated NLRP3. |
Altered morphology, decreased function; increased ROS. |
| Bioenergetic failure | Decreased ATP production; impaired mitochondrial respiration; shift to anaerobic metabolism. | Decreased ATP production; impaired mitochondrial respiration; shift to anaerobic metabolism. | Decreased ATP production; impaired mitochondrial respiration; shift to anaerobic metabolism. |
| Cellular metabolic sensors |
HIF-1α activation; AMPK inhibition; altered mTOR and PI3K–Akt signaling. | HIF-1α and PI3K–Akt–mTOR axis activation; energy reprogramming. | mTOR and PI3K–Akt upregulation. |
| Pathway | SARS-CoV-2 | Influenza virus | RSV |
| Glutamine/glutamate | Increased glutaminolysis | Increased glutaminolysis | Increased glutaminolysis |
| Arginine metabolism | Arginine depletion; arginase upregulation; decreased NO production. | Arginine depletion; arginase upregulation; decreased NO production. | Arginine depletion; arginase upregulation; decreased NO production. |
| Tryptophan–kynurenine | Strongly activated; increased IDO1 activity; correlates with severity | Moderate activation; role in immune modulation | Upregulated IDO1 in severe cases; less characterized |
| Cysteine metabolism | Decreased cysteine availability. | Decreased cysteine availability. | Decreased cysteine availability. |
| Nucleotide metabolism | Increased biosynthesis and salvage pathways | Increased biosynthesis and salvage pathways | Increased biosynthesis and salvage pathways |
| Pathway | SARS-CoV-2 | Influenza virus | RSV |
| Glutathione | Increased glutahione oxidation. | Transient increased glutahione oxidation. | Increased glutahione oxidation. |
| Superoxide dismutase | Decreased activity in plasma and tissues; associated with disease severity. | Variable: Decreased activity in severe disease; oxidative burden overwhelms the enzyme. | Variable: Decreased activity in severe disease; oxidative burden overwhelms the enzyme. |
| Catalase | Decreased activity. | Decreased activity. | Decreased activity. |
| Paraoxonase 1 | Markedly decreased activity. | Less characterized. | Less characterized. |
| Metabolic pathway | Proposed drug | Targeted virus |
| Viral entry | Camostat mesylate Statins Chloroquine |
SARS-CoV-2 Influenza SARS-CoV-2 |
| Lipid metabolism | Orlistat Statins Omega-3 fatty acids |
SARS-CoV-2, Influenza, RSV SARS-CoV-2 SARS-CoV-2 |
| Glycolysis | 2-deoxy-D-glucose KAN0438757 3PO |
SARS-CoV-2 Influenza Influenza |
| Mitochondrial dynamics Bioenergetics |
Mdivi 1 Melatonin Coenzyme Q10 Metformin Szeto-Schiller peptides |
Other viruses SARS-CoV-2, Influenza, RSV Not tested on viruses SARS-CoV-2 Not tested on viruses |
| Endogenous antioxidants | N-acetylcysteine Melatonin Tempol Ebselen |
SARS-CoV-2, Influenza SARS-CoV-2, Influenza, RSV SARS-CoV-2 SARS-CoV-2 |
| Kynurenine pathway | Epacadostat, navoximod | Not tested on viruses |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





