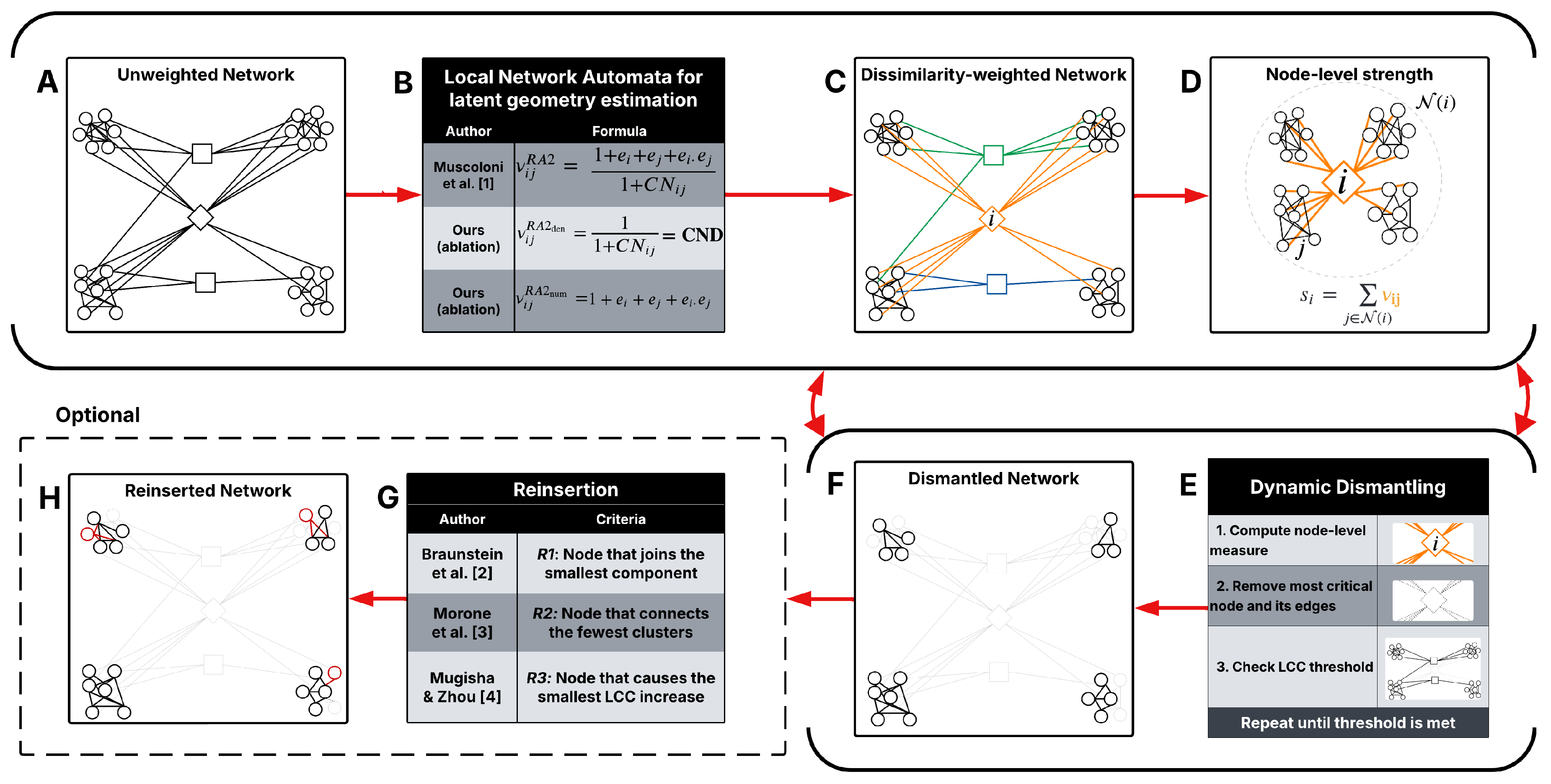
Appendix A. Latent Geometry Estimators
Table A1.
Comparison of latent geometry estimators and their variants. is the weight of the link between nodes i and j; and denote the number of external links of nodes i and j, respectively; is the number of common neighbors shared by i and j. Information Locality denotes the type of structural information required to assign a score to each node for dismantling. Time Complexity denotes the time complexity for dynamic dismantling using each estimator on sparse graphs, without reinsertion. N: number of nodes. m: number of links.
Table A1.
Comparison of latent geometry estimators and their variants. is the weight of the link between nodes i and j; and denote the number of external links of nodes i and j, respectively; is the number of common neighbors shared by i and j. Information Locality denotes the type of structural information required to assign a score to each node for dismantling. Time Complexity denotes the time complexity for dynamic dismantling using each estimator on sparse graphs, without reinsertion. N: number of nodes. m: number of links.
| Estimator |
Author |
Year |
Formula |
Information Locality |
Time Complexity |
| Repulsion Attraction 2 |
Muscoloni et al. [1] |
2017 |
|
Local |
|
| RA2 denominator-ablation (CND) |
Ours |
2025 |
|
Local |
|
| RA2 numerator-ablation |
Ours |
2025 |
|
Local |
|
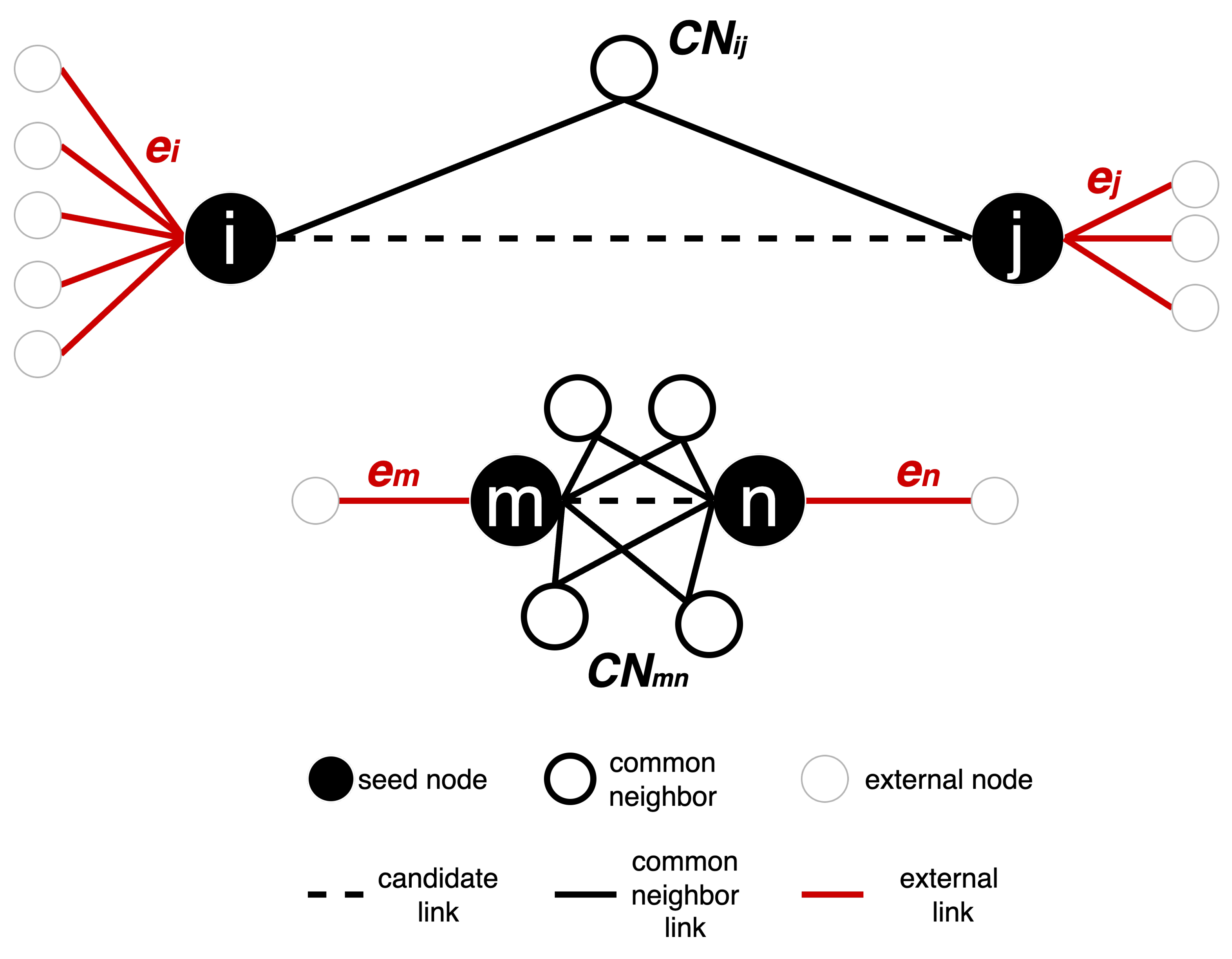
Figure A1.
Illustration of how RA2 measures are computed on two toy networks. Seed nodes are shown in black; common neighbors (CN) are shown in white with a black border, and external nodes are white with a grey border. The dashed line is the edge that is being assigned a weight. External links e denote the number of edges connecting a node to nodes outside its CN set, here in red. In black, the links to common neighbors. For the link in the top network, , , and . For the link in the bottom network, , , and .
Figure A1.
Illustration of how RA2 measures are computed on two toy networks. Seed nodes are shown in black; common neighbors (CN) are shown in white with a black border, and external nodes are white with a grey border. The dashed line is the edge that is being assigned a weight. External links e denote the number of edges connecting a node to nodes outside its CN set, here in red. In black, the links to common neighbors. For the link in the top network, , , and . For the link in the bottom network, , , and .
Figure A1 illustrates how RA2-based network automata rules assign edge weights by estimating geometric distances using only local topological features. The two toy subnetworks demonstrate how the RA2 rule and its variants distinguish between geometrically distant and close node pairs. In the top subnetwork, nodes
i and
j have only one common neighbor and are each connected to many external nodes (
,
), indicating a weak integration in a local community and stronger connectivity to distinct parts of the network. According to the Repulsion-Attraction rule, this suggests a larger latent distance due to high repulsion and low attraction. In contrast, in the bottom subnetwork, nodes
m and
n share four common neighbors and have only one external link each (
,
). This pattern indicates a stronger local community and a higher likelihood that the nodes are geometrically close in the latent space, with a lower dissimilarity score. These examples highlight how latent geometry-driven RA2-based network automata rules estimate hidden distances: fewer common neighbors and more external links suggest geometrical separation, while many common neighbors and few external links imply proximity in the latent manifold.
In geometric networks, nearby nodes form dense, closed neighborhoods: they share many common neighbors (high ) and have few “external” links (small e). Distant node pairs show the opposite pattern. The Repulsion-Attraction rule 2 (RA2) captures these patterns in their formulation: any RA variant that decreases with and increases with external connectivity, e is therefore monotonic with latent distance: small values indicate proximity; large values indicate separation. Crucially, this relies on topological proximity, not a specific geometric space, so it applies across hyperbolic, Euclidean, or elliptic latent geometries. The only assumption to apply this estimator of underlying geometry is that the topology displays:
node heterogeneity (meaning that the node degree distribution displays a standard deviation different from zero).
homophily (similar nodes link together), for instance geometric proximity in latent space causes nodes that are geometrically close have overlapping neighborhoods.
We can aggregate RA2 to score node criticality by summing its pairwise RA2 to neighbors. This turns local edge-level “distance” into a node-level bridging load:
Few adjacent nodes (neighbors) with mostly short links yields a small RA2. This node is peripheral and non-critical; removal has little global effect.
Many neighbors with mostly short links still yields a modest RA2 (short links contribute little). The node is locally redundant; removal is buffered by community structure.
Many neighbors with a mix of short and long links yields a large RA2 because long links carry high RA2. The node simultaneously anchors a local community and bridges distant regions; removing it is likely to disconnect communities and degrade global connectivity.
Few neighbors with many long links (rare under geometric attachment) still yields a large RA2; such nodes are likewise critical inter-community hubs.
As a result, RA2 encodes latent separation from purely local topology. Summing RA2 over a node’s incident edges ranks nodes by how much long-range connectivity they support. Dismantling the highest-scoring nodes precisely targets those bridges whose removal most effectively fragments the network.
We analyze the time complexity for the full dynamic dismantling process (excluding reinsertion) for the latent geometry-driven network automata rules in
Table A1, where dynamic means recomputing the dismantling measure after each node removal. For RA2 and its variants, the dominant operation is the computation of the common neighbor (CN) matrix. This operation has a time complexity of
for dense graphs and
for sparse graphs, where
N is the number of nodes and
m is the number of links. Assuming
N dismantling steps in the worst-case scenario, the overall time complexity becomes
for sparse graphs. The assumption of
N dismantling steps applies to all the time complexity analyses of dynamic dismantling methods.
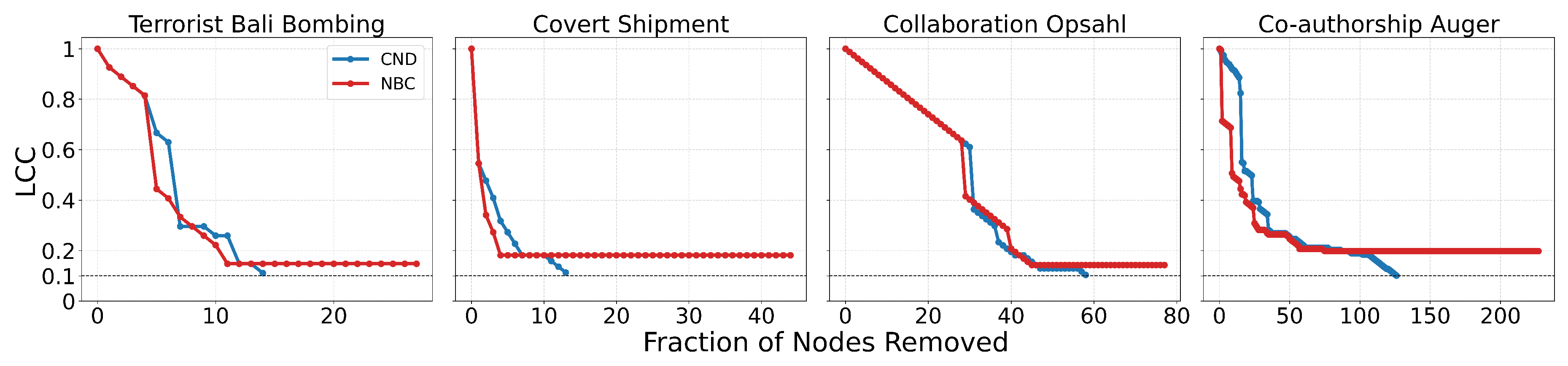
Figure A2.
Dynamic dismantling process on example networks comparing local network automata rule RA2 and its variants versus NBC, when the former outperforms NBC in terms of AUC. The plot shows the normalized size of the largest connected component (LCC) as a function of the fraction of nodes removed, with a target LCC threshold of 10%. The final evaluation metric is the Area Under the Curve (AUC) of the LCC trajectory.
Figure A2.
Dynamic dismantling process on example networks comparing local network automata rule RA2 and its variants versus NBC, when the former outperforms NBC in terms of AUC. The plot shows the normalized size of the largest connected component (LCC) as a function of the fraction of nodes removed, with a target LCC threshold of 10%. The final evaluation metric is the Area Under the Curve (AUC) of the LCC trajectory.
Appendix B. Topological Centrality Measures
Table A2.
Comparison of topological centrality measures and the associated time complexity for dynamic dismantling using each centrality measure. Information Locality denotes the type of structural information required to assign a score to each node. Time Complexity denotes the time complexity for dynamic dismantling using each centrality measure on sparse graphs, without reinsertion. N: number of nodes. m: number of links.
Table A2.
Comparison of topological centrality measures and the associated time complexity for dynamic dismantling using each centrality measure. Information Locality denotes the type of structural information required to assign a score to each node. Time Complexity denotes the time complexity for dynamic dismantling using each centrality measure on sparse graphs, without reinsertion. N: number of nodes. m: number of links.
| Measure |
Author |
Year |
Type |
Information Locality |
Time Complexity |
| Degree |
|
|
Degree-based |
Local |
|
| Eigenvector |
Bonacich [40] |
1972 |
Walks-based |
Global |
|
| Node Betweenness (NBC) |
Freeman [21] |
1977 |
Shortest path-based |
Global |
|
| PageRank (PR) |
Page et al. [41] |
1999 |
Random walk-based |
Global |
|
| Resilience |
Zhang et al. [33] |
2020 |
Resilience-based |
Global |
|
| Domirank |
Engsig et al. [22] |
2024 |
Fitness-based |
Global |
|
| Fitness |
Servedio et al. [23] |
2025 |
Fitness-based |
Global |
|
We analyze the time complexity of dynamic dismantling (excluding reinsertion) for the topological centrality measures used in our experiments, summarized in
Table A2. As before, the analysis assumes
N dismantling steps in the worst-case scenario. For degree, the score update after each removal is local and can be done in
time using a binary heap. For NBC, we use Brandes’ algorithm [
45], which computes betweenness centrality in
time per step for unweighted networks. Eigenvector, PageRank, Resilience, Domirank, and Fitness all rely on matrix-vector multiplications, which has a time complexity of
. We also add the term
N, which represents the overhead of looping over nodes to update or normalize the resulting vector at each iteration. This leads to a total per-step cost of
. We also omit the constant
k for Eigenvector, PageRank, and Fitness centrality, which represents the number of iterations these methods perform. In practice, reaching full convergence to a single optimal solution is often computationally infeasible; this is why a fixed number of
k iterations is typically defined.
Appendix D. Reinsertion Methods
Reinsertion was originally introduced in the context of immunization as a reverse process: starting from a fully dismantled network, nodes are reinserted one by one, each time selecting the node whose addition causes the smallest increase in the largest connected component (LCC) [
46]. This reversed sequence then defines an effective dismantling order. In subsequent studies, reinsertion has been used as a post-processing step to improve dismantling outcomes [
6]: the network is first dismantled by a given method, and nodes are reinserted until the LCC reaches the dismantling threshold. This reduces the dismantling cost while preserving the original attack target.
In this work, dismantling cost is defined as the number of nodes removed from the network. The reinsertion step aims to directly minimize this cost by reintroducing nodes that were initially removed but found to be unnecessary for achieving the dismantling objective. It’s important to note that while reinsertion reduces the number of physical removals, it does introduce a higher computational cost as it’s a post-processing step performed after the initial dismantling. However, the primary objective is to minimize this physical intervention, as in many real-world scenarios, the logistical and financial implications of physically removing network components (e.g., infrastructure) far outweigh the computational resources expended during the optimization phase. This is why we compare all methods with and without the reinsertion step.
Several reinsertion criteria have been proposed: Braunstein et al. [
2] select the node that ends up in the smallest resulting component after reinsertion; Morone et al. [
3] choose the node that reconnects the fewest components; Mugisha and Zhou [
4] select the node that causes the smallest LCC increase. See
Table A4 for a full comparison.
Reinsertion can greatly enhance dismantling performance. However, recent work shows that this step can overpower the dismantling algorithm itself, allowing weak methods to appear effective when paired with reinsertion [
44]. To address this, we enforce two constraints to ensure fair comparisons and prevent reinsertion from dominating the dismantling process:
Reinsertion must stop once the LCC exceeds the dismantling threshold. Recomputing a new dismantling order by reinserting all nodes is not allowed.
Ties in the reinsertion criterion must be broken by reversing the dismantling order: nodes removed later are prioritized.
These rules ensure that reinsertion complements rather than overrides the dismantling process, preserving the integrity of the original method.
In our experiments, we implement three reinsertion methods, adapted from prior work, here we explain which part of their method we change for our experiments. Those changes are marked with an asterisk (*) in
Table A4.:
R1 [
2]: We replace their original tiebreak (smallest node index) with reverse dismantling order.
R2 [
3]: We apply the LCC stopping condition. Originally, all nodes are reinserted to compute a new dismantling sequence.
R3 [
4]: We apply reverse dismantling order as the tiebreak, as no rule is defined in their paper, and their code is unavailable.
R3 is the most similar to the reverse immunization method proposed by Schneider et al. [
46], where nodes are added back one by one based on minimal LCC growth. In their original method, ties are broken by selecting the node with the fewest connections to already reinserted nodes; if multiple candidates remain, one is chosen at random.
We note that reinsertion typically reduces the number of removals but does not always lead to a lower AUC. Since the trajectory of the LCC changes with reinsertion, the dismantling process may reach the threshold faster, improving AUC. However, this is not guaranteed, as we see in the first two subplots of
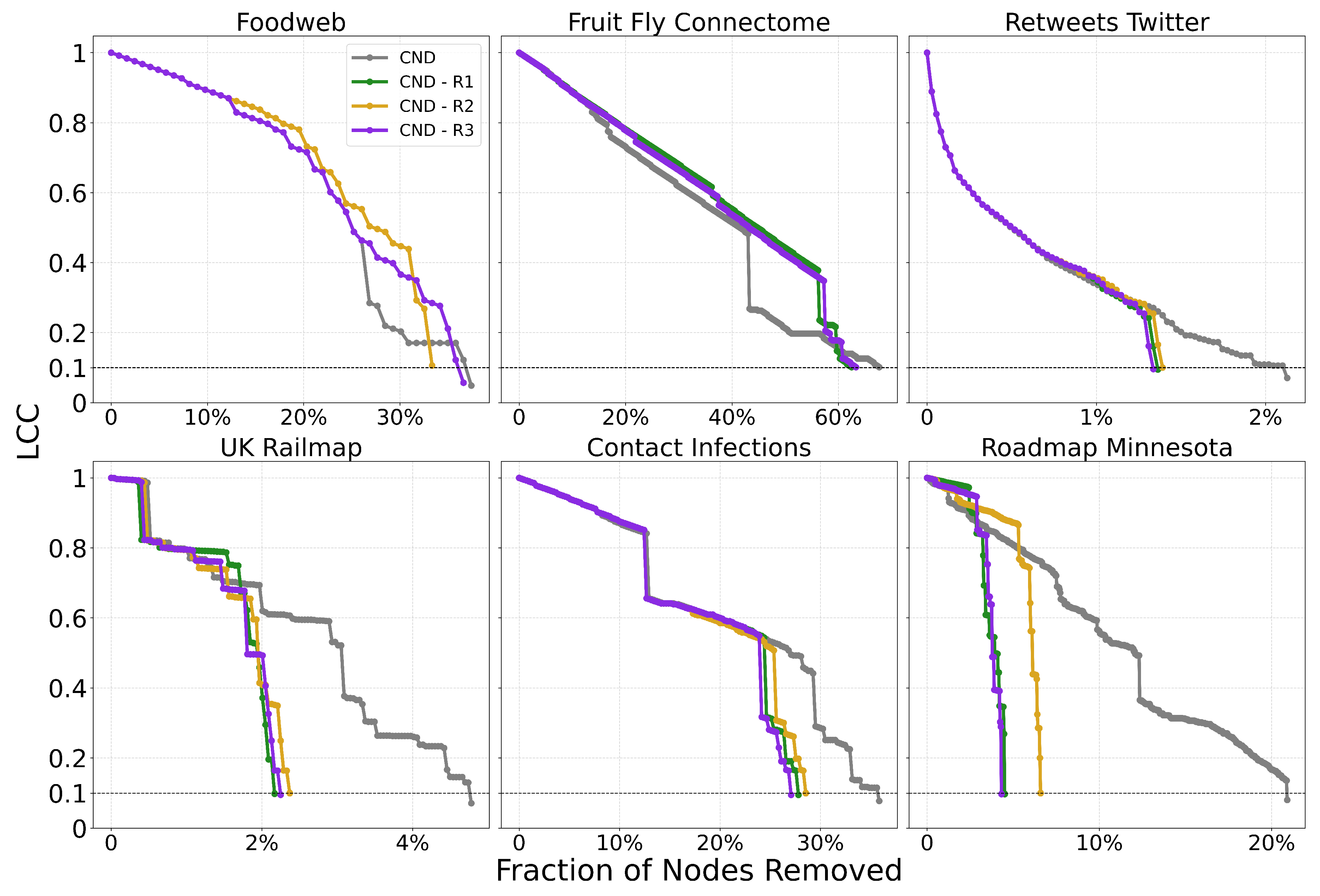
Figure A4 for the Foodweb and Fruit Fly Connectome networks. The methods with reinsertion arrive at the dismantling threshold in fewer number of removals, but the change in the LCC curve results in a worse final AUC.
We also see that the reduction in AUC is not proportional to the reduction in the number of removals, as seen in
Figure A5 for CND. Indeed, reinsertion, by definition, reinserts nodes that were ultimately unnecessary for the dismantling process to reach its target.
We report the total time complexity of each reinsertion method over the full reinsertion process in
Table A4, assuming all dismantled nodes are considered for reinsertion for every step and that all nodes are reinserted. Candidates for reinsertion are denoted as
r. As a result, we multiply the per-step cost of updating for each method by the total number of reinsertion candidates.
is the maximum number of components a node can connect to, equal to the maximum degree in the original graph, and
is the maximum size of any connected component during the reinsertion phase. For R1, the candidate node that ends up in the smallest resulting component is selected. Reinserting a node may merge up to
components, each of size at most
, requiring an update of at most
nodes. These updates are tracked in a binary heap of size
r, where at maximum
nodes have to be updated, giving a cost of
per update. The per-step cost is therefore
. R2 selects the node that connects the fewest existing components. Unlike R1, it requires inspecting not only the components merged by the candidate node, but also the neighbors of the affected neighbors. This increases the complexity by a factor of
, resulting in a per-step time complexity of
. R3 evaluates each candidate by explicitly computing the resulting LCC size after reinsertion. Each evaluation requires a graph traversal to recompute connected components, which takes
time on sparse graphs. This has to be done for each reinsertion candidate, at every step, so
,
Table A4.
Comparison of reinsertion methods.
Criteria defines the criterion for selecting which node to reinsert.
Tiebreak specifies how ties are resolved.
LCC Condition indicates whether all dismantled nodes are reinserted or if reinsertion stops once the predefined LCC threshold is reached.
Time Complexity denotes the time complexity of each reinsertion method on sparse graphs, for the whole reinsertion process.
N: number of nodes.
m: number of links.
r: set of reinsertion candidates.
: maximum degree in the original graph
G.
: maximum size of any connected component during the reinsertion phase.
Used In lists the methods that use each method, in bold, the dismantling method that originally proposed that reinsertion method. An asterisk (*) marks components of the reinsertion method that were modified in our study, as detailed in
Appendix D.
Table A4.
Comparison of reinsertion methods.
Criteria defines the criterion for selecting which node to reinsert.
Tiebreak specifies how ties are resolved.
LCC Condition indicates whether all dismantled nodes are reinserted or if reinsertion stops once the predefined LCC threshold is reached.
Time Complexity denotes the time complexity of each reinsertion method on sparse graphs, for the whole reinsertion process.
N: number of nodes.
m: number of links.
r: set of reinsertion candidates.
: maximum degree in the original graph
G.
: maximum size of any connected component during the reinsertion phase.
Used In lists the methods that use each method, in bold, the dismantling method that originally proposed that reinsertion method. An asterisk (*) marks components of the reinsertion method that were modified in our study, as detailed in
Appendix D.
| Name |
Author |
Year |
Criteria |
Tiebreak |
LCC Condition |
Time Complexity |
Used In |
| R1 |
Braunstein et al. [2] |
2016 |
Node that ends up in the smallest component |
Reverse dismantling order* |
Yes |
|
MS, CoreGDM, CoreHD, GDM, GND |
| R2 |
Morone et al. [3] |
2016 |
Node that connects to the fewest clusters |
Reverse dismantling order |
Yes* |
|
CI |
| R3 |
Mugisha and Zhou [4] |
2016 |
Node that causes the smallest increase in LCC size |
Reverse dismantling order* |
Yes |
|
BPD |
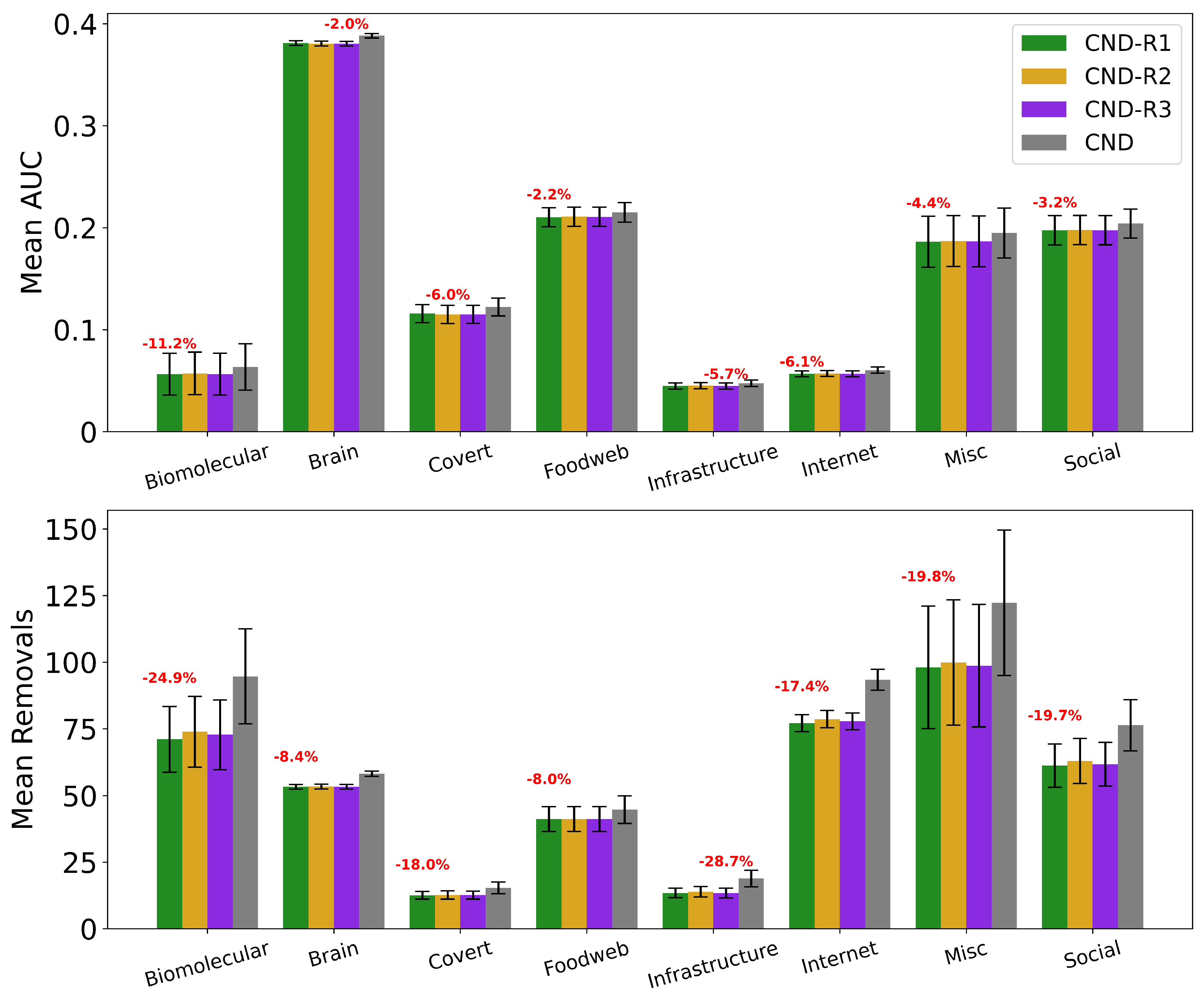
Figure A5.
Mean AUC and number of removals by field for CND without reinsertion and with each reinsertion method (R1, R2, R3) ( for all methods). Error bars represent the standard error of the mean (SEM). Red text indicates the percentage improvement achieved by using the best-performing reinsertion method for each field.
Figure A5.
Mean AUC and number of removals by field for CND without reinsertion and with each reinsertion method (R1, R2, R3) ( for all methods). Error bars represent the standard error of the mean (SEM). Red text indicates the percentage improvement achieved by using the best-performing reinsertion method for each field.
Table A5.
Full summary statistics of the ATLAS networks used in this study, averaged by field: number of subfields and networks, average number of nodes
, number of edges
, density
, mean degree
, characteristic path length
, assortativity
[
47], transitivity
, mean local clustering coefficient
, maximum
k-core
, average
k-core
, LCP-corr
[
48], and modularity
[
49].
Table A5.
Full summary statistics of the ATLAS networks used in this study, averaged by field: number of subfields and networks, average number of nodes
, number of edges
, density
, mean degree
, characteristic path length
, assortativity
[
47], transitivity
, mean local clustering coefficient
, maximum
k-core
, average
k-core
, LCP-corr
[
48], and modularity
[
49].
| Field |
Biomolecular |
Brain |
Covert |
Foodweb |
Infrastructure |
Internet |
Misc |
Social |
Total |
| Subfields |
5 |
1 |
2 |
1 |
7 |
1 |
8 |
7 |
32 |
| Networks |
27 |
529 |
89 |
71 |
314 |
206 |
38 |
201 |
1,475 |
|
2,997 |
97 |
107 |
117 |
664 |
5,708 |
2,880 |
3,267 |
|
11,855 |
1,535 |
266 |
1,087 |
1,332 |
19,601 |
19,921 |
53,977 |
|
|
0.01 |
0.34 |
0.17 |
0.16 |
0.07 |
0.01 |
0.07 |
0.11 |
|
6.7 |
28.3 |
5.7 |
15.2 |
4.9 |
7.5 |
14.1 |
26.9 |
|
|
4.4 |
1.7 |
3 |
2.2 |
9.9 |
3.4 |
3.5 |
3.5 |
|
|
-0.21 |
-0.03 |
-0.15 |
-0.28 |
-0.52 |
-0.22 |
-0.07 |
-0.05 |
|
|
0.06 |
0.55 |
0.39 |
0.19 |
0.06 |
0.11 |
0.22 |
0.29 |
|
|
0.13 |
0.63 |
0.46 |
0.22 |
0.11 |
0.31 |
0.34 |
0.36 |
|
|
10.6 |
20.1 |
5.9 |
12.8 |
4.9 |
25 |
21.6 |
25.7 |
|
|
3.6 |
17.5 |
4.2 |
9.2 |
3 |
4 |
8.3 |
15.4 |
|
|
0.66 |
0.97 |
0.76 |
0.67 |
0.15 |
0.94 |
0.85 |
0.77 |
|
|
0.59 |
0.25 |
0.48 |
0.26 |
0.46 |
0.5 |
0.49 |
0.5 |
|
Table A6.
Number and size of real-world networks tested by dismantling algorithms. N denotes the number of nodes, E the number of edges.
Table A6.
Number and size of real-world networks tested by dismantling algorithms. N denotes the number of nodes, E the number of edges.
| Algorithm |
Year |
Networks |
|
|
| Collective Influence (CI) [3] |
2016 |
2 |
14M |
51M |
| CoreHD [30] |
2016 |
12 |
1.7M |
11M |
| Explosive Immunization (EI) [31] |
2016 |
5 |
50K |
344K |
| Min-Sum (MS) [2] |
2016 |
2 |
1.1M |
2.9M |
| Generalized Network Dismantling (GND) [32] |
2019 |
10 |
5K |
17K |
| Resilience Centrality [33] |
2020 |
4 |
1K |
14K |
| Graph Dismantling Machine (GDM) [34] |
2021 |
57 |
1.4M |
2.8M |
| CoreGDM [35] |
2023 |
15 |
79K |
468K |
| Domirank Centrality [22] |
2024 |
6 |
24M |
58M |
| Fitness Centrality [23] |
2025 |
5 |
297 |
4K |
| LGD-NA |
2025 |
1,475 |
23K |
507K |
Appendix F. Engineering Network Robustness
Here, we would like to comment on the feasibility of our suggested network modifications in practical scenarios. In the case of PPI networks, recent advances in structural modeling of molecules using AlphaFold 3 (AF3) [
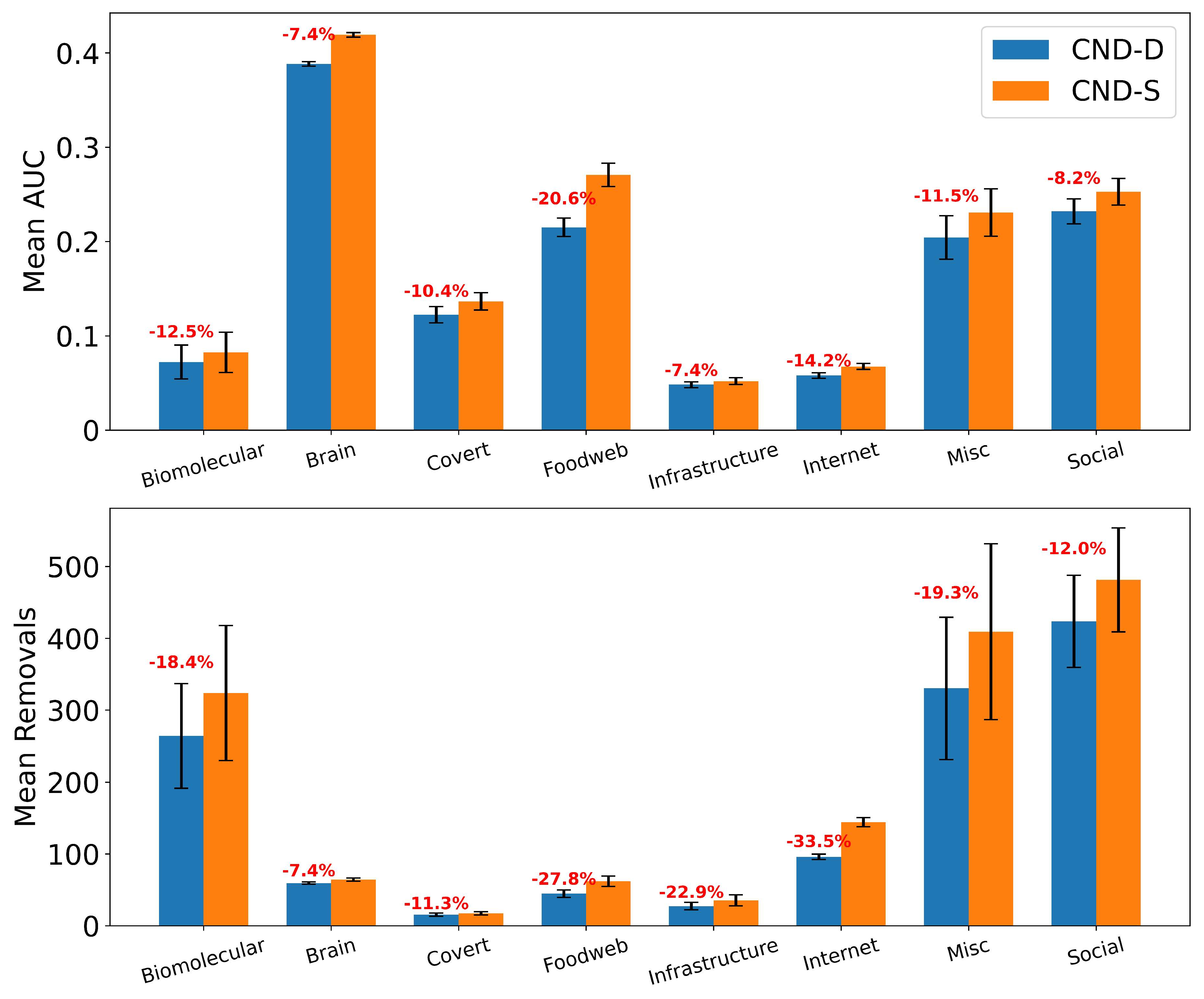
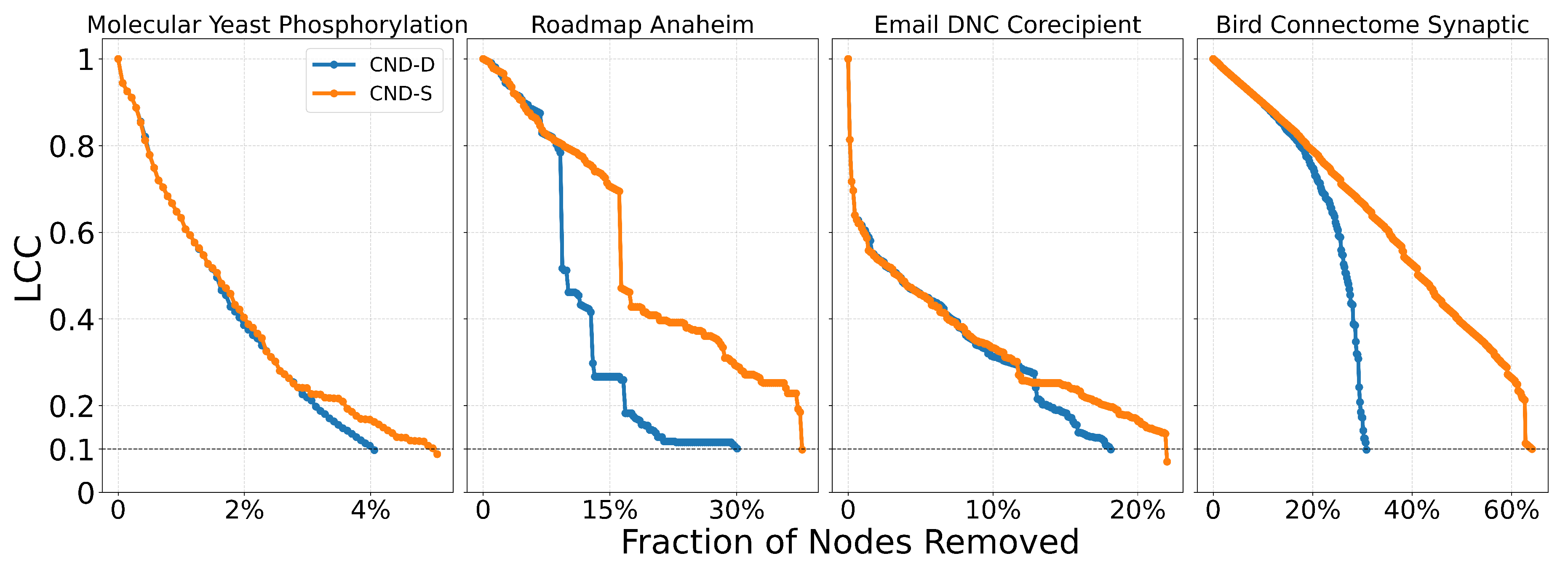
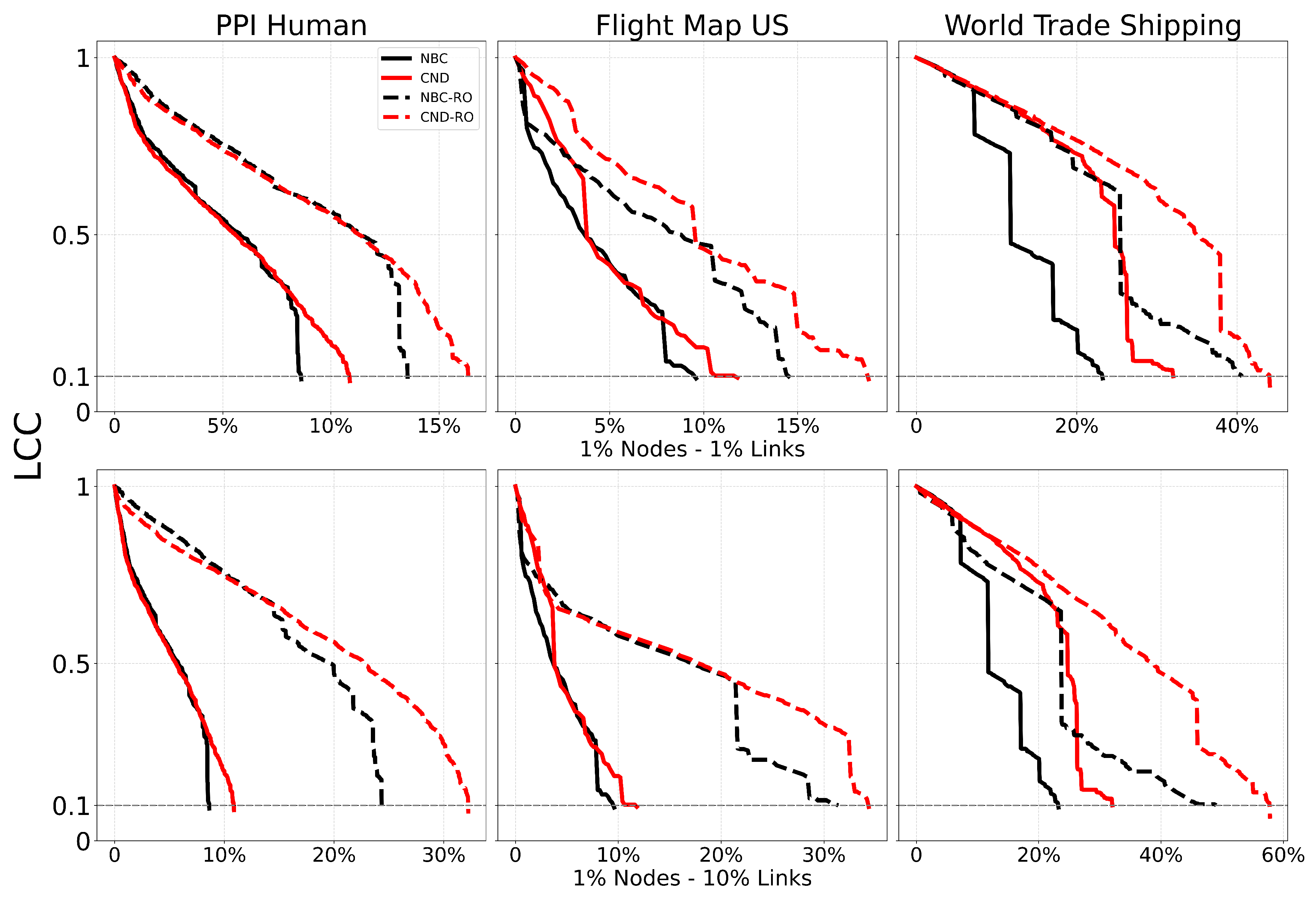
50] have reduced the longstanding limitations in testing and engineering arbitrary proteins. Coupling our dismantling predictions with AF3 could impact drug repositioning and drug design. For example, in the case of antibiotic-resistant bacteria, knowledge of how to dismantle the bacterial PPI network could be used to identify which proteins to target with repositioned, modified, or newly designed antibiotics. Meanwhile, to minimize side effects of newly designed drugs that target critical proteins in bacterial PPI networks, our reinforcement strategy based on common-neighbor generation could be applied to predict which protective bindings to promote in the human PPI network, thereby reducing the destructive impact of a drug on a critical human protein whose impairment could cause side effects. Finally, the application of the proposed common-neighbor reinforcement strategy to increase the robustness of flight and shipping networks is straightforward, as it can indicate which critical nodes should be reinforced by adding links between their adjacent nodes.
We validate our reinforcement strategy explained in
Section 4.6 on three types of real-world networks considered critical: a human PPI network (biological), a flight map network (transportation with social and geographic constraints), and a shipping trade network (transportation with economic and geographic constraints). We select the top 1% of highest-scoring nodes according to the chosen measure (NBC or CND) and randomly add either 1% or 10% of the potential links between their respective adjacent nodes, thereby modifying the network topology according to the explainable rule discovered via CND.
Table A7.
Area Under the dismantling Curve (AUC) for NBC and CND on the original network and "reinforced" networks, by adding 1% and 10% of the links between common neighbours of the top 1% of nodes. In parentheses the increase in the AUC compared to the original network, representing the reduction in the dismantling effectiveness.
Table A7.
Area Under the dismantling Curve (AUC) for NBC and CND on the original network and "reinforced" networks, by adding 1% and 10% of the links between common neighbours of the top 1% of nodes. In parentheses the increase in the AUC compared to the original network, representing the reduction in the dismantling effectiveness.
| Human PPI |
0% Links |
1% Links |
10% Links |
|
| NBC-baseline |
0.051 |
|
|
|
| NBC-reinforced |
|
0.093 (+84%) |
0.159 (+214%) |
|
| CND-baseline |
0.055 |
|
|
|
| CND-reinforced |
|
0.098 ((+79%)) |
0.198 ((+259%)) |
|
| Flight Map US |
0% Links |
1% Links |
10% Links |
|
| NBC-baseline |
0.042 |
|
|
|
| NBC-reinforced |
|
0.067 (+61%) |
0.122 (+193%) |
|
| CND-baseline |
0.055 |
|
|
|
| CND-reinforced |
|
0.098 ((+95%)) |
0.172 ((+241%)) |
|
| Trade Shipping |
0% Links |
1% Links |
10% Links |
|
| NBC-baseline |
0.138 |
|
|
|
| NBC-reinforced |
|
0.236 (+71%) |
0.240 (+74%) |
|
| CND-baseline |
0.220 |
|
|
|
| CND-reinforced |
|
0.298 ((+36%)) |
0.350 ((+59%)) |
|
Figure A8.
Dismantling curve on the original (solid) and reinforced networks (dashed). The plot shows the normalized size of the largest connected component (LCC) as a function of the fraction of nodes removed, with a target LCC threshold of 10%. Performance is evaluated using the Area Under the Curve (AUC) of the LCC trajectory.
Figure A8.
Dismantling curve on the original (solid) and reinforced networks (dashed). The plot shows the normalized size of the largest connected component (LCC) as a function of the fraction of nodes removed, with a target LCC threshold of 10%. Performance is evaluated using the Area Under the Curve (AUC) of the LCC trajectory.
Appendix G. RA Measures in Hyperbolic Networks
Here, we explore the relevance of the RA2 measure as an estimator for latent geometry in hyperbolic networks. As previously mentioned, the RA measures were introduced to serve as pre-weighting strategies for approximating angular distances associated with node similarities in hyperbolic network embeddings [
1].
To investigate this, we generate networks using the non-uniform Popularity-Similarity Optimization (nPSO) model [
37]. The nPSO model is built on the principle that radial coordinates represent hierarchy (popularity) while angular coordinates represent similarity. It produces networks that are both scale-free (power-law degree distribution, meaning a network has a few highly connected hubs while the majority of nodes have few links) and clustered with distinct communities, closely mimicking the structure of many real-world complex systems.
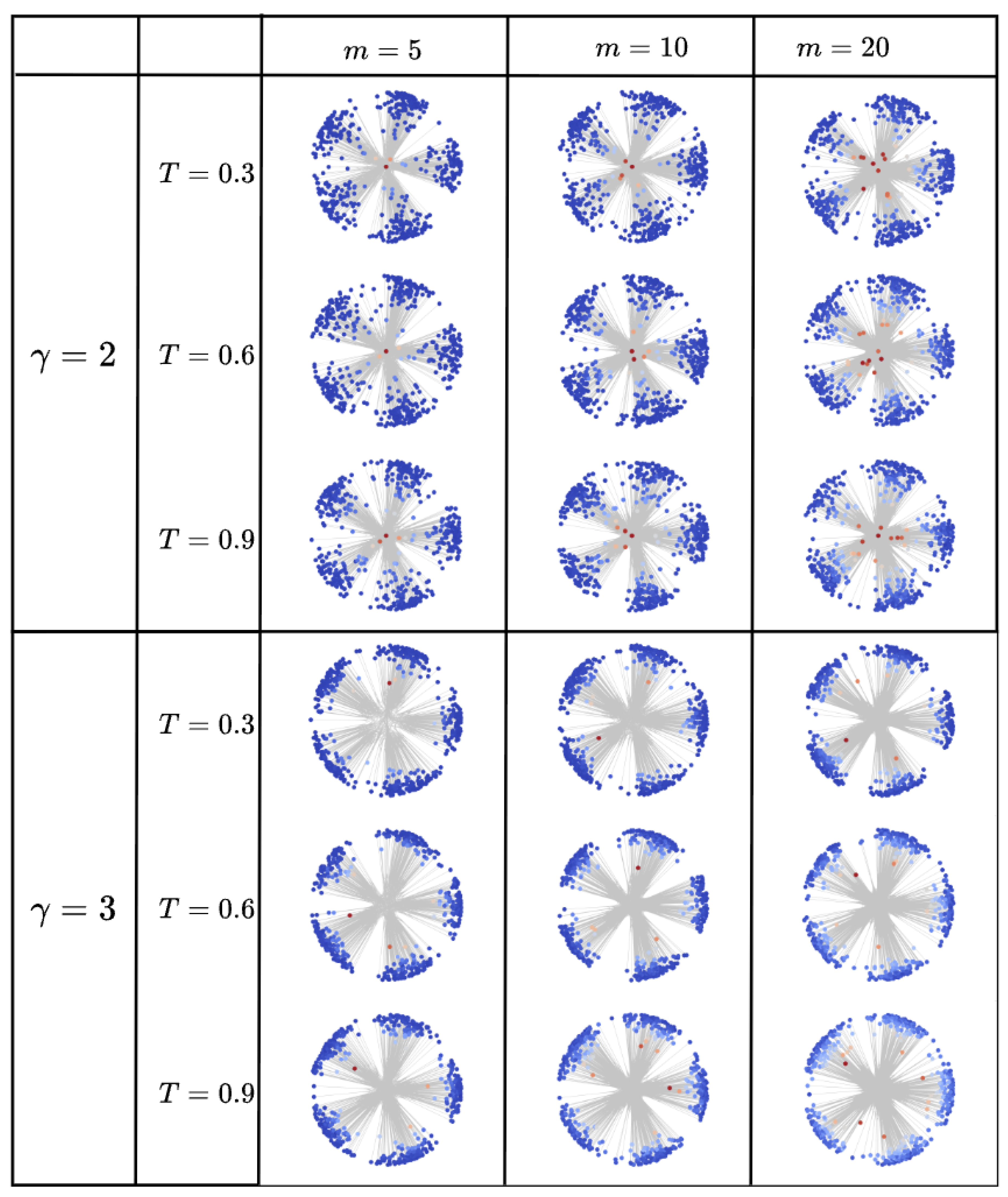
In
Figure A9, we visualize various nPSO networks in the hyperbolic space, keeping the number of nodes (
) and communities (
) fixed. We test different network topologies by varying:
The power-law exponent , which represent common lower and upper bounds for real-world scale-free networks.
The number of nodes a new node will connect to when being added to the network, , and represents approximately half of the average node degree, making the network more or less connected.
The temperature , which controls the level of clustering (lower temperatures yield stronger clustering).
Nodes in each network are colored according to their RA2 value. The results show that the RA2 measure effectively captures the network’s hierarchical structure. Nodes with higher RA2 values are consistently those that are more radially central (i.e., the hubs). This relationship is particularly evident for , where the skewed degree distribution creates a clearer distinction between central hubs and peripheral nodes. The trend persists for , though it is less pronounced due to the presence of fewer "super-hubs." This confirms that RA2 serves as a reliable proxy for the radial coordinates that encode centrality in hyperbolic network embeddings.
Figure A9.
nPSO model networks visualized in the hyperbolic space. Fixed parameters are the number of nodes, N=500, and the number of communities, C=5. Nodes are colored according to their RA2 measure.
Figure A9.
nPSO model networks visualized in the hyperbolic space. Fixed parameters are the number of nodes, N=500, and the number of communities, C=5. Nodes are colored according to their RA2 measure.
Appendix H. GPU Acceleration
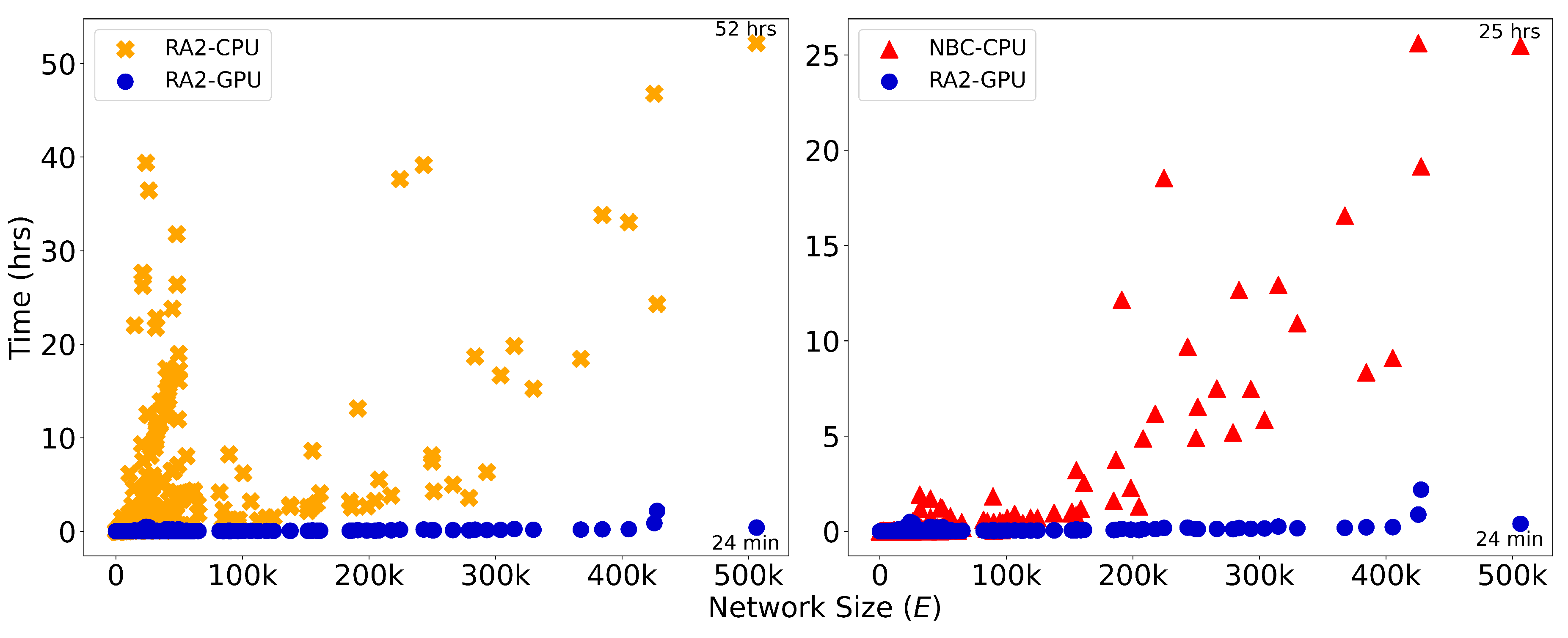
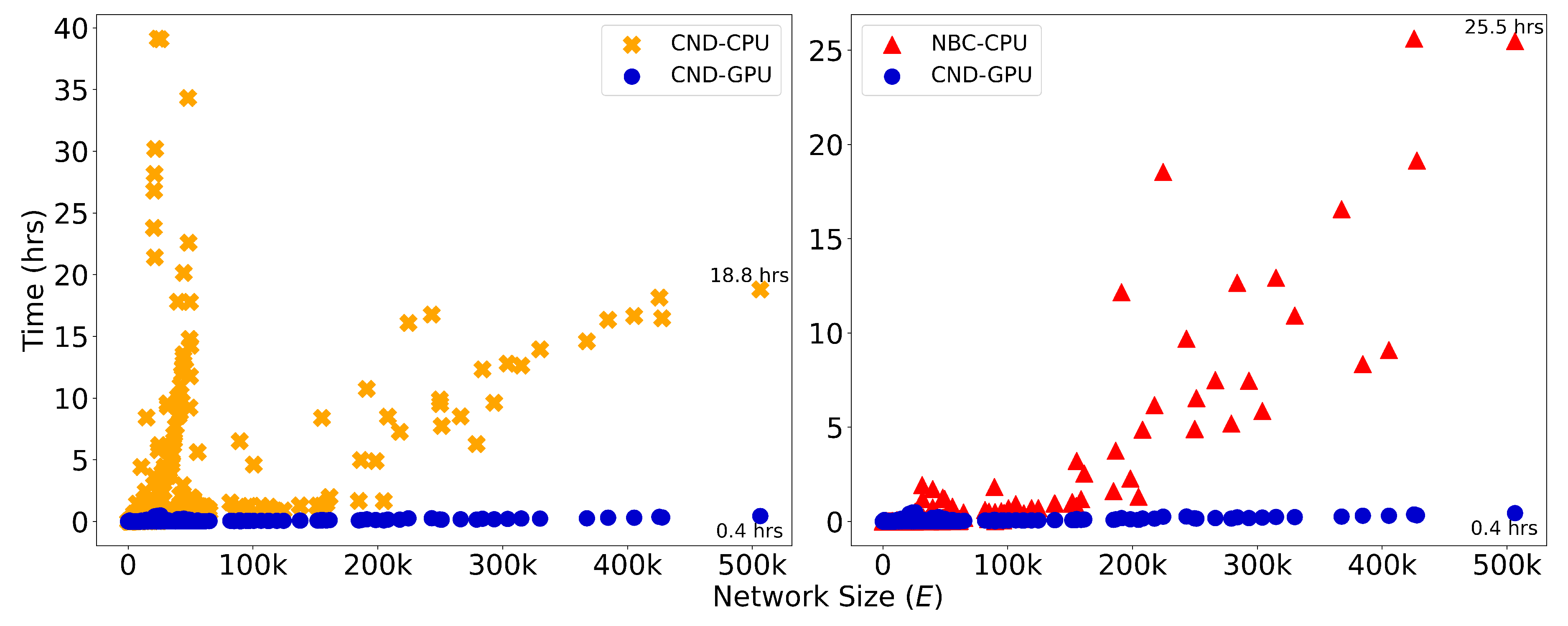
Figure A10.
Runtime (in hours) is plotted against network size, measured by the number of edges, E, for dynamic dismantling. The annotated time indicates the runtime for the largest network. Evaluated on networks of up to 23,000 nodes and 507,000 edges ().
Figure A10.
Runtime (in hours) is plotted against network size, measured by the number of edges, E, for dynamic dismantling. The annotated time indicates the runtime for the largest network. Evaluated on networks of up to 23,000 nodes and 507,000 edges ().
Since the difference in running time between the three LGD-NA methods is not relevant, neither for CPU nor GPU, we report the running time of the original RA2 in the main text (
Figure 3) and the CND in the Appendix (
Figure A10). When comparing CND and NBC, on the largest network, GPU-accelerated CND is over 46 times faster than its CPU counterpart and also over 63 times faster than NBC running on CPU.
Section 4.5 details how we implement the RA-based measures for GPU, using matrix multiplication. We end up with the following formula for RA2:
and for CND:
In our experiments, the CPU-based RA2 and CND implementation uses Python’s NumPy (implemented in C) while the GPU implementation uses Python’s CuPy (implemented in C++/CUDA).
As mentioned earlier, we report only the CPU running time for NBC, as its GPU implementation did not yield any speedup. While some studies report GPU implementations of NBC with improved performance [
51,
52,
53,
54,
55,
56], these are often limited by hardware-specific optimizations, data-specific assumptions (e.g., small-world, social, or biological networks), and using heuristics that are tailored to specific settings rather than offering general solutions. Moreover, publicly available code is rare, making these approaches difficult to reproduce or integrate. Overall, NBC is not naturally suited for GPU implementation, as it does not rely on matrix multiplication, but is based on computing shortest path counts between all node pairs. In our experiments, the CPU-based NBC implementation from Python’s
graph_tool (implemented in C++), based on Brandes’ algorithm [
45] with time complexity
for unweighted graphs, outperformed the GPU version from Python’s
cuGraph (implemented for C++/CUDA).
Appendix I. Experimental Setup
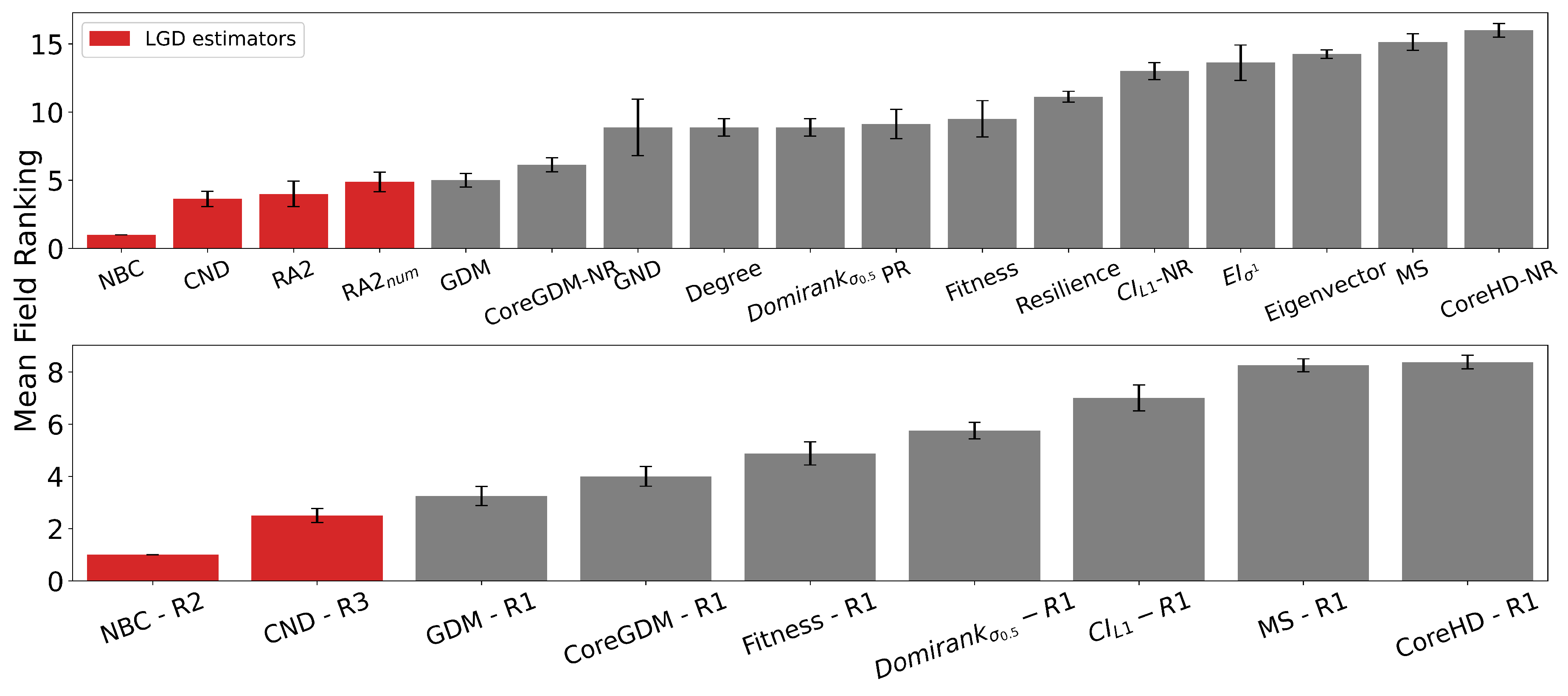
We selected centrality measures to cover diverse categories: shortest path-based (NBC), degree-based (degree), walk-based (eigenvector), random walk-based (PageRank), resilience-based (Resilience), and fitness-based (Domirank and Fitness centrality). We also tested closeness and load centrality, but both performed worse than NBC and rely on the same shortest-path principle; thus, we retained NBC. Similarly, Katz centrality underperformed compared to eigenvector centrality and is also based on spectral properties of the adjacency matrix. For DomiRank, we tested three values for the numerator in the parameter formula: 0.1, 0.5, and 0.9. While the original study sometimes performs a grid search to find the optimal per network, this is not feasible for our large-scale evaluation. Instead, we selected a representative range and found that yielded the best performance, and we report that value. The parameter controls the balance between local degree-based dominance and global network-structure-based competition. As , the scores approximate degree centrality. As increases, nodes are evaluated in increasingly competitive environments, where centrality depends more on non-local structural dominance than individual connectivity. For Fitness centrality, we capped the number of iterations at . Without this cap, the method took a prohibitively long time to converge, especially on large networks.
We selected the best-performing and most widely adopted dismantling algorithms from the literature [
6]. As mentioned earlier, we did not include BPD and FINDER in our experiments due to unavailable and outdated code, respectively. For Collective Influence (CI), we tested values
, where
ℓ defines the radius of a ball centered at node
i, and
is the frontier at distance
ℓ (i.e., nodes exactly
ℓ hops away). We found that
performed best across our benchmarks and report this setting, while
performed the worst. For Explosive Immunization (EI), we evaluated both scores
and
. The
score targets high-connectivity nodes to rapidly fragment the network early on. The
score aims to prevent large cluster merges near the percolation threshold by avoiding the connection of separate components. We found that
consistently outperformed
, and thus we use it in our final experiments. For eigenvector centrality, we capped the number of power iterations at
to avoid long or unbounded runtimes, since convergence can be very slow in large networks. For PageRank, we used a convergence tolerance of
, as the algorithm runs until the change in scores falls below this threshold.
We did not apply reinsertion techniques on all networks included in the initial dismantling experiments. In some cases, certain methods performed so poorly that applying reinsertion became prohibitively slow. To ensure consistency, we excluded these networks from the reinsertion analysis for all methods. Specifically, we imposed a cutoff: networks were excluded if any method required more than 800 node removals to reach the dismantling threshold. Based on this criterion, 59 networks were excluded.
A lower number of removals does not always imply a lower AUC. Our AUC metric rewards methods that fragment the network early, even if they require more steps to reach the dismantling threshold. As shown in
Figure A11, we show cases where a method that reaches the threshold with more removals can still achieve a lower AUC, due to earlier damage to the network structure.
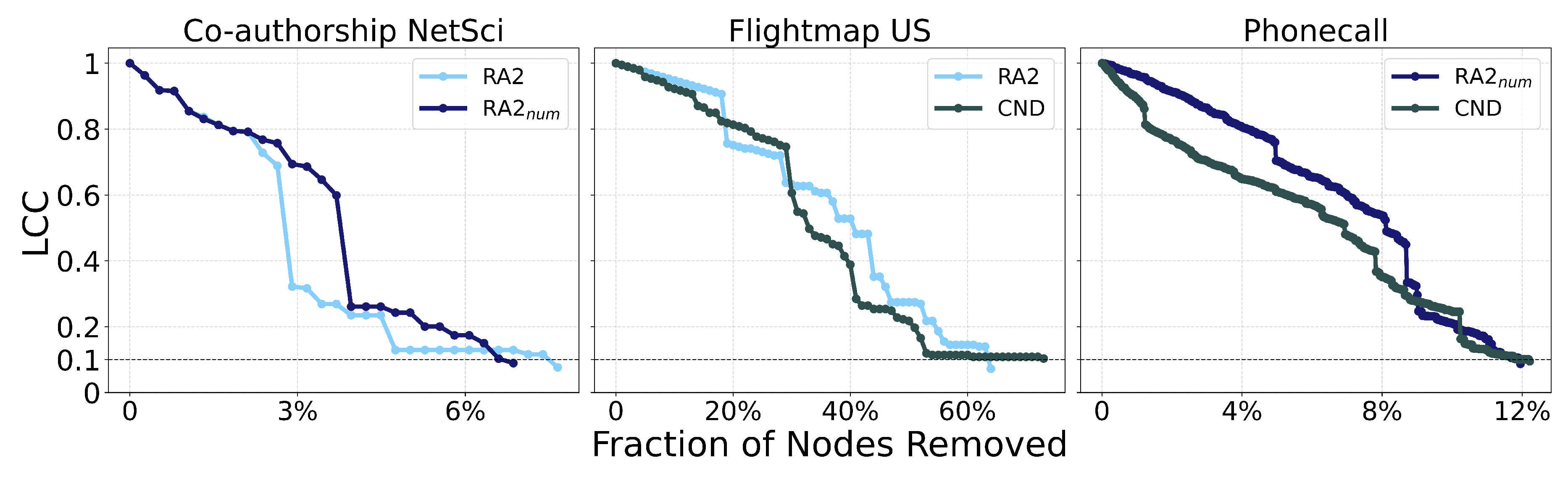
Figure A11.
Dynamic dismantling process on example networks comparing CND, RA2 and , showing a lower removal number does not necessarily mean a lower Area Under the Curve (AUC) of the LCC trajectory. The plot shows the normalized size of the largest connected component (LCC) as a function of the fraction of nodes removed, with a target LCC threshold of 10%.
Figure A11.
Dynamic dismantling process on example networks comparing CND, RA2 and , showing a lower removal number does not necessarily mean a lower Area Under the Curve (AUC) of the LCC trajectory. The plot shows the normalized size of the largest connected component (LCC) as a function of the fraction of nodes removed, with a target LCC threshold of 10%.
All our methods are implemented in our codebase, available in the supplementary material. We implement RA2, RA2
num, CND, NBC, as well as degree and eigenvector centrality. We also adapt and integrate the original implementations of
Domirank centrality and
Fitness centrality. Since no code was available for Resilience centrality, we implemented it ourselves based on the original description in the paper. Instructions for running these methods and reproducing the experiments are included in the supplementary material. We also provide a representative network from the ATLAS dataset for testing purposes. For GDM, CoreGDM, GND, CI, EI, MS, and CoreHD, we use the publicly available
code from Artime et al. (2024)’s review.
All experiments were conducted on a machine equipped with an AMD Ryzen Threadripper PRO 3995WX CPU (64 cores), 251 GiB of RAM, and a single NVIDIA RTX A4000 GPU with 16 GiB of memory. All code was implemented in Python, with dependencies and library versions specified in the supplementary material to ensure full reproducibility.
All results are in Tables S1–S13 in the Supplementary Material.
Appendix J. Discussion, Limitations, and Future Work
The robustness of our LGD-NA methods to missing or manipulated information, such as missing neighbor data or adversarial modifications to neighborhood structure, is a crucial question for practical applications of any network analysis method, especially those relying on local information. Our LGD-NA methods, by design, are inherently more robust to global missing or manipulated information compared to methods that require a complete global view of the network (e.g., exact NBC). Since our methods rely solely on local neighborhood information, random missing data across the network would primarily affect only the scores of directly impacted nodes, rather than propagating errors throughout the entire graph. Similarly, adversarial modifications would need to target specific local neighborhoods to significantly alter a node’s dismantling score, making large-scale, coordinated attacks more challenging than for global metrics. However, we acknowledge that direct adversarial manipulation of a node’s immediate neighborhood could indeed impact its calculated score.
While the prohibitive computational cost of the highly effective Node Betweenness Centrality (NBC) metric, a global estimator of latent geometry, has led to the development of numerous approximators [
57,
58,
59,
60], our proposed LGD-NA methods are founded on a fundamentally different principle. Existing NBC approximators primarily focus on creating faster implementations of a global metric. In contrast, our approach leverages purely local topological network information to directly estimate pairwise distances in the latent metric space. A crucial distinction is that our method bypasses the need for the global knowledge or complex sampling strategies upon which many NBC approximators still depend. Although these approximation techniques can be faster than calculating the exact NBC, their reliance on sampling or broader computations is inherently less efficient than our strictly local approach. Furthermore, the imperfections introduced by sampling can degrade dismantling performance. Such sampling of global information is also not robust to missing data and remains vulnerable to adversarial attacks. The strength of our method, therefore, lies in its ability to achieve high dismantling performance by directly utilizing local geometric insights, rather than attempting to approximate a computationally intensive, global metric like NBC.
A limitation of this study is the mismatch between theoretical and observed runtimes, which can vary across methods. These differences stem from factors such as the programming language used, hardware acceleration, and implementation-level optimizations. However, all experiments were run on the same CPU and GPU to ensure a fair comparison, and we made a strong effort to optimize all methods both in terms of runtime and dismantling performance. For example, we tested different parameters for Domirank, CI, and EI, and evaluated multiple variants of shortest path-based and spectral partitioning-based centrality measures. Furthermore, we were unable to test on extremely large networks due to hardware constraints and the high computational cost of running a broad set of dismantling methods. However, we are confident that our results would generalize to larger networks, given the diversity of the 1,475 networks tested, spanning a wide range of domains and sizes from very small to large. Another limitation relates to the parameter tuning required by some baseline methods, especially machine learning-based approaches and Domirank. Due to the scale of our experimental setup—both in the number and size of networks—we were unable to perform extensive tuning. Although targeted tuning could enhance performance for specific methods on individual networks, it would compromise consistency across the wide range of complex systems domains considered. In contrast, LGD-NA requires no parameter tuning and consistently achieves strong, generalizable performance across all tested networks.
Future research could further explore latent geometry, particularly how to effectively combine local and global information in dismantling strategies. Improving the scalability of matrix-based computations, especially for very large and sparse networks, is another important direction. There is also a need for more cost-efficient dynamic dismantling strategies that reduce the overhead of recomputing scores after every node removal without significantly sacrificing performance. In addition, edge dismantling remains a relatively underexplored area compared to node-based dismantling, and it would be valuable to investigate whether latent geometry-driven principles can also guide the efficient removal of links in complex networks. Targeting edges can be just as important as targeting nodes, and in many real-world systems, such as transportation networks (railroads, roads, subways, or shipping trade routes), edge removal may represent the more realistic and sensible threat scenario, making it highly relevant for dismantling strategies. Finally, our work can also be of interest to Explainable AI (XAI) for models like GNNs and Reservoir Computers (RC). By identifying critical (dismantling) or unimportant (percolation) nodes in a network, we can conduct "perturbation experiments" to assess their impact on an ML model’s performance. This process reveals which parts of the graph are most crucial for the model’s predictions or computations, effectively opening the model’s “black box” and providing crucial insights into its internal decision-making, robustness, and vulnerabilities.