Submitted:
25 May 2025
Posted:
26 May 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
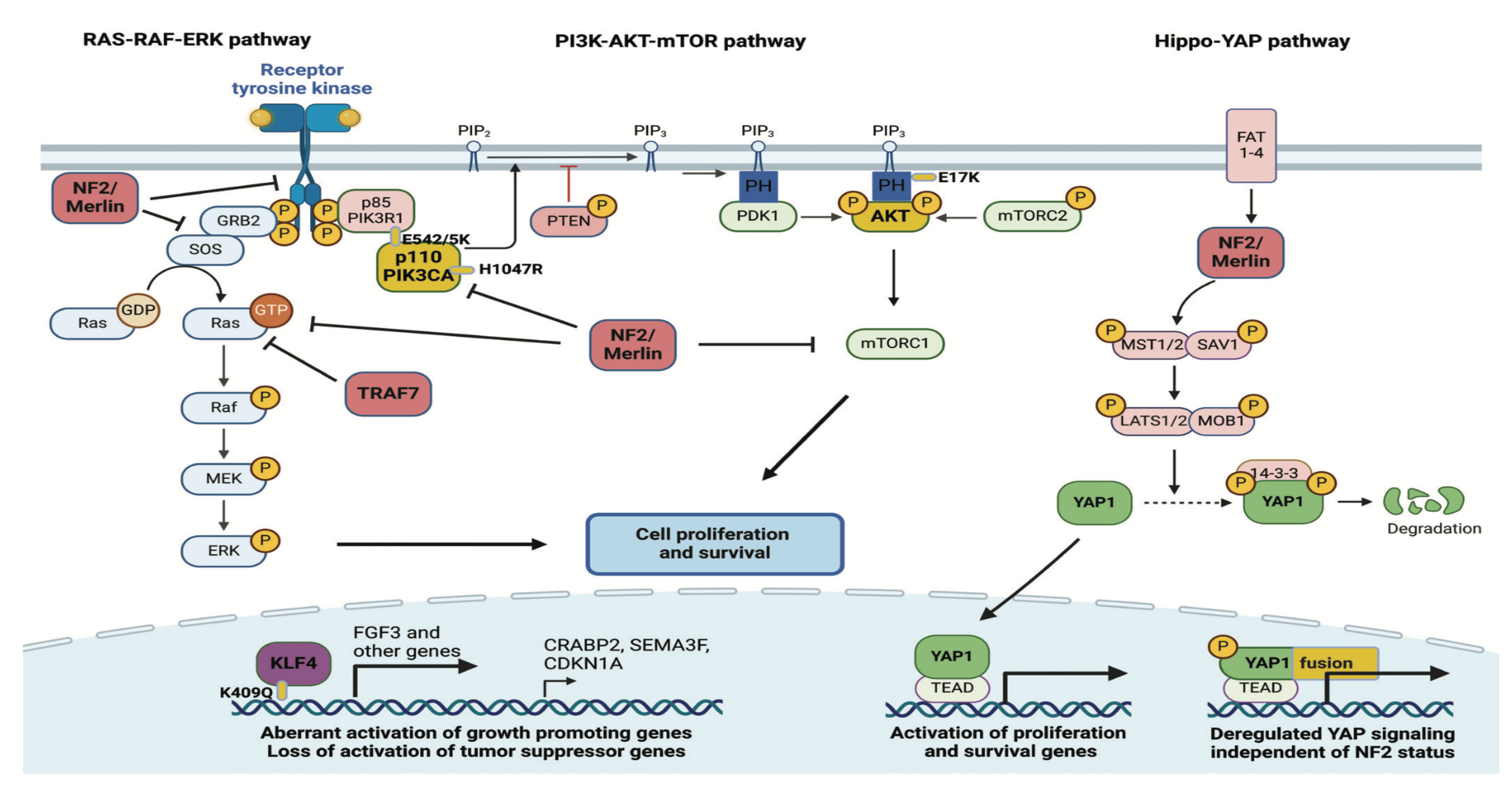
1.1. Meningioma
1.1.1. World Health Organization (WHO) Classification
1.1.2. Historical perspective
began to lose his mind gradually over a two year period, to such an extent that finally he was completely stupefied and did nothing rationally. He had no desire for food and ate only when forcibly fed. ... Finally after matters had gone on thus for six months, he died.
a round fleshy tumor, like an acorn. It was hard and full of holes and was as large as a medium-sized apple. It was covered with its own membrane and was entwined with veins. However, it was free of all connections with the matter of the brain, so much so that when it was removed by hand, it left behind a remarkable cavity.
1.1.3. Pathophysiology
1.2. Ion Channels
1.3. The Discovery of Aquaporins and the 2003 Nobel Committee’s Decisions
In other words the child we first detected was recognized as having the predicted qualities only in 1992. In 1993 CHIP28 was renamed aquaporin 1. Looking in retrospect, asking the crucial question, when was the first water channel protein, aquaporin 1, discovered, a fair and clear cut answer would be: the first water channel protein, now called aquaporin 1, was identified or "seen" in situ in the human RBC membrane by Benga and coworkers in 1986. It was again "seen" when it was by chance purified by Agre and coworkers in 1988 and was again identified when its main feature, the water transport property was found by Agre and coworkers in 1992.
Agre’s group did include a reference to Benga’s work in their Science paper; but this reference was only to a 1983 paper on protease resistance of "water channels" (which was relevant) and not the pertinent 1986 Biochemistry paper, or even the subsequent publications. The report of the recent exciting finding of possible involvement of aquaporins in epilepsy, published in 2005 in Proc Natl Acd Sci USA by a group including Agre failed to cite Benga and Morariu’s novel and startling report in Nature in 1977.
1.4. Aquaporin 3 (AQP-3)
1.5. Insight Behind the Work
1.6. Combinatorial Search Problem and a Possible Solution
2. Methods
2.1. Static Data from [1]
2.2. Design for Static Data from [1]
2.3. Translating the Pipeline Into Code of Execution in R
2.4. Regarding Biological Insight
3. Results & Discussion
3.1. AQP3 Related Synergies
3.1.1. AQP3—Individual Genes
4. Conclusions
Author Contributions
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patel, A.J.; Wan, Y.W.; Al-Ouran, R.; Revelli, J.P.; Cardenas, M.F.; Oneissi, M.; Xi, L.; Jalali, A.; Magnotti, J.F.; Muzny, D.M.; et al. Molecular profiling predicts meningioma recurrence and reveals loss of DREAM complex repression in aggressive tumors. Proceedings of the National Academy of Sciences 2019, 116, 21715–21726. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S. Machine learning ranking of plausible (un) explored synergistic gene combinations using sensitivity indices of time series measurements of Wnt signaling pathway. Integrative Biology 2024, 16, zyae020. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro-oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Torp, S.H.; Solheim, O.; Skjulsvik, A.J. The WHO 2021 Classification of Central Nervous System tumours: a practical update on what neurosurgeons need to know—a minireview. Acta Neurochirurgica 2022, 164, 2453–2464. [Google Scholar] [CrossRef]
- Czarnetzki, A.; Schwaderer, E.; Pusch, C.M. Fossil record of meningioma. The Lancet 2003, 362, 408. [Google Scholar] [CrossRef]
- Okonkwo, D.O.; Laws Jr, E.R. Meningiomas: historical perspective. In Meningiomas; Springer, 2009; pp. 3–10.
- Paterniti, S. Meningiomas surgery in Italy in the nineteenth century: Historical review. Austin Neurosurg Open Access 2015, 2, 1030. [Google Scholar]
- Pecchioli, Z. Soria di un fungo della dura madre, operato coll’estirpazione dal Professor Zanobi Pecchioli. Nuovo Giornale de’Letterati—Scienze 1838, 36, 39–44. [Google Scholar]
- Cushing, H. The meningiomas (dural endotheliomas): their source, and favoured seats of origin. Brain 1922, 45, 282–316. [Google Scholar] [CrossRef]
- contributors, W. Meningioma — Wikipedia, The Free Encyclopedia, 2024. Online; accessed 11-April-2025.
- Pamir, M.N.; Black, P.M.; Fahlbusch, R. Meningiomas : a comprehensive text; Elsevier Health Sciences, 2010.
- Szulzewsky, F.; Thirimanne, H.N.; Holland, E.C. Meningioma: current updates on genetics, classification, and mouse modeling. Upsala Journal of Medical Sciences 2024, 129, 10–48101. [Google Scholar] [CrossRef]
- Verkman, A.; Mitra, A.K. Structure and function of aquaporin water channels. American Journal of Physiology-Renal Physiology 2000, 278, F13–F28. [Google Scholar] [CrossRef]
- Benga, G.; Popescu, O.; Pop, V.I.; Holmes, R.P. p-(Chloromercuri) benzenesulfonate binding by membrane proteins and the inhibition of water transport in human erythrocytes. Biochemistry 1986, 25, 1535–1538. [Google Scholar] [CrossRef] [PubMed]
- Denker, B.M.; Smith, B.L.; Kuhajda, F.P.; Agre, P. Identification, purification, and partial characterization of a novel Mr 28,000 integral membrane protein from erythrocytes and renal tubules. Journal of biological chemistry 1988, 263, 15634–15642. [Google Scholar] [CrossRef] [PubMed]
- Preston, G.M.; Carroll, T.P.; Guggino, W.B.; Agre, P. Appearance of water channels in Xenopus oocytes expressing red cell CHIP28 protein. Science 1992, 256, 385–387. [Google Scholar] [CrossRef] [PubMed]
- Agre, P.; Preston, G.M.; Smith, B.L.; Jung, J.S.; Raina, S.; Moon, C.; Guggino, W.B.; Nielsen, S. Aquaporin CHIP: the archetypal molecular water channel. American Journal of Physiology-Renal Physiology 1993, 265, F463–F476. [Google Scholar] [CrossRef]
- Benga, G. Birth of water channel proteins—the aquaporins. Cell Biology International 2003, 27, 701–709. [Google Scholar] [CrossRef]
- Knepper, M.A.; Nielsen, S. Peter Agre, 2003 Nobel Prize winner in chemistry. Journal of the American Society of Nephrology 2004, 15, 1093–1095. [Google Scholar] [CrossRef]
- Kuchel, P. The story of the discovery of aquaporins: convergent evolution of ideas—but who got there first. Cell Mol Biol (Noisy-le-grand) 2006, 52, 2–5. [Google Scholar]
- Ishibashi, K.; Sasaki, S.; Fushimi, K.; Uchida, S.; Kuwahara, M.; Saito, H.; Furukawa, T.; Nakajima, K.; Yamaguchi, Y.; Gojobori, T. Molecular cloning and expression of a member of the aquaporin family with permeability to glycerol and urea in addition to water expressed at the basolateral membrane of kidney collecting duct cells. Proceedings of the National Academy of Sciences 1994, 91, 6269–6273. [Google Scholar] [CrossRef]
- Ishibashi, K.; Sasaki, S.; Saito, F.; Ikeuchi, T.; Marumo, F. Structure and chromosomal localization of a human water channel (AQP3) gene. Genomics 1995, 27, 352–354. [Google Scholar] [CrossRef]
- Inase, N.; Fushimi, K.; Ishibashi, K.; Uchida, S.; Ichioka, M.; Sasaki, S.; Marumo, F. Isolation of human aquaporin 3 gene. Journal of Biological Chemistry 1995, 270, 17913–17916. [Google Scholar] [CrossRef]
- Mulders, S.; Olde Weghuis, D.; Van Boxtel, J.; Geurts van Kessel, A.; Echevarria, M.; Van Os, C.; Deen, P. Localization of the human gene for aquaporin 3 (AQP3) to chromosome 9, region p21→ p12, using fluorescent in situ hybridization. Cytogenetic and Genome Research 1996, 72, 303–305. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Song, Y.; Yang, B.; Gillespie, A.; Carlson, E.J.; Epstein, C.J.; Verkman, A. Nephrogenic diabetes insipidus in mice lacking aquaporin-3 water channels. Proceedings of the National Academy of Sciences 2000, 97, 4386–4391. [Google Scholar]
- Roudier, N.; Ripoche, P.; Gane, P.; Le Pennec, P.Y.; Daniels, G.; Cartron, J.P.; Bailly, P. AQP3 deficiency in humans and the molecular basis of a novel blood group system, GIL. Journal of Biological Chemistry 2002, 277, 45854–45859. [Google Scholar] [CrossRef] [PubMed]
- Tsitsikov, E.N.; Hameed, S.; Tavakol, S.A.; Stephens, T.M.; Tsytsykova, A.V.; Garman, L.; Bi, W.L.; Dunn, I.F. Specific gene expression signatures of low grade meningiomas. Frontiers in Oncology 2023, 13, 1126550. [Google Scholar]
- Joachims, T. Training linear SVMs in linear time. In Proceedings of the Proceedings of the 12th ACM SIGKDD international conference on Knowledge discovery and data mining. ACM, 2006, pp. 217–226.
- Pujol, G.; Iooss, B.; Janon, A.; Boumhaout, K.; Da Veiga, S.; Fruth, J.; Gilquin, L.; Guillaume, J.; Le Gratiet, L.; Lemaitre, P.; et al. Sensitivity: global sensitivity analysis of model outputs. R package version 2017, 1. [Google Scholar]
- Huang, H.; Liu, Y.; Yuan, M.; Marron, J. Statistical significance of clustering using soft thresholding. Journal of Computational and Graphical Statistics 2015, 24, 975–993. [Google Scholar] [CrossRef]
- Sinha, S. Hilbert-Schmidt and Sobol sensitivity indices for static and time series Wnt signaling measurements in colorectal cancer-part A. BMC systems biology 2017, 11, 120. [Google Scholar] [CrossRef]
- Team, R.C. R language definition. Vienna, Austria: R foundation for statistical computing 2000, 3, 116. [Google Scholar]
| Ranking Individual members VS AQP3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ranking of Individual members w.r.t AQP3 | |||||||||
| laplace | linear | rbf | jansen | laplace | linear | rbf | jansen | ||
| USH1C - AQP3 | 245 | 205 | 200 | 15 | MCOLN3 - AQP3 | 235 | 194 | 202 | 1 |
| ERBB3 - AQP3 | 189 | 191 | 168 | 19 | GAL - AQP3 | 180 | 161 | 134 | 161 |
| ST6GALNAC2 - AQP3 | 177 | 163 | 158 | 63 | CAPN6 - AQP3 | 201 | 177 | 148 | 34 |
| IGSF9 - AQP3 | 49 | 154 | 137 | 156 | ABCB1 - AQP3 | 249 | 176 | 187 | 45 |
| PTHLH - AQP3 | 155 | 174 | 227 | 38 | PITPNM3 - AQP3 | 148 | 178 | 201 | 174 |
| TLL2 - AQP3 | 204 | 175 | 189 | 62 | NTSR1 - AQP3 | 194 | 200 | 205 | 12 |
| MSLN - AQP3 | 227 | 202 | 203 | 14 | ANXA13 - AQP3 | 221 | 167 | 157 | 149 |
| RAPGEFL1 - AQP3 | 173 | 72 | 149 | 189 | PRMT8 - AQP3 | 184 | 193 | 173 | 59 |
| IL1RL1 - AQP3 | 176 | 170 | 164 | 10 | IRF6 - AQP3 | 198 | 148 | 169 | 126 |
| PLPPR4 - AQP3 | 214 | 189 | 197 | 29 | CNNM1 - AQP3 | 219 | 100 | 144 | 218 |
| PCSK2 - AQP3 | 212 | 201 | 209 | 2 | KLK8 - AQP3 | 242 | 206 | 198 | 26 |
| PRRG3 - AQP3 | 163 | 190 | 165 | 6 | BICDL1 - AQP3 | 196 | 197 | 170 | 68 |
| FXYD6 - AQP3 | 164 | 160 | 208 | 49 | EGF - AQP3 | 168 | 88 | 163 | 184 |
| MAEL - AQP3 | 208 | 203 | 235 | 135 | ILDR2 - AQP3 | 255 | 243 | 210 | 81 |
| SUSD4 - AQP3 | 188 | 183 | 190 | 17 | GADL1 - AQP3 | 203 | 192 | 154 | 28 |
| SLC26A7 - AQP3 | 236 | 220 | 242 | 85 | CPAMD8 - AQP3 | 152 | 142 | 69 | 152 |
| COX6B2 - AQP3 | 161 | 210 | 185 | 13 | SGPP2 - AQP3 | 207 | 180 | 138 | 61 |
| GABRG1 - AQP3 | 202 | 165 | 250 | 9 | NPY1R - AQP3 | 145 | 173 | 177 | 77 |
| HCN1 - AQP3 | 232 | 214 | 152 | 67 | DEFB1 - AQP3 | 266 | 276 | 221 | 47 |
| WNK2 - AQP3 | 256 | 179 | 229 | 51 | PHYHIPL - AQP3 | 273 | 256 | 260 | 229 |
| SLC16A9 - AQP3 | 182 | 248 | 222 | 129 | PPP1R36 - AQP3 | 265 | 265 | 248 | 141 |
| CACNB2 - AQP3 | 226 | 299 | 289 | 148 | SPINT1 - AQP3 | 303 | 273 | 278 | 132 |
| SCN3B - AQP3 | 248 | 240 | 274 | 158 | USP54 - AQP3 | 283 | 267 | 301 | 230 |
| SERPINB8 - AQP3 | 286 | 307 | 286 | 296 | CCDC68 - AQP3 | 222 | 215 | 186 | 40 |
| PLEKHA7 - AQP3 | 274 | 264 | 297 | 274 | ANPEP - AQP3 | 263 | 249 | 287 | 133 |
| MYO1A - AQP3 | 247 | 209 | 257 | 114 | SGSM1 - AQP3 | 297 | 304 | 293 | 232 |
| GPD1 - AQP3 | 190 | 287 | 194 | 136 | DNAAF3 - AQP3 | 251 | 241 | 249 | 159 |
| HRASLS5 - AQP3 | 227 | 222 | 217 | 37 | BDKRB2 - AQP3 | 233 | 232 | 232 | 24 |
| PHYHIP - AQP3 | 199 | 212 | 243 | 32 | NSG1 - AQP3 | 254 | 279 | 262 | 162 |
| PCSK9 - AQP3 | 224 | 223 | 228 | 27 | RAB3B - AQP3 | 239 | 251 | 245 | 137 |
| MUC3A - AQP3 | 191 | 207 | 223 | 11 | OTUD7A - AQP3 | 261 | 298 | 267 | 255 |
| DLGAP1 - AQP3 | 268 | 271 | 211 | 131 | HS6ST2 - AQP3 | 179 | 219 | 214 | 22 |
| Ranking Individual members VS AQP3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ranking of Individual members w.r.t AQP3 | |||||||||
| laplace | linear | rbf | jansen | laplace | linear | rbf | jansen | ||
| CDC42BPG - AQP3 | 305 | 226 | 285 | 213 | KCNK3 - AQP3 | 223 | 235 | 226 | 36 |
| LPAR3 - AQP3 | 238 | 216 | 236 | 70 | ANO5 - AQP3 | 244 | 258 | 263 | 78 |
| CFAP46 - AQP3 | 279 | 270 | 281 | 278 | ANGPTL7 - AQP3 | 299 | 233 | 280 | 263 |
| ISG20 - AQP3 | 302 | 269 | 276 | 172 | MBOAT1 - AQP3 | 294 | 229 | 305 | 270 |
| TPSAB1 - AQP3 | 170 | 246 | 246 | 58 | SPTLC3 - AQP3 | 272 | 277 | 303 | 287 |
| BPGM - AQP3 | 267 | 268 | 300 | 206 | CSDC2 - AQP3 | 230 | 259 | 247 | 44 |
| RCAN2 - AQP3 | 288 | 263 | 302 | 226 | RSPH9 - AQP3 | 253 | 261 | 265 | 199 |
| CES4A - AQP3 | 271 | 274 | 288 | 201 | PIFO - AQP3 | 282 | 296 | 290 | 195 |
| LINGO2 - AQP3 | 187 | 260 | 234 | 130 | SLCO2A1 - AQP3 | 262 | 244 | 283 | 209 |
| SLC35G1 - AQP3 | 277 | 282 | 396 | 256 | MYO1D - AQP3 | 220 | 234 | 266 | 75 |
| FAM91A1 - AQP3 | 301 | 306 | 271 | 252 | KCNA2 - AQP3 | 269 | 247 | 292 | 125 |
| LRRN4CL - AQP3 | 285 | 278 | 255 | 279 | GPR4 - AQP3 | 264 | 288 | 298 | 264 |
| RPRM - AQP3 | 195 | 218 | 179 | 52 | ERN1 - AQP3 | 287 | 257 | 307 | 234 |
| TMEM125 - AQP3 | 241 | 286 | 270 | 171 | WSCD1 - AQP3 | 281 | 275 | 295 | 275 |
| SYNM - AQP3 | 260 | 290 | 240 | 116 | CAPN12 - AQP3 | 304 | 245 | 296 | 186 |
| RIPK4 - AQP3 | 240 | 213 | 237 | 56 | FAM162B - AQP3 | 231 | 237 | 239 | 87 |
| ALDH1A3 - AQP3 | 216 | 227 | 224 | 80 | CCSER1 - AQP3 | 289 | 228 | 219 | 185 |
| BEX5 - AQP3 | 292 | 255 | 218 | 74 | OSBP2 - AQP3 | 259 | 293 | 241 | 120 |
| SDR42E1 - AQP3 | 211 | 231 | 277 | 124 | MAFF - AQP3 | 280 | 280 | 258 | 222 |
| RBM11 - AQP3 | 278 | 285 | 238 | 233 | TCN2 - AQP3 | 284 | 305 | 304 | 258 |
| LSAMP - AQP3 | 229 | 292 | 216 | 110 | PCLO - AQP3 | 215 | 238 | 233 | 41 |
| INSIG1 - AQP3 | 307 | 297 | 284 | 306 | NF2 - AQP3 | 275 | 291 | 264 | 181 |
| BCR - AQP3 | 246 | 302 | 268 | 257 | MPPED1 - AQP3 | 209 | 224 | 213 | 3 |
| FOXI2 - AQP3 | 186 | 252 | 225 | 4 | KRT16 - AQP3 | 228 | 230 | 231 | 35 |
| NCR3LG1 - AQP3 | 290 | 266 | 261 | 246 | PLA2G2A - AQP3 | 141 | 239 | 199 | 5 |
| FAM78B - AQP3 | 250 | 262 | 299 | 219 | FAT4 - AQP3 | 234 | 236 | 256 | 98 |
| SLC6A9 - AQP3 | 298 | 284 | 259 | 154 | LAMB3 - AQP3 | 293 | 281 | 279 | 109 |
| TPSB2 - AQP3 | 276 | 217 | 252 | 99 | ENPP1 - AQP3 | 291 | 295 | 251 | 153 |
| AFDN-DT - AQP3 | 296 | 303 | 291 | 280 | DCAF12L2 - AQP3 | 185 | 204 | 206 | 21 |
| SH3BGRL2 - AQP3 | 306 | 253 | 275 | 140 | ANKRD35 - AQP3 | 300 | 301 | 282 | 281 |
| SREBF2 - AQP3 | 270 | 289 | 294 | 282 | COL15A1 - AQP3 | 295 | 283 | 269 | 238 |
| C9orf129 - AQP3 | 252 | 272 | 273 | 96 | SEC14L6 - AQP3 | 237 | 294 | 244 | 107 |
| Unexplored combinatorial hypotheses | |
|---|---|
| Individual members w.r.t AQP3 | |
| USH1C / MCOLN3 / ERBB3 / GAL / ST6GALNAC2... | |
| CAPN6 / IGSF9 / ABCB1 / PTHLH / PITPNM3... | |
| TLL2 / NTSR1 / MSLN / ANXA13 / RAPGEFL1... | |
| PRMT8 / IL1RL1 / IRF6 / PLPPR4 / CNNM1... | |
| PCSK2 / KLK8 / PRRG3 / BICDL1 / FXYD6... | |
| EGF / MAEL / ILDR2 / SUSD4 / GADL1... | |
| SLC26A7 / CPAMD8 / COX6B2 / SGPP2 / GABRG1... | |
| NPY1R / HCN1 / DEFB1 / WNK2 / PHYHIPL... | |
| SLC16A9 / PPP1R36 / CACNB2 /SPINT1... | |
| SCN3B / USP54 / SERPINB8 / CCDC68 / PLEKHA7... | |
| ANPEP / MYO1A / SGSM1 / GPD1 / DNAAF3... | |
| HRASLS5 / BDKRB2 / PHYHIP / NSG1 / PCSK9... | |
| RAB3B / MUC3A / OTUD7A / DLGAP1 / HS6ST2... | |
| CDC42BPG / KCNK3 / LPAR3 / ANO5 / CFAP46... | |
| ANGPTL7 / ISG20 / MBOAT1 / TPSAB1 / SPTLC3... | |
| BPGM / CSDC2 / RCAN2 / RSPH9 / CES4A / PIFO... | |
| LINGO2 / SLCO2A1 / SLC35G1 / MYO1D / FAM91A1... | |
| KCNA2 / LRRN4CL / GPR4 / RPRM / ERN1 / TMEM125... | |
| WSCD1 / SYNM / CAPN12 / RIPK4 / FAM162B... | |
| ALDH1A3 / CCSER1 / BEX5 / OSBP2 / SDR42E1... | |
| MAFF / RBM11 / TCN2 / LSAMP / PCLO / INSIG1... | |
| NF2 / BCR / MPPED1 / FOXI2 / KRT16 / NCR3LG1... | |
| PLA2G2A / FAM78B / FAT4 / SLC6A9 / LAMB3 / TPSB2... | |
| ENPP1 / AFDN-DT / DCAF12L2 / SH3BGRL2 / ANKRD35... | |
| SREBF2 / COL15A1 / C9orf129 / SEC14L6 | AQP3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





