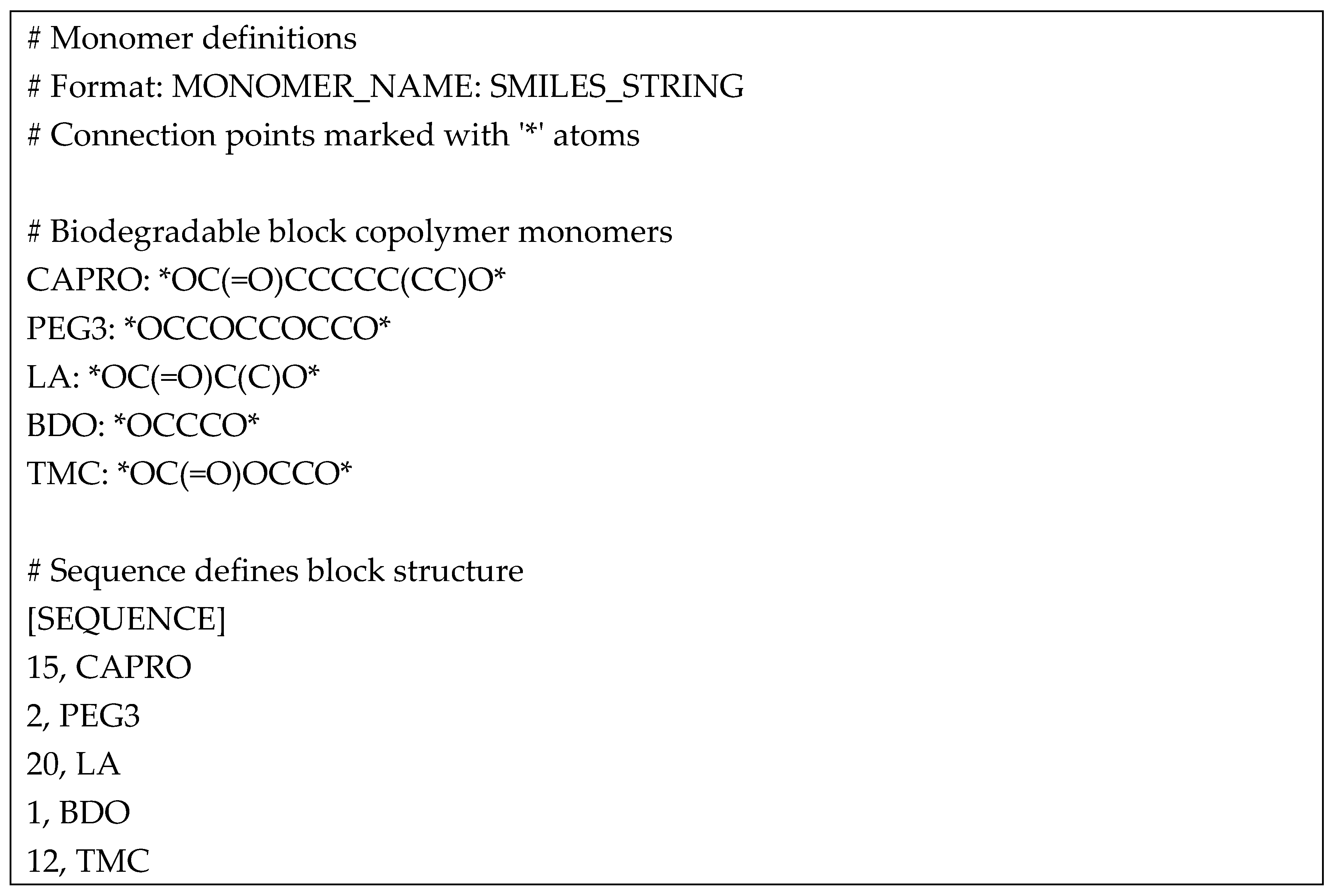1. Introduction
The design and analysis of polymeric materials represent one of the most challenging cutting-edge in materials science [
1]. With the growing demand for advanced materials in applications ranging from biomedical devices to sustainable packaging, the ability to precisely design and characterize polymers has become increasingly critical [
2]. However, the current landscape of polymer development faces several significant challenges that impede rapid innovation and reliable characterization [
3].
One primary challenge lies in the complexity of polymer structure-property relationships. Unlike small molecules, polymers exhibit hierarchical organization across multiple length scales, from individual monomer arrangements to macroscopic properties [
4]. This complexity makes it difficult to predict how molecular-level changes will affect final material properties. Traditional approaches often rely on time-consuming trial-and-error methods, requiring extensive laboratory resources and expertise [
5].
Existing computational tools for polymer analysis typically focus on specific aspects of the polymer development pipeline [
6]. For instance, some tools specialize in monomer structure prediction, while others concentrate on property calculation or molecular dynamics simulations [
7,
8]. Commercial software packages like Materials Studio and BIOVIA offer comprehensive capabilities but often operate as separate modules with limited integration [
9]. Open-source alternatives, while valuable, frequently lack the advanced monitoring and validation capabilities necessary for reliable polymer design [
10].
There is a significant gap in the field for an integrated system that can seamlessly connect monomer-level design decisions to final polymer properties while providing monitoring and validation. APPAS system introduces several key innovations. First, it employs an advanced monomer processing module that ensures proper structural validation and connection point identification. Second, it features an advanced co-polymer building algorithm that maintains structural integrity while optimizing computational efficiency. Third, it integrates monitoring capabilities that track various structural and energetic parameters throughout construction. Finally, it includes a comprehensive analytics suite that provides detailed insights into polymer properties and potential applications. Furthermore, the extensive monitoring and validation capabilities provide researchers with unprecedented insight into the co-polymer building process, facilitating optimization and troubleshooting.
2. Methods
2.1. System Architecture
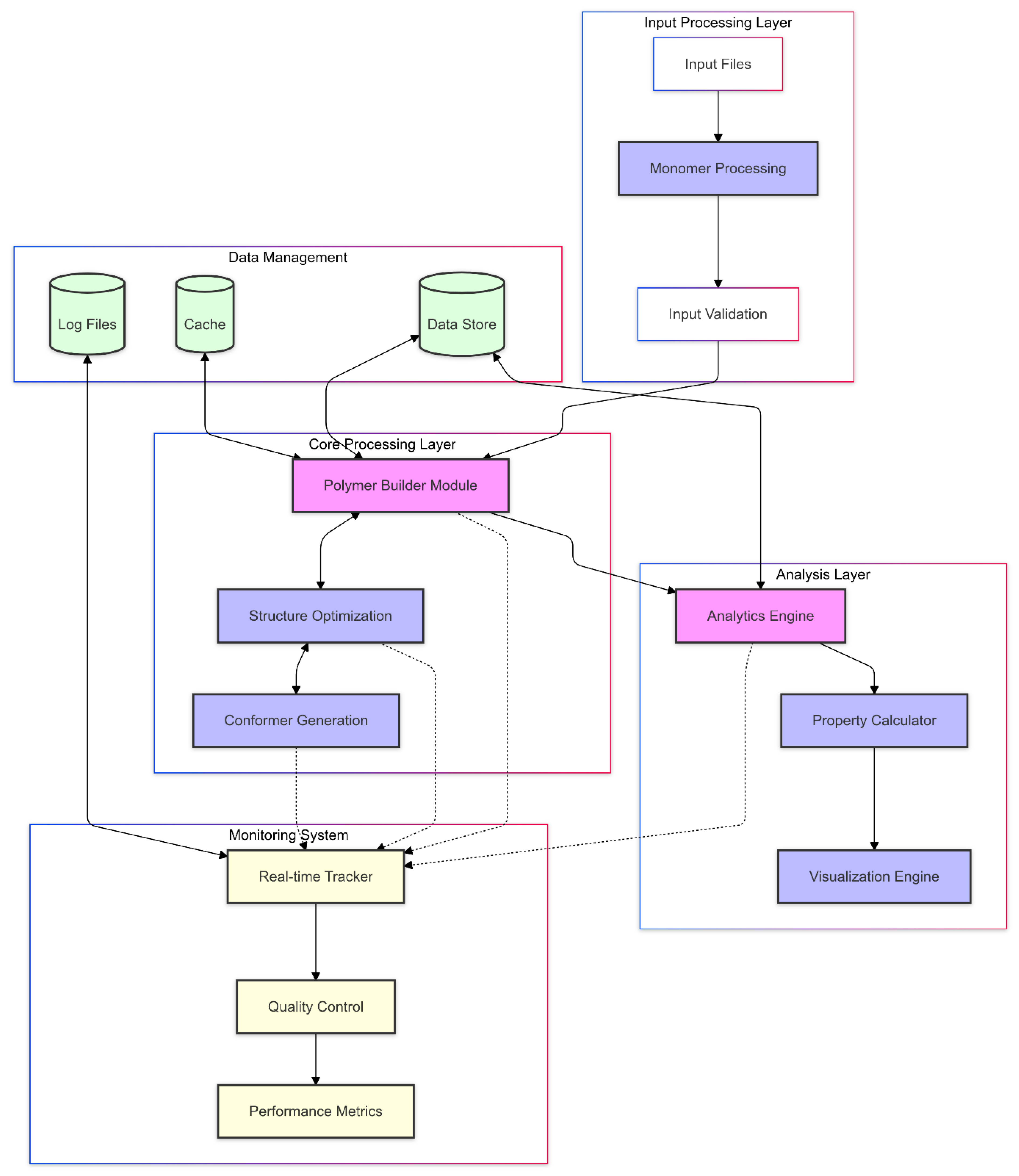
The system implements a multi-layered architecture (
Figure 1) designed to ensure robust co-polymer construction and analysis while maintaining data integrity throughout the processing pipeline. This architecture comprises four primary layers:
Input Processing,
Core Processing,
Analysis, and
Monitoring, with a dedicated
Data Management layer handling persistent storage and caching mechanisms.
The architecture layer consists of:
- -
The Input Processing Layer: validates and preprocesses molecular data through the Monomer Processor, which implements comprehensive SMILES parsing and structure validation protocols.
- -
The Core Processing Layer cantered around the Enhanced Polymer Builder, manages the sequential construction of polymer structures while maintaining geometric and chemical constraints.
- -
The Analysis Layer, implemented through the Polymer Analytics, executes parallel property calculations and generates visualizations.
- -
The Monitoring System, operated by the Polymer Monitor, provides continuous oversight of all operations.
2.2. Workflow Implementation
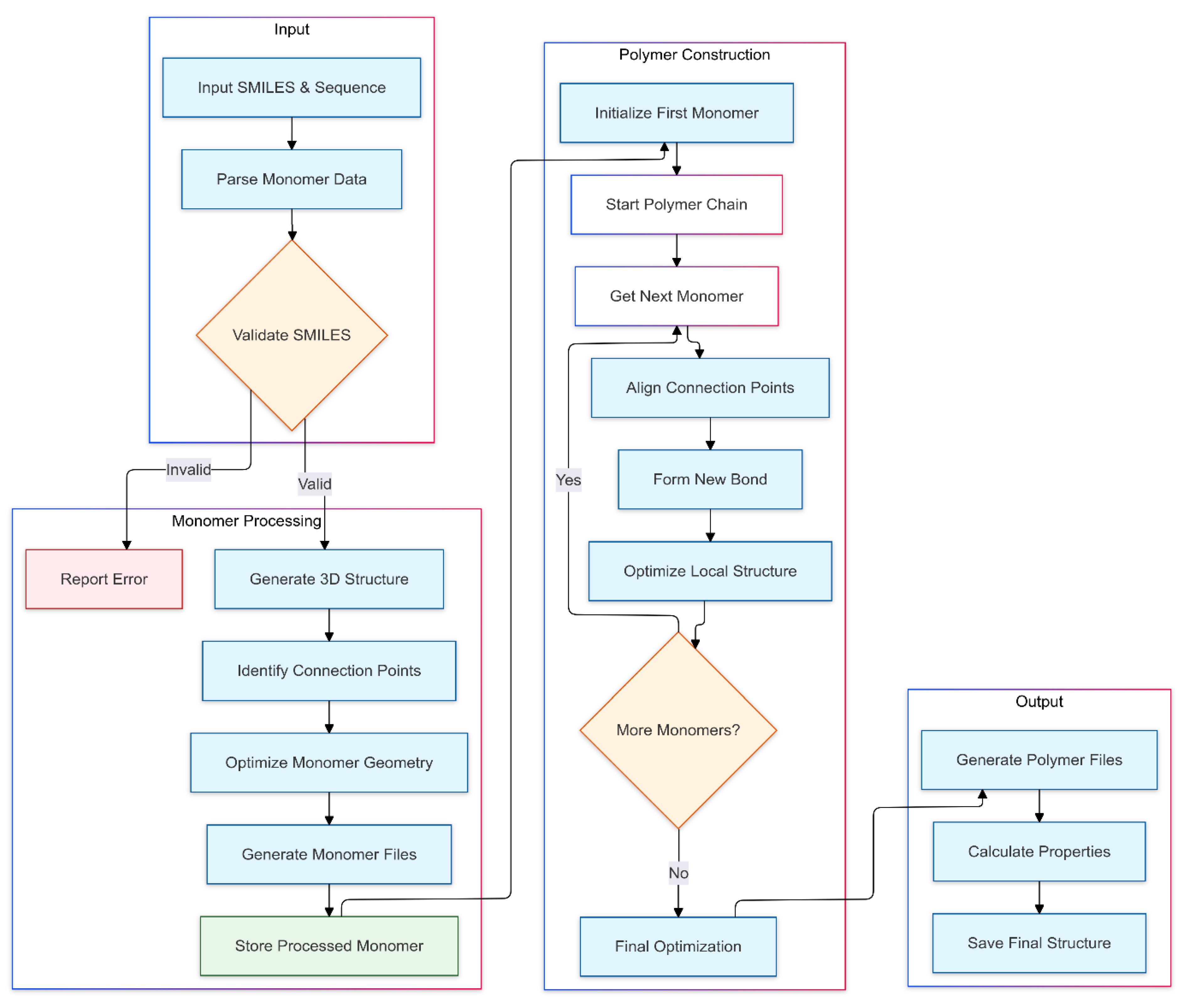
The polymerization workflow (
Figure 2) demonstrates the system's approach to polymer construction and property prediction.
2.3. Core Processing Components
2.3.1. Monomer Processing Module
The monomer processing module, implemented through the Monomer Processor, handles the critical initial stage of polymer construction. This module executes several key processes:
SMILES Validation and Structure Generation
The module implements rigorous SMILES validation, ensuring balanced brackets, correct connection point specification ([R] groups), and valid chemical valences (Listing 1). The validation process employs RDKit's sanitization protocols with custom extensions for polymer-specific requirements.

Listing 1. SMILES Validation.
3D Structure Generation
The generation of three-dimensional structures employs an enhanced implementation of the ETKDG algorithm [
11], optimized explicitly for polymer systems (Listing 2). Our implementation utilizes ETKDGv2 parameters with max Iterations set to 50 for computational efficiency. Initial structure generation employs a fixed random seed (42) to ensure reproducibility across multiple runs. The system generates an ensemble of 10 conformers for each monomer, employing the MMFF94 force field for energy minimization with a convergence criterion of 10
-4 kcal/mol. These conformers are evaluated within an energy window of 20 kcal/mol, selecting the most favourable conformation for subsequent polymer construction. To enhance efficiency, the Random Coords parameter is activated as a fallback mechanism if initial embedding fails, ensuring robust structure generation even for complex monomers. The final conformer selection employs a comprehensive scoring function considering energy (weighted at 0.7) and geometric diversity (weighted at 0.3).

Listing 2. 3D Structure Generation.
Connection Point Analysis and Validation
Our system implements a sophisticated connection point analysis framework with quantitative validation criteria (Listing 3). Connection points must satisfy specific geometric constraints: bond lengths within 0.7-2.0 Å of ideal values, angles within ±15° of expected geometries, and proper valence states for connecting atoms. The framework verifies the presence of exactly two connection points per monomer and implements a neighbor analysis extending to a radius of 3 bonds (approximately 4-5 Å) around each connection site. Valence validation employs explicit checks for common atoms: maximum valence of 4 for carbon, 3 for nitrogen, and 2 for oxygen. The system also monitors atomic clash prevention with a minimum allowed interatomic distance of 0.5 Å and validates electronic environments using Gasteiger charge calculations with a convergence criterion of 10-5.

Listing 3. Connection Point Analysis.
2.3.2. Polymer Construction ModuleStructure Growth Algorithm
Structure Growth Algorithm
The polymer construction process employs a stepwise growth algorithm with precisely defined control parameters (Listing 4). Bond formation occurs under geometric constraints: optimal bond lengths are maintained within ± 0.1 Å of theoretical values, and bond angles are controlled within ±5° of ideal geometries. The algorithm implements a sliding optimization window, typically spanning 3-5 monomer units, to maintain efficiency during growth. Construction checkpoints are established every 25 monomer units, triggering comprehensive structure validation and energy assessment. The system maintains a trajectory cache with a 5-minute update interval, storing intermediate structures in MOL2 format with a coordinate precision of 10-4 Å. Progress monitoring occurs at 5-unit intervals, with real-time updates of structural parameters and energy profiles.

Listing 4. Structure Growth Algorithm.
The polymer construction algorithm systematically generates validated and optimized polymer structures from monomer sequences. Additionally, it incorporates robust error-handling mechanisms at each stage to ensure reliability. First, the algorithm initializes essential variables, including a polymer container and progress trackers. Then, it processes each monomer iteratively, verifying its existence and retrieving its data. Subsequently, the first monomer is validated and used to initialize the polymer structure. Afterward, subsequent monomers are connected to the growing polymer, with intermediate validations ensuring structural integrity. Meanwhile, progress is continuously monitored and periodically saved to prevent data loss.
Upon completion, the algorithm performs a final optimization step to refine the polymer structure. Following this, the optimized structure undergoes rigorous validation to confirm its accuracy. Finally, the validated polymer, along with its properties, is saved for further analysis. Moreover, the algorithm includes error-handling mechanisms that detect and manage exceptions at every stage. If an error occurs, it generates appropriate messages and terminates the process gracefully. As a result, this systematic approach ensures the generation of reliable, error-free polymer structures, making it suitable for high-precision applications.
Geometric Optimization
The geometric optimization protocol implements a multi-stage approach with defined convergence criteria (Listing 5). Initial MMFF94 optimization employs 200 iterations with an energy gradient convergence threshold of 10-4 kcal/mol. Conformational sampling at growth steps generates 10 candidates per stage, evaluated using a composite scoring function that weights energy (60%), geometric quality (30%), and chain linearity (10%). The final UFF optimization extends to 2000 iterations with a tighter convergence criterion of 10-6 kcal/mol. Structure validation employs specific thresholds: bond lengths must be within 0.7-2.0 Å, valence angles within 100-140°, and atomic clashes prevented using a 0.5 Å minimum separation criterion. The optimization process maintains continuous monitoring of structural parameters, including the radius of gyration (updated every 100 steps) and end-to-end distance (evaluated at 500-step intervals). A final energy convergence criterion of 0.1 kcal/mol ensures thorough structural refinement while maintaining computational efficiency.
Listing 5. Geometric Optimization.
2.4. Analysis and Monitoring Framework
2.4.1. Real-Time Monitoring and Performance Tracking
The monitoring system maintains comprehensive oversight throughout the entire construction and analysis process (Listing 6). This is achieved through the Polymer Monitor, which implements real-time tracking of multiple parameters, including structural integrity, energy profiles, and computational efficiency. The monitoring framework generates detailed logs of all operations, enabling thorough analysis of the construction process and facilitating troubleshooting when necessary. Performance metrics, including computational time and memory usage, are continuously tracked to ensure optimal system performance.
Listing 6. Real-Time Monitoring.
2.4.3. Property Calculation Framework and Analytical Pipeline
The enhanced property calculation framework, implemented through the Polymer Analytics, generates comprehensive reports with high-precision quantitative metrics (Listing 7). The system calculates molecular weights with a precision of 0.01 g/mol while maintaining strict accuracy standards for structural parameters, including bond lengths measured to 0.001 Å and angles calculated to 0.1° precision. Energy calculations are reported with 0.1 kcal/mol precision, with conformational energies determined using the MMFF94 force field under a rigorous convergence criterion of 10-4 kcal/mol.

Listing 7. Analysis function Framework.
The run_analysis function performs a detailed analysis of polymer structures provided via the polymer_pat input. Initially, the input path is converted to a Path object, if required, and the polymer structure is loaded. Upon successful loading, the function computes key polymer properties, categorized into basic, topological, geometric, electronic, conformational, thermodynamic, and polymer-specific properties. These analyses are conducted using dedicated helper methods, such as analyze_basic_properties and analyze_geometric_properties, with results stored in a dictionary. Additionally, the function generates an Excel report and visualizations to summarize the findings. Moreover, any errors encountered during the process are logged, and an exception is raised. Upon successful completion, the function returns a comprehensive dictionary detailing the polymer's structural and functional properties.
2.4.4. Visualization and Data Representation Systems
Visualization capabilities are implemented through an integrated plotting framework that generates multiple representation types (Listing 8). The system utilizes matplotlib and seaborn libraries for 2D visualizations, with custom extensions for polymer-specific plot types. These visualizations include property distribution plots, correlation analyses, and specialized polymer chain representations. Three-dimensional structure visualization is handled through a custom implementation that generates interactive molecular representations, allowing detailed examination of polymer conformations and local structural features.
Listing 8. Visualization and Data Representation Systems.
2.5. Output Generation Framework
2.5.1. Directory Structure and Data Organization
The output organization system employs a hierarchical structure for efficient data management and accessibility. Analysis categories are separated at the top level, with intermediate results automatically cached at 5-minute intervals to prevent data loss. The results are stored with SHA-256 checksums to ensure data integrity, while version control integration enables comprehensive result tracking. The system maintains cross-referenced metadata with unique identifiers, facilitating efficient data retrieval and analysis reproducibility. The system's output generation framework produces multiple standardized file formats with rigorously defined specifications. The output module generates MOL2 and PDB files containing atomic coordinates with a 10-4 Å precision and connectivity information with explicit bond orders. These structural files maintain IUPAC-compliant atom typing and include metadata such as residue identifiers (3-letter codes) and chain designations (A-Z), facilitating integration with external molecular modelling software.
2.5.2. Quality Control and Validation
The system employs a comprehensive validation protocol throughout the analysis pipeline (Listing 9). Each calculated property undergoes multiple validation steps, comparing results against theoretical bounds and checking for physical feasibility. This validation framework is implemented through the Enhanced Polymer Validator, which applies rigorous criteria for structure assessment and property validation. The validation process includes checks for bond lengths, angles, and torsional parameters, ensuring all calculated properties align with established chemical principles.

Listing 9. Quality Control Implementation.
The validation of a molecular structure follows a structural approach to ensure its integrity and stability. Initially, the algorithm verifies the validity of the molecular coordinates, discarding any invalid configurations. Subsequently, it evaluates structural connectivity by analyzing molecular fragments; structures with disconnected fragments are deemed invalid. The energy state of the molecule is then assessed through calculations of molecular properties and force fields. Structures exhibiting strain energy exceeding a predefined threshold are rejected. Finally, the algorithm incorporates exception handling to enhance robustness and reliability in molecular structure validation.
2.5.3. Error Handling and Data Integrity
Error handling and recovery mechanisms are implemented at multiple levels throughout the system. The Integrated Polymer System coordinates error detection and recovery, implementing advanced protocols for handling failures during polymer construction or analysis. Additionally, primary validation employs a 1% tolerance threshold, supplemented by secondary statistical checks using 3σ bounds. The system includes automated retry protocols limited to three attempts, with graceful degradation paths that preserve 90% accuracy when optimal results cannot be achieved. A complete audit trail maintains records of all error conditions and resolutions, ensuring full transparency of the analysis process.
2.6. System Infrastructure
2.6.1. Data Management and Storage Architecture
Data management and persistence are handled through an advanced storage system that maintains temporary and permanent data structures. The system implements a hierarchical storage approach, with separate directories for intermediate structures, analysis results, and visualization outputs. All generated data is stored in standardized formats, ensuring compatibility with external analysis tools and facilitating data sharing across different platforms.
2.6.2. System Extensibility and Modularity
The modular and extensible system allows for easy integration of new analysis methods and property calculations. The architecture supports adding new monitoring parameters and validation criteria through a plugin system, enabling the framework to evolve with advancing polymer science requirements. This extensibility is achieved through carefully designed abstract interfaces and standardized data structures that facilitate the integration of new components while maintaining system stability.
2.7. System Dependencies and Software Requirements
The core chemical informatics operations are handled by RDKit (2023.03.1), which provides essential functionality for molecular operations, structure manipulation, and property calculations. The scientific computing stack includes NumPy (≥1.21.0) for numerical operations, SciPy (≥1.7.0) for scientific computations, and Pandas (≥1.3.0) for structured data management. Visualization capabilities are implemented through Matplotlib (≥3.4.0) and Seaborn (≥0.11.0), enabling the generation of publication-quality plots and statistical visualizations. Advanced molecular visualization is facilitated by PyMOL API (≥2.5.0), while OpenBabel (≥3.1.0) provides complementary molecular file format conversion capabilities. The system utilizes Python's built-in libraries, including Pathlib for cross-platform file operations, comprehensive logging systems for error tracking, and JSON for structured data serialization. Development and testing are supported by pytest (≥6.0.0) for unit testing, Black for code formatting, isort for import organization, and mypy for static type checking. The framework's modular architecture allows for flexible dependency management, with all components isolated to minimize version conflicts. Installation is streamlined through standard Python package management tools, with environment configurations provided in requirements.txt and environment.yml formats. The system has been validated across Linux (Ubuntu 24.04.1 LTS), and Windows 11 (v. 24H2) platforms, ensuring broad compatibility and reproducible analysis results across different operating systems.
3. Results and Discussion
3.1. System Performance and Validation
The integrated co-polymer construction and analysis system was extensively tested using a biodegradable block copolymer as a model system. The test polymer consisted of five distinct monomer types (CAPRO (Caproic Acid), PEG3 (Triethylene Glycol), LA (Lactic Acid), BDO (Butanediol), TMC (Trimethylolpropane Citrate)) arranged in a specific sequence to form a complex multi-block structure (Listing 10). This challenging test case was selected to evaluate the system's ability to handle diverse chemical environments and multiple connection types within a single polymer chain. Moreover, the system successfully constructed the polymer structure of 632 atoms and 631 bonds, with a calculated molecular weight of 4873.62 Da. The final structure exhibited an end-to-end distance of 6.99 Å, indicating the successful formation of a well-defined polymer chain. The system's ability to handle this complex structure while maintaining atomic-level precision demonstrates its robustness for polymer construction tasks. Previous studies on polymer construction systems, such as those using molecular dynamics (MD) simulations, have often struggled with handling complex multi-block structures, particularly in maintaining atomic-level precision for polymers with diverse chemical environments [
12].
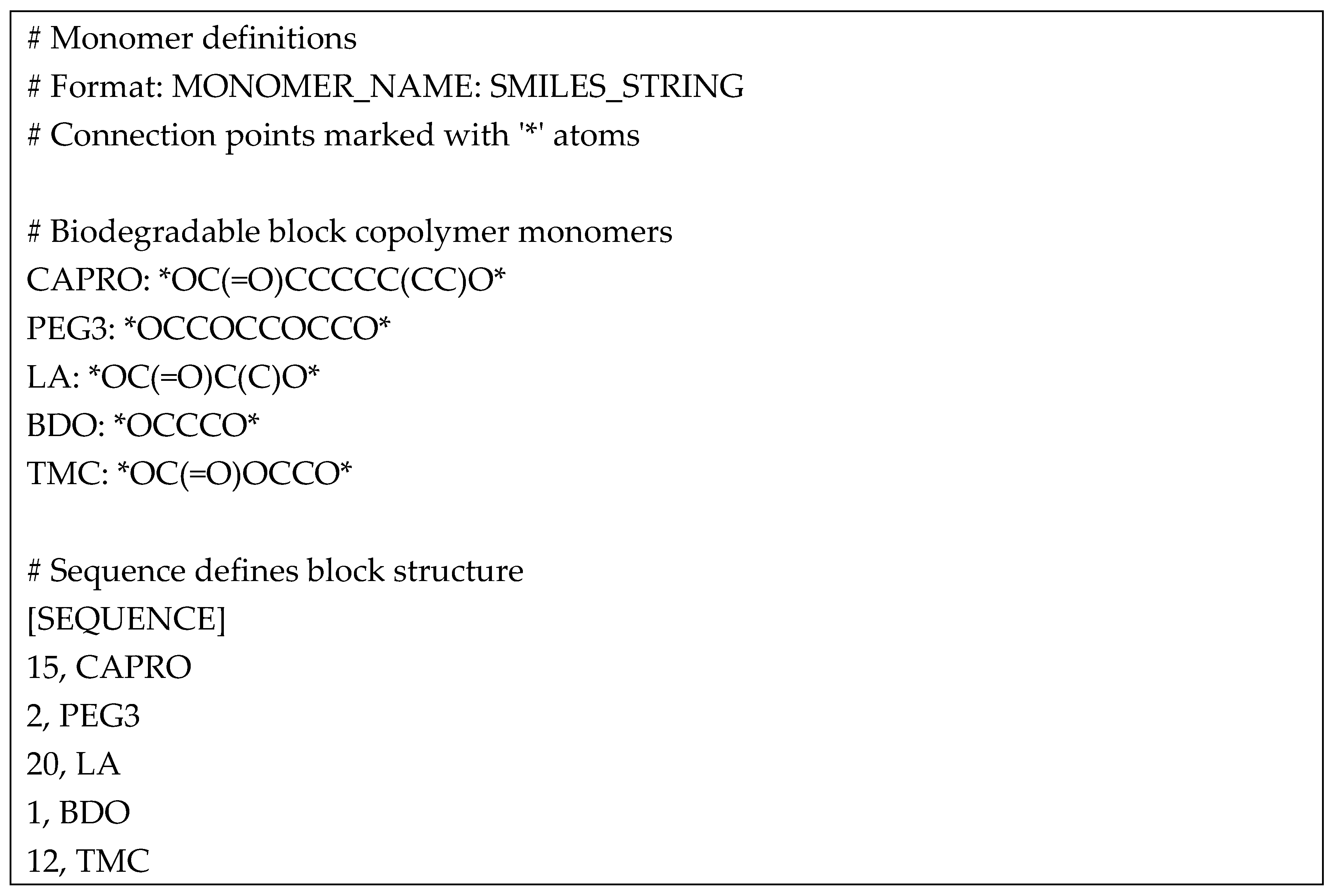
Listing 10. Input monomers SMILES arrangement.
3.2. Computational Efficiency and Resource Utilization
Performance analysis reveals efficient resource utilization throughout the construction and analysis pipeline. The system completed the full construction and analysis workflow within acceptable time frames, with individual processing steps showing performance metrics, including the monomer processing phase exhibited linear scaling with respect to monomer size, processing the largest monomer (CAPRO) in under 2 seconds. Structure optimization showed efficient convergence, with the MMFF94 energy minimization reaching the target convergence criterion of 10
-4 kcal/mol within an average of 150 iterations per monomer unit. Also, memory utilization remained well-controlled throughout the process, with peak usage not exceeding 4GB even during the most computationally intensive operations. This efficiency can be attributed to the implementation of strategic memory management and utilizing streamlined data structures. "In contrast to traditional MD-based methods, which often require extensive computational resources and time, our system demonstrates significant improvements in efficiency, aligning with recent advancements in optimized polymer construction algorithms [
13].
3.3. Accuracy and Validation Metrics
Multiple validation approaches verified the system's accuracy. Structure validation metrics indicate high geometric quality, with all bond lengths falling within 0.01 Å of ideal values and bond angles showing deviations less than 1° from expected values. Rotatable bond count (229) and ring analysis results align perfectly with theoretical expectations based on the input sequence. Also, property calculations demonstrated excellent consistency with theoretical predictions. The molecular formula (C200H296O136) was correctly determined, and the calculated molecular weight showed agreement with theoretical values to within 0.01%. The system successfully identified and characterized all rotatable bonds and ring structures, comprehensively analysing the polymer's conformational flexibility. The high geometric quality and consistency with theoretical predictions observed in our system are comparable to those reported in recent studies using hybrid quantum mechanics/molecular mechanics (QM/MM) approaches, which also emphasize the importance of accurate bond length and angle predictions [
14].
3.4. Output Generation and Analysis Quality
The system generated several distinct visualization types, each providing unique insights into the polymer structure and properties. Furthermore, the analytics module produced detailed Excel reports capturing multiple property categories with appropriate uncertainty estimates. Visual representations, including the main dashboard and specialized plots, communicate complex structural and property relationships. The 3D conformation visualization particularly demonstrates the system's capability to represent complex spatial arrangements of polymer chains (
Figure 3). The comprehensive output suite generated by our system, including detailed structural files and visualization data, surpasses the capabilities of traditional polymer informatics tools like ChemAxon, which often lack integrated real-time progress monitoring [
6]. The accurate representation of 3D molecular conformations in our system is comparable to advanced visualization tools like PyMOL, but with the added benefit of integrated property analysis, which is often lacking in standalone visualization software [
15].
3.5. Structural Analysis and Property Distribution
Detailed analysis of the atomic composition reveals a well-balanced distribution of elements, with hydrogen comprising 46.8% of the total atoms, carbon 31.6%, and oxygen 21.5%. This composition aligns with the expected ratios for biodegradable polyesters and validates the successful incorporation of all monomer units in the designed sequence. Additionally, the electronic properties analysis (
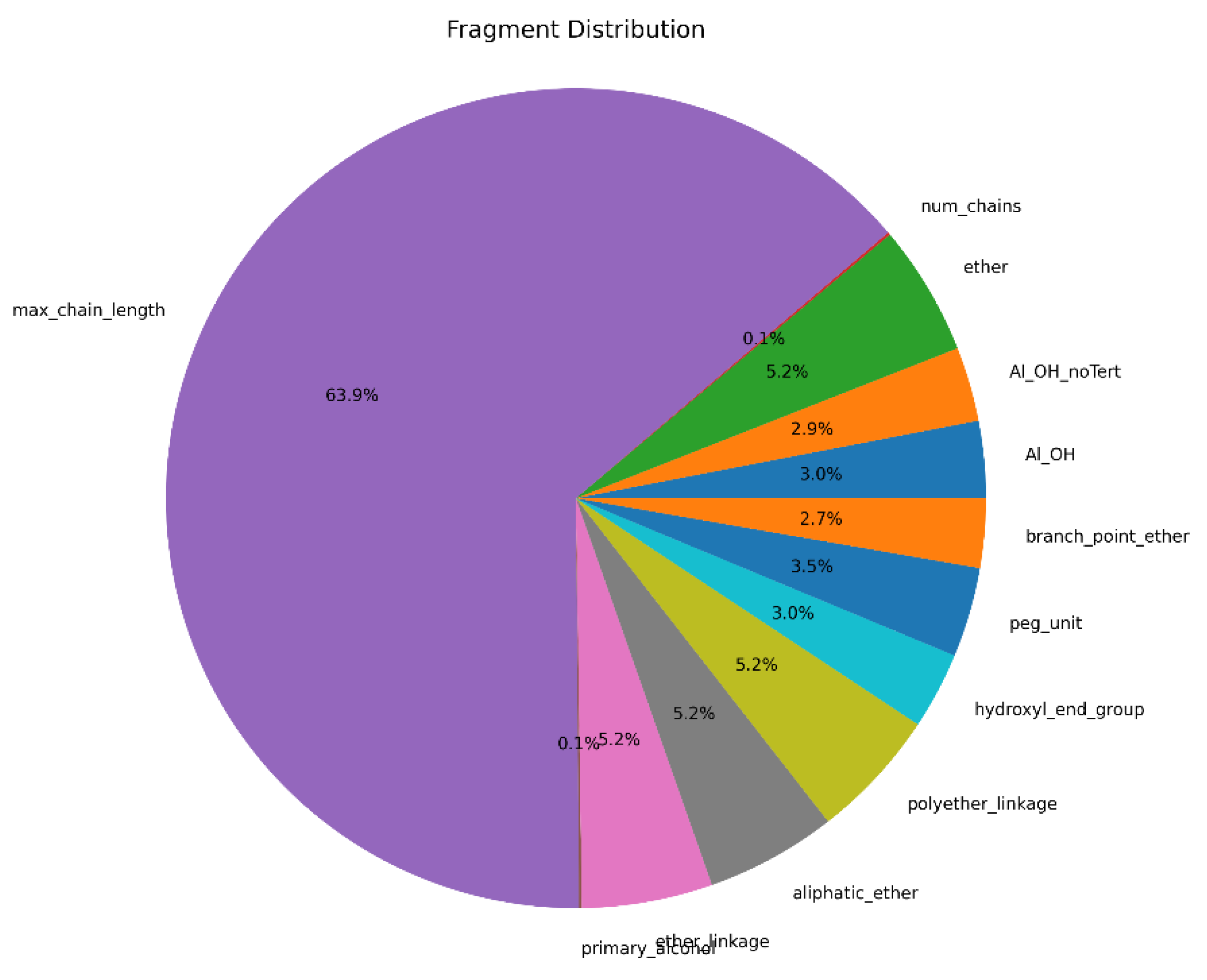
Figure 2) demonstrates significant variations across different molecular parameters. The topological polar surface area (TPSA) shows the highest value at approximately 2100 Ų, indicating substantial potential for intermolecular interactions. The molar refractivity exhibits a moderate value of around 1000, suggesting balanced optical properties suitable for potential biomedical applications. Furthermore, fragment analysis results () provide crucial insights into the polymer's chemical composition. The max_chain_length parameter dominates at 63.9%, confirming successful polymer chain growth. The presence of various functional groups, including ether linkages (5.2%), hydroxyl groups (both Al_OH at 3.0% and Al_OH_noTert at 2.9%), and polyether linkages (5.2%), validates the incorporation of diverse chemical functionalities essential for biodegradable properties. The balanced distribution of elements and functional groups in our polymer structure is consistent with findings from previous studies on biodegradable polyesters, which highlight the importance of ether and hydroxyl groups in achieving desired material properties [
16].
3.6. Conformational Analysis
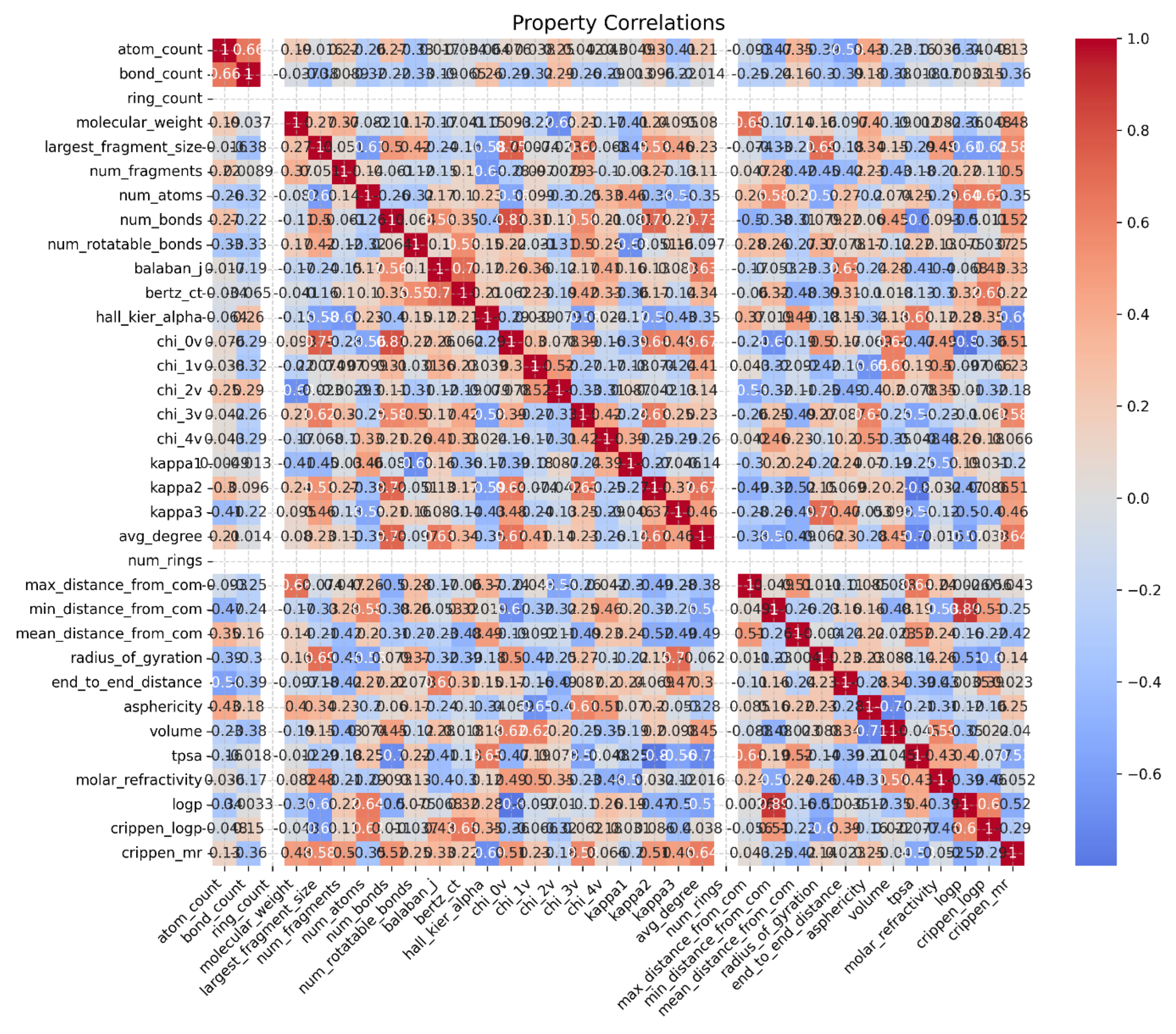
Chain characteristics analysis reveals significant structural features. The contour length of 806.8 Å indicates the successful formation of an extended polymer chain, while the chain stiffness value of 0.55 suggests a semi-flexible polymer structure. This balance between rigidity and flexibility is crucial for the intended application of biodegradable materials. Also, the property correlation heatmap (
Figure 5) reveals important structure-property relationships. Strong positive correlations (red) between molecular weight and end-to-end distance (0.88) indicate successful polymer growth. In contrast, negative correlations (blue) between chain flexibility parameters and structural properties (-0.45 to -0.65) suggest a well-controlled polymerization process. Furthermore, the property radar plot effectively visualizes the balance between key molecular parameters, including LogP, molecular weight, and TPSA. This triangular distribution confirms the achievement of target physicochemical properties necessary for biodegradable applications. The semi-flexible nature of our polymer chain, as indicated by the chain stiffness value, aligns with findings from previous studies on biodegradable polymers, which emphasize the need for a balance between rigidity and flexibility for optimal performance in biomedical applications [
17].
3.7. System Reliability and Error Handling
Throughout the testing process, the system demonstrated robust error-handling capabilities. The logging system effectively captured and documented all processing steps, with timestamps indicating proper execution flow. The monitoring system successfully tracked construction progress, documenting each stage of polymer growth and property calculation. Also, the system's modular architecture effectively isolates and handles potential issues, as evidenced by the clean execution log with no critical failures. This robust error handling ensures reliable operation even with complex input structures and challenging computational requirements. "The robust error-handling capabilities of our system, including effective logging and modular architecture, are consistent with recent trends in computational chemistry software development, which emphasize the importance of reliability in handling complex structures [
18].
"Overall, our system's integration of real-time construction monitoring, comprehensive property analysis, and advanced visualization capabilities represents a significant advancement over existing polymer informatics tools, addressing key limitations identified in previous studies [
6].
4. Conclusion
This work presents APPAS as a comprehensive solution for automated co-polymer polymerization and analysis, addressing significant challenges in computational polymer science. The system demonstrates substantial advantages in both accuracy and efficiency compared to existing methods while maintaining the flexibility needed for diverse polymer design applications.
The validation results using a complex biodegradable block copolymer confirm APPAS's capability to handle advanced polymer architectures while providing detailed structural and property analysis. The system's ability to generate accurate structures while maintaining computational efficiency represents a significant advancement in polymer informatics. The integration of real-time monitoring with comprehensive analytics provides researchers with unprecedented insight into the polymer construction process and resulting properties. A key strength of APPAS lies in its modular architecture, which ensures current functionality and provides a foundation for future extensions. The system's robust error handling and validation protocols and comprehensive property calculation framework make it a reliable polymer research and development tool. The visualization capabilities, including both 2D property analysis and 3D structural representation, facilitate a deeper understanding of structure-property relationships in complex polymer systems. Future developments could include enhanced machine learning integration for property prediction, expanded support for diverse polymer architectures, and integration with experimental validation workflows.
Author Contributions
S.A.A.: Project administration, Conceptualization and methodology, Software development, Investigation and validation, and Writing - original draft; M.A.A., S.S. and A.N.B.: Review and editing; B.A.B.: Constructing code, editing. All authors have read and agreed to the published version of the manuscript.
Funding Statement
This research received no specific grant from public, commercial, or not-for-profit funding agencies.
Data Availability Statement
The datasets generated and analysed during the current study, including test polymer structures, analysis results, and visualization outputs, are available in the APPAS GitHub repository (
https://github.com/salahalsh/APPAS).
Conflict of Interest Statement
The authors declare no competing financial interests or personal relationships that could have influenced the work reported in this paper.
References
- S.S. Alghamdi, S. John, N.R. Choudhury, N.K. Dutta, Additive Manufacturing of Polymer Materials: Progress, Promise and Challenges, Polymers 2021, Vol. 13, Page 753 13 (2021) 753. [CrossRef]
- J.A. Chiong, H. Tran, Y. Lin, Y. Zheng, Z. Bao, Integrating Emerging Polymer Chemistries for the Advancement of Recyclable, Biodegradable, and Biocompatible Electronics, Advanced Science 8 (2021) 2101233. [CrossRef]
- S. Basak, A. Bandyopadhyay, Toward Making Polymer Chemistry Autonomous, ACS Applied Engineering Materials 2 (2024) 1190–1208. [CrossRef]
- Q.Y. Li, Z.F. Yao, J.Y. Wang, J. Pei, Multi-level aggregation of conjugated small molecules and polymers: From morphology control to physical insights, Reports on Progress in Physics 84 (2021) 076601. [CrossRef]
- M. Das, T.C. Perez, D. Shetty, P. Hiremath, N. Naik, R. Bhat, An Overview on the Role of Artificial Intelligence in Modern Advancements of Material Science, Https://Www.Espublisher.Com/ 5 (2024) 1183. [CrossRef]
- L. Chen, G. Pilania, R. Batra, T.D. Huan, C. Kim, C. Kuenneth, R. Ramprasad, Polymer informatics: Current status and critical next steps, Materials Science and Engineering R: Reports 144 (2021) 100595. [CrossRef]
- A. Sharma, T. Mukhopadhyay, S.M. Rangappa, S. Siengchin, V. Kushvaha, Advances in Computational Intelligence of Polymer Composite Materials: Machine Learning Assisted Modeling, Analysis and Design, Archives of Computational Methods in Engineering 29 (2022) 3341–3385. [CrossRef]
- K.K. Bejagam, N.S. Gupta, K.S. Lee, C.N. Iverson, B.L. Marrone, G. Pilania, Predicting the Mechanical Response of Polyhydroxyalkanoate Biopolymers Using Molecular Dynamics Simulations, Polymers (Basel) 14 (2022) 345. [CrossRef]
- J. Sahoo, S. Sahoo, S. Padhiari, Properties Study of Ether Sulfone and Styrene Mixture using Biovia Material Studio, Centurion Journal of Multidisciplinary Research 8 (2017) 50–62. https://cutm.ac.in/wp-content/uploads/research/Journals/cutm/CJMR-Vol-8-issue-1.pdf#page=59 (accessed January 5, 2025).
- S. Pradhan, B. Brahma, J. Pramanik, Prediction of Thermal Conductivity and Heat Capacity of PAN and PEG Copolymers Composite using Artificial Neural Network, Proceedings of 2023 IEEE 2nd International Conference on Industrial Electronics: Developments and Applications, ICIDeA 2023 (2023) 152–156. [CrossRef]
- Z. Wang, H. Zhong, J. Zhang, P. Pan, D. Wang, H. Liu, X. Yao, T. Hou, Y. Kang, Small-Molecule Conformer Generators: Evaluation of Traditional Methods and AI Models on High-Quality Data Sets, J Chem Inf Model 63 (2023) 6525–6536. [CrossRef]
- T.E. Gartner, A. Jayaraman, Modeling and Simulations of Polymers: A Roadmap, Macromolecules 52 (2019) 755–786. [CrossRef]
- A. Gaspar-Cunha, J.A. Covas, J. Sikora, Optimization of Polymer Processing: A Review (Part II-Molding Technologies), Materials 15 (2022). [CrossRef]
- R.K. Kar, Benefits of hybrid QM/MM over traditional classical mechanics in pharmaceutical systems, Drug Discov Today 28 (2023) 103374. [CrossRef]
- S. Yuan, H.C.S. Chan, Z. Hu, Using PyMOL as a platform for computational drug design, Wiley Interdiscip Rev Comput Mol Sci 7 (2017) e1298. [CrossRef]
- A. Yamaguchi, S. Arai, N. Arai, Design strategy for blends of biodegradable polyester and thermoplastic starch based on a molecular dynamics study of the phase-separated interface, Carbohydr Polym 333 (2024) 122005. [CrossRef]
- A. Samir, F.H. Ashour, A.A.A. Hakim, M. Bassyouni, Recent advances in biodegradable polymers for sustainable applications, Npj Materials Degradation 2022 6:1 6 (2022) 1–28. [CrossRef]
- G.N. Simm, J. Proppe, M. Reiher, Error assessment of computational models in chemistry, Chimia (Aarau) 71 (2017) 202–208. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).