Submitted:
20 May 2025
Posted:
22 May 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
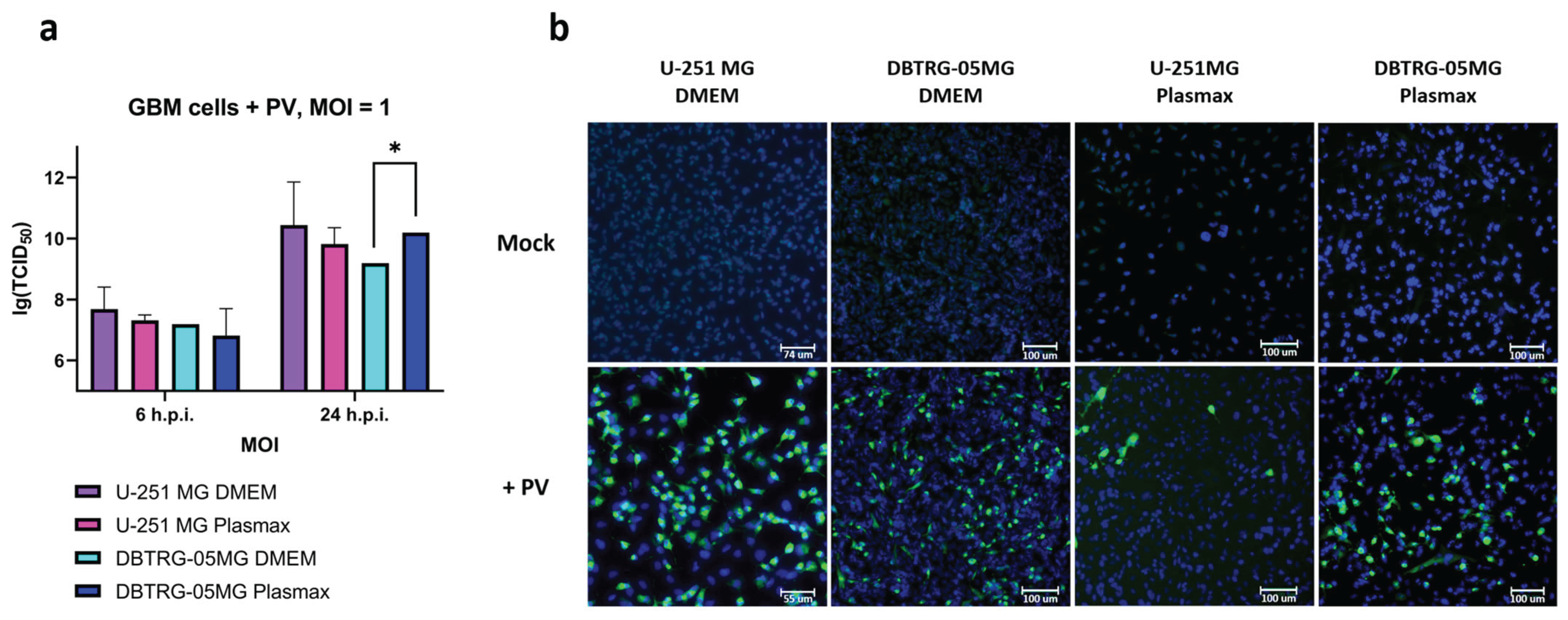
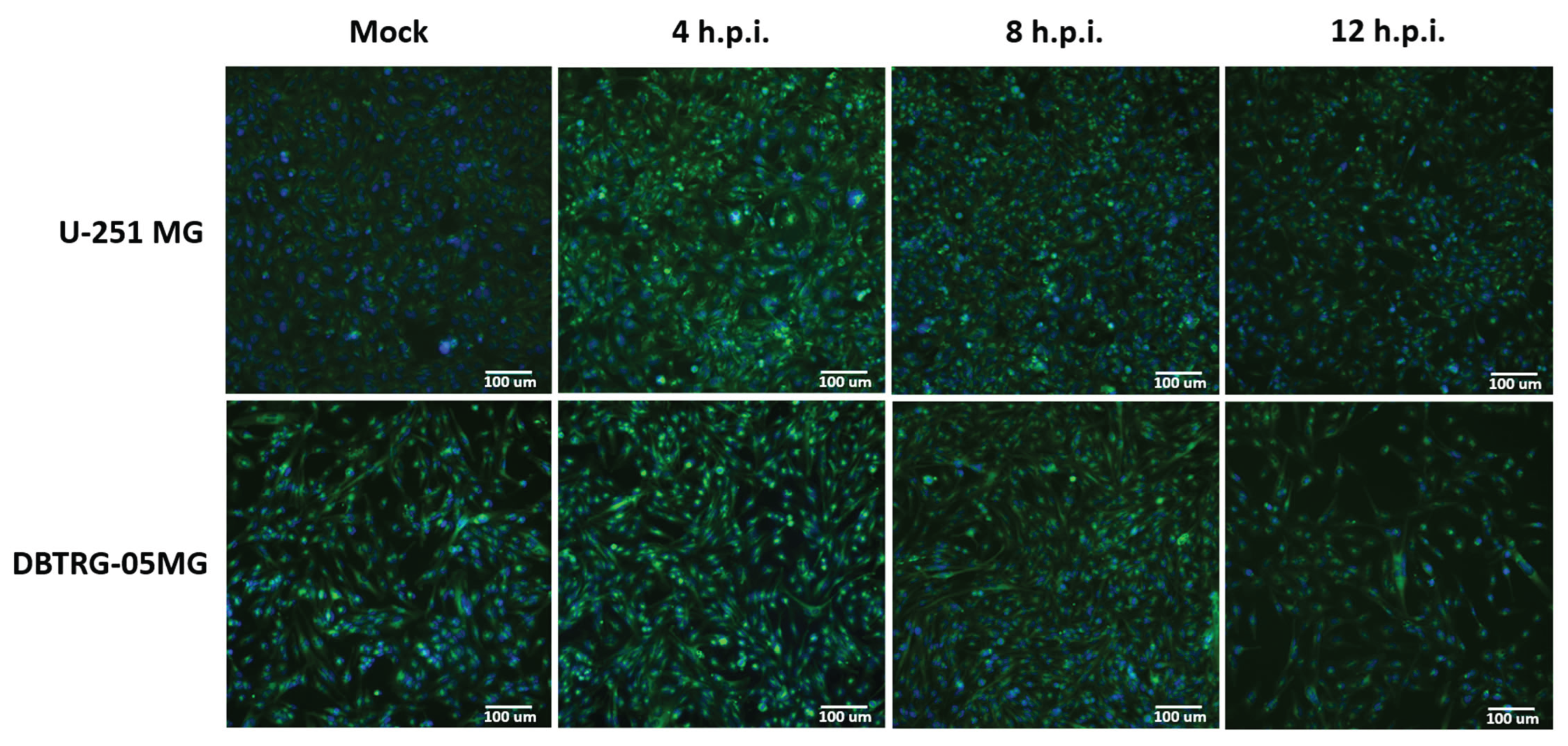
2.1. Kinetics of Poliovirus Replication in Glioblastoma Cells Maintained in Various Media
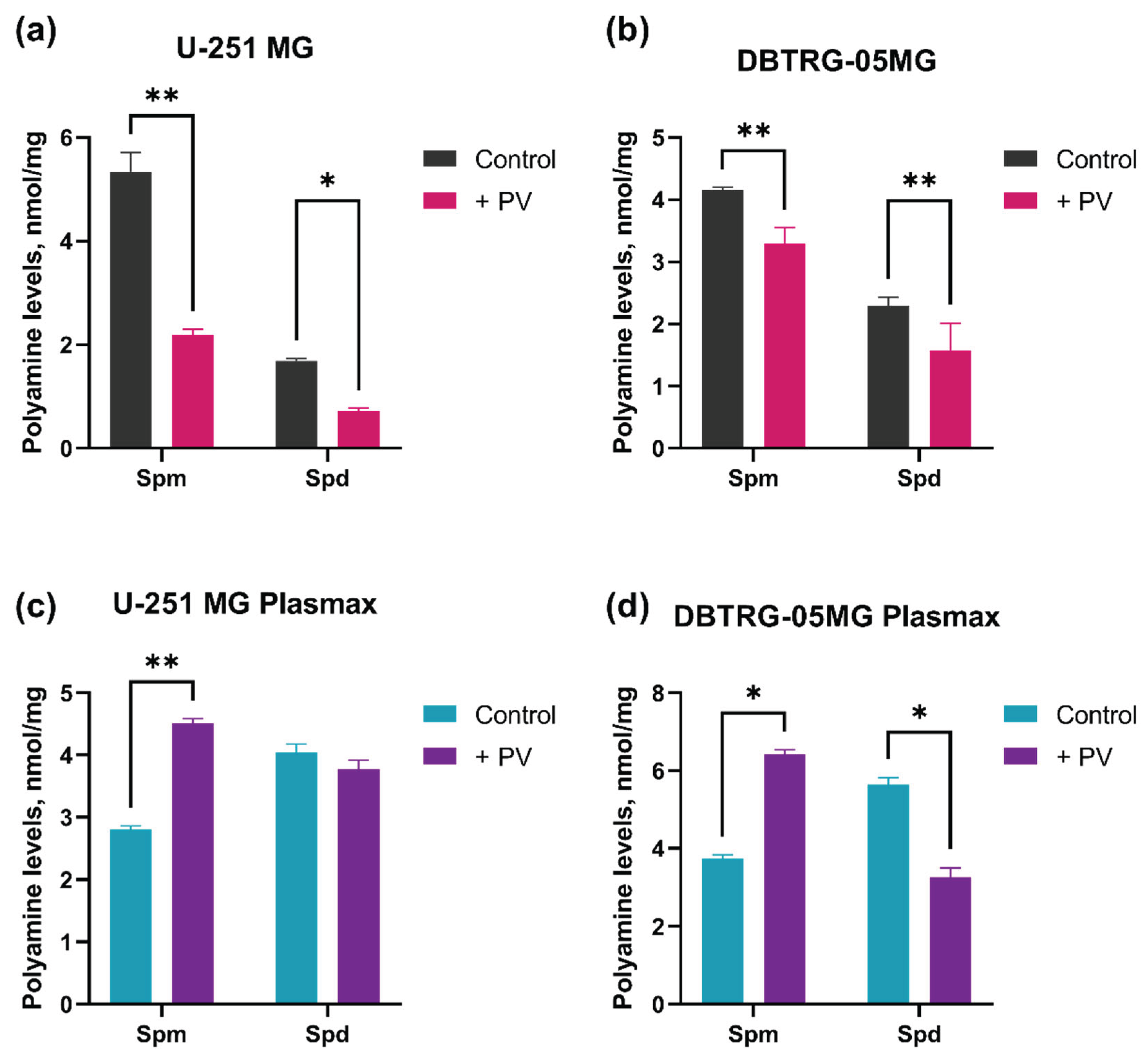
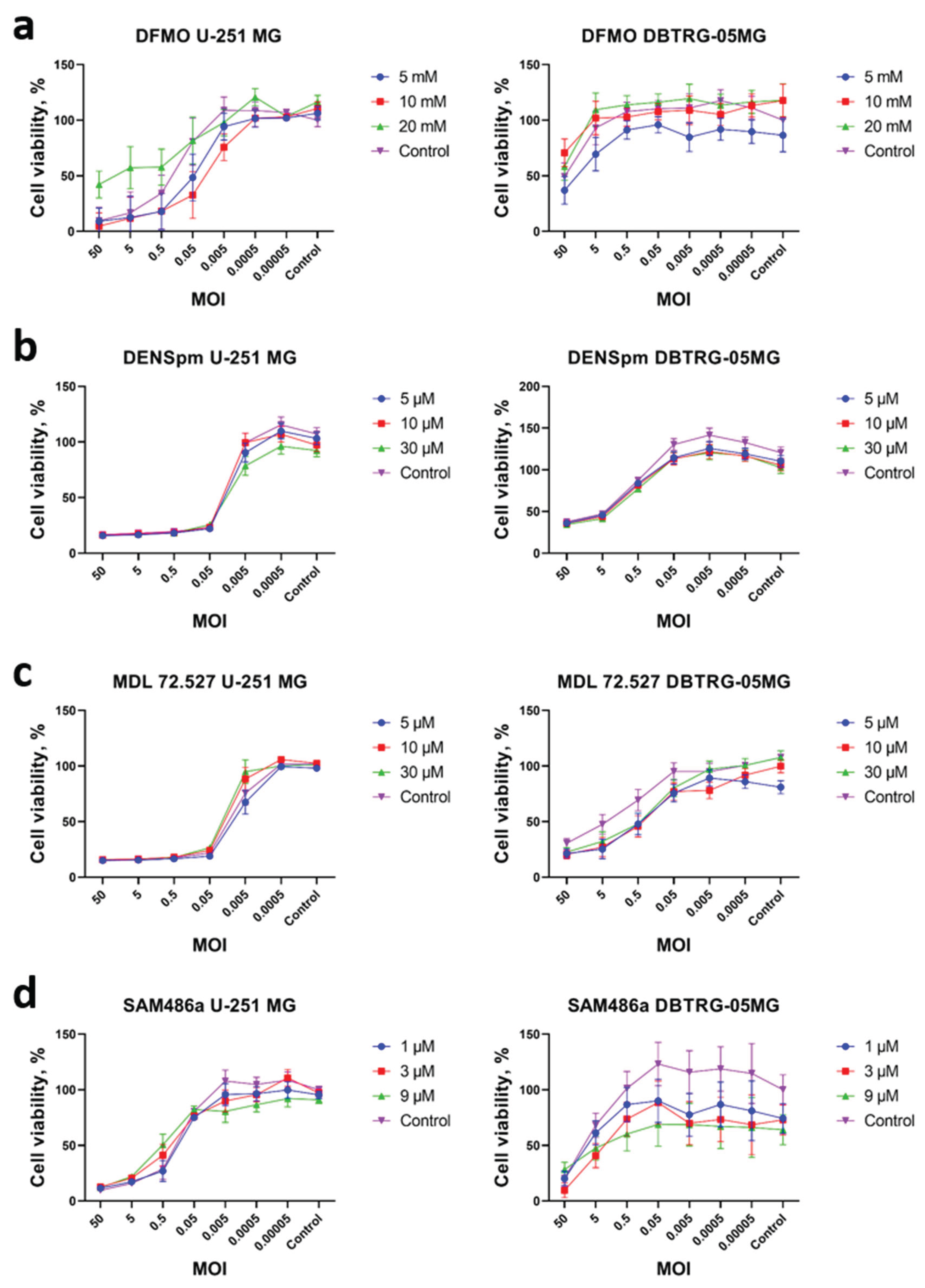
2.2. Changes in Metabolism of Biogenic Polyamines During Poliovirus Infection Are Dispensable for Virus Replication
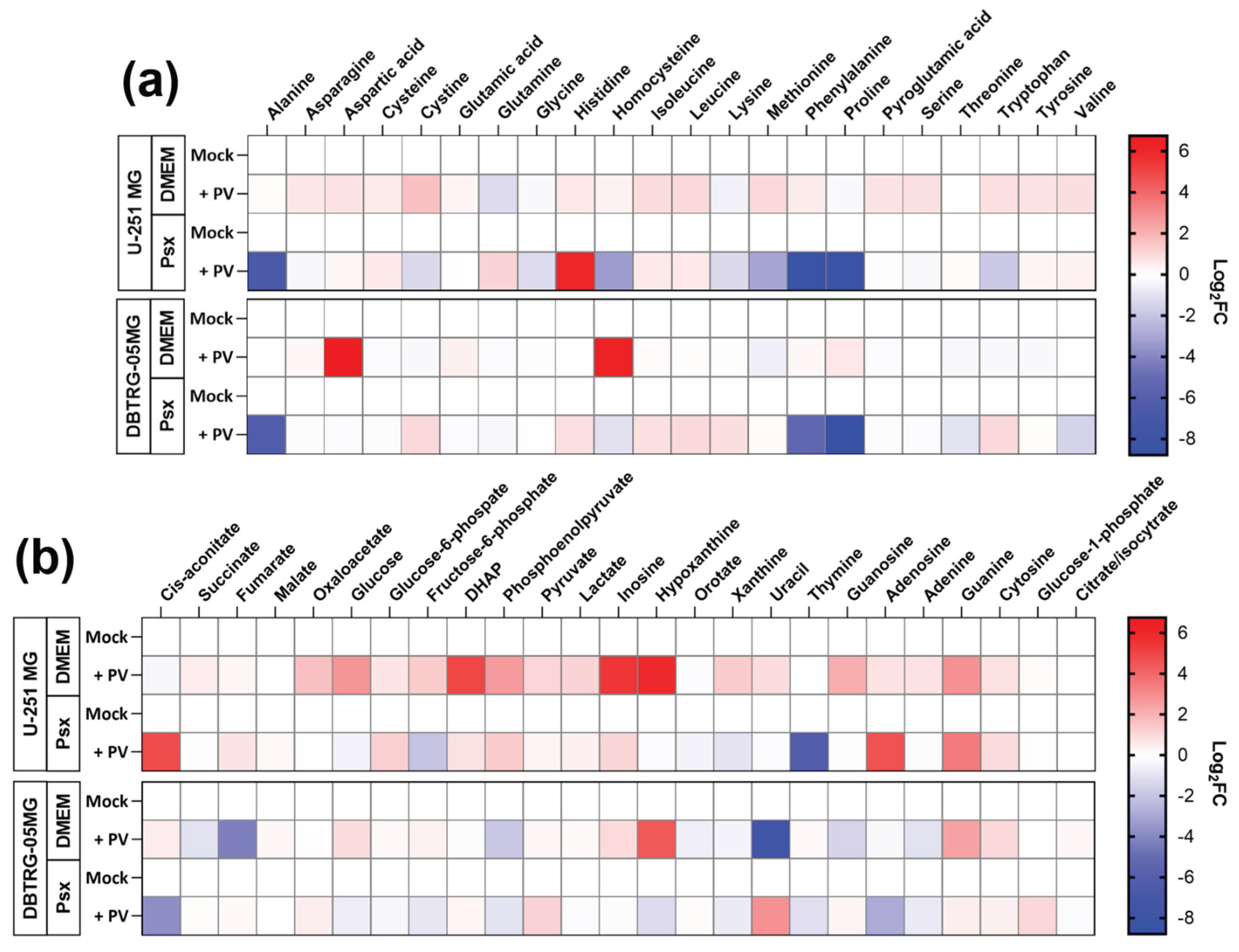
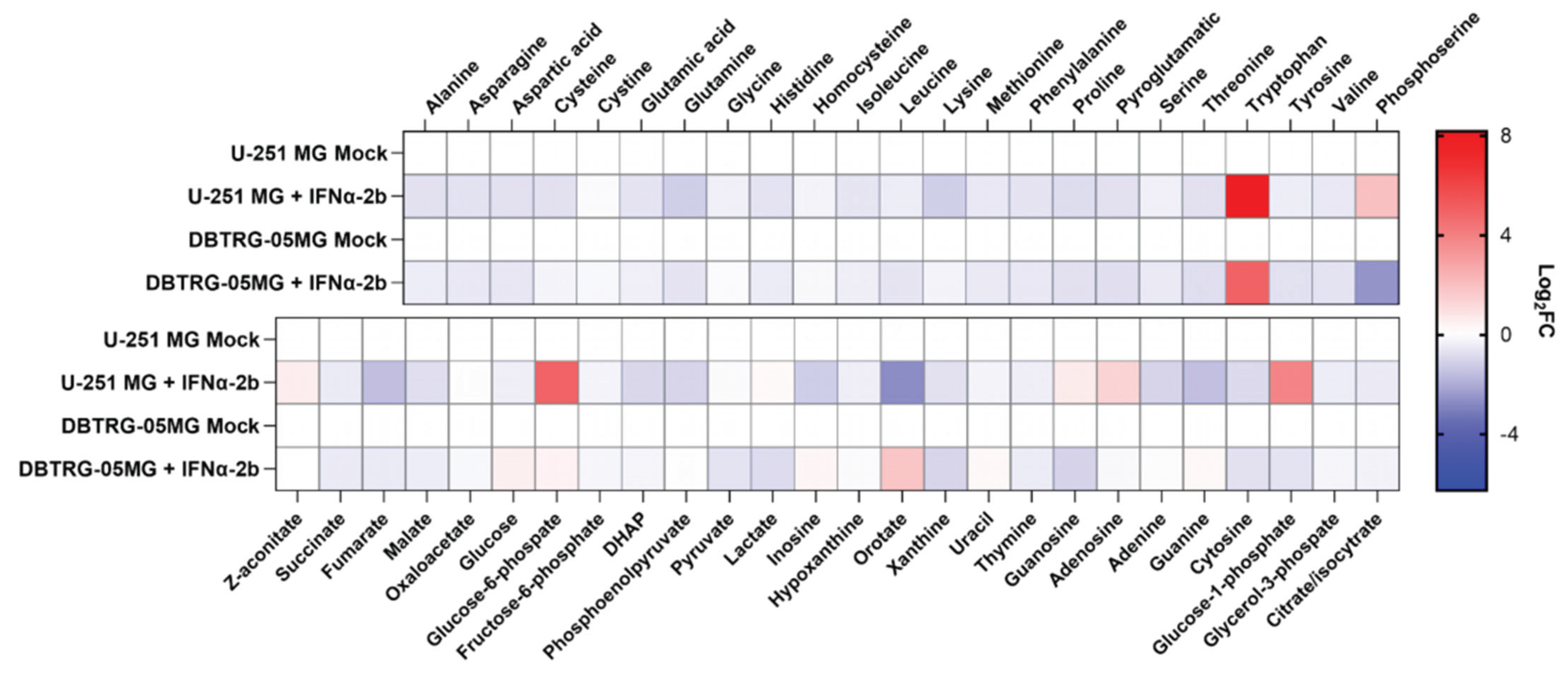
2.3. Poliovirus Interferes with Central Carbon Metabolism in a Cell Line-Specific Manner
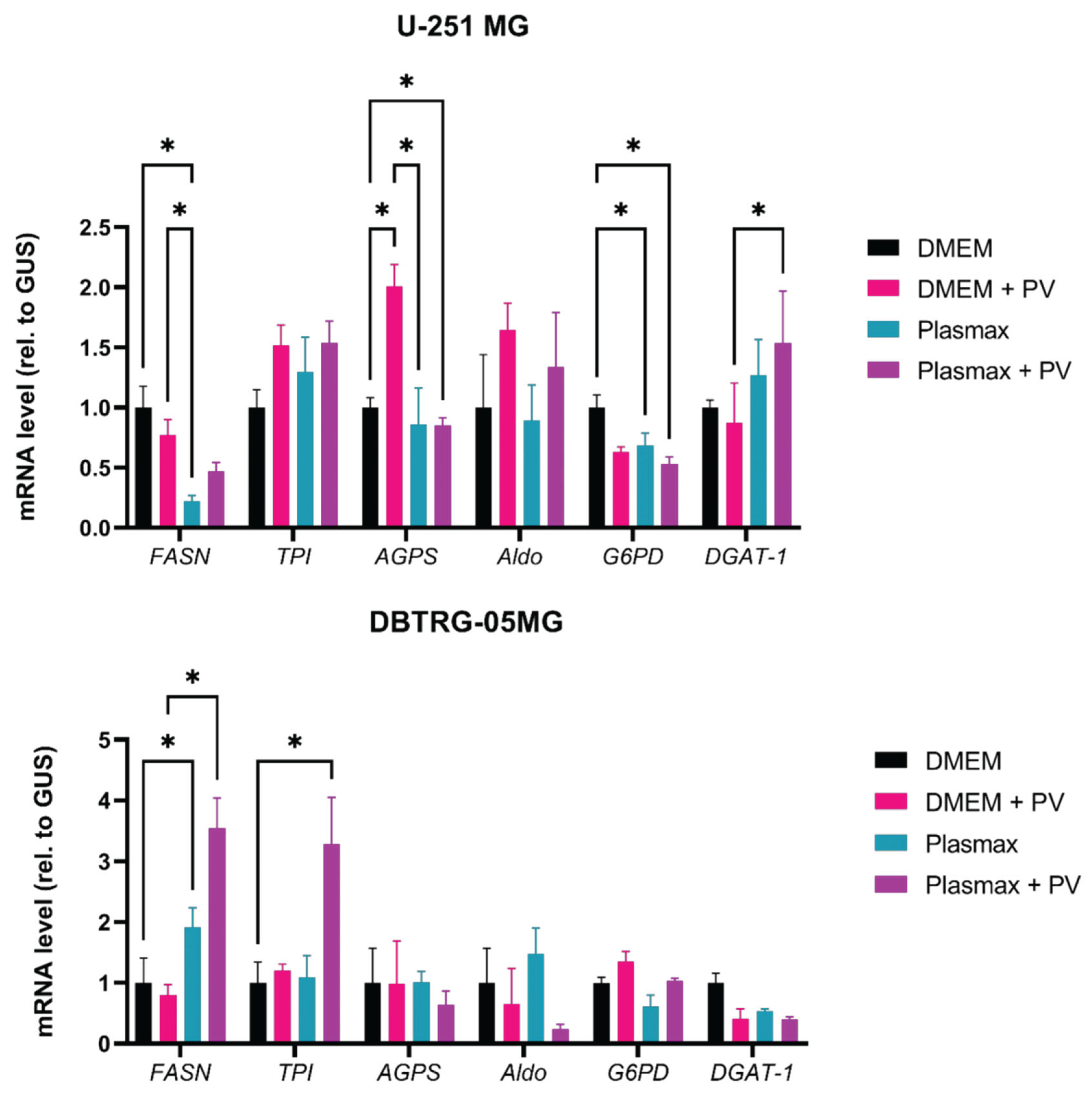
2.4. Poliovirus Infection Induces Accumulation of Lipid Droplets in GBM Cells
2.5. Metabolic Changes in Glioblastoma Cells by Poliovirus Are Not Mediated by Interferon Production
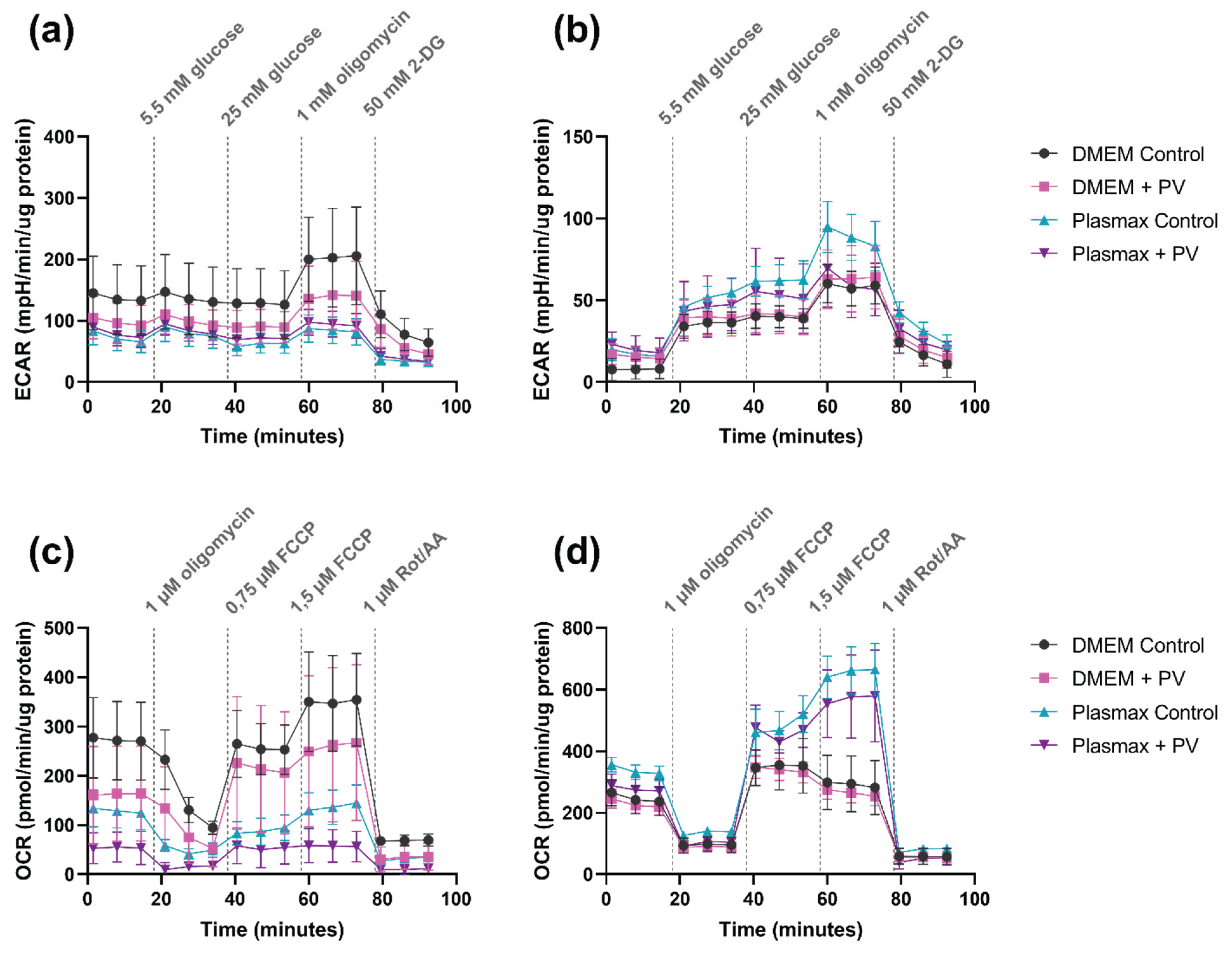
2.6. Poliovirus Suppresses Glycolysis and Mitochondrial Respiration
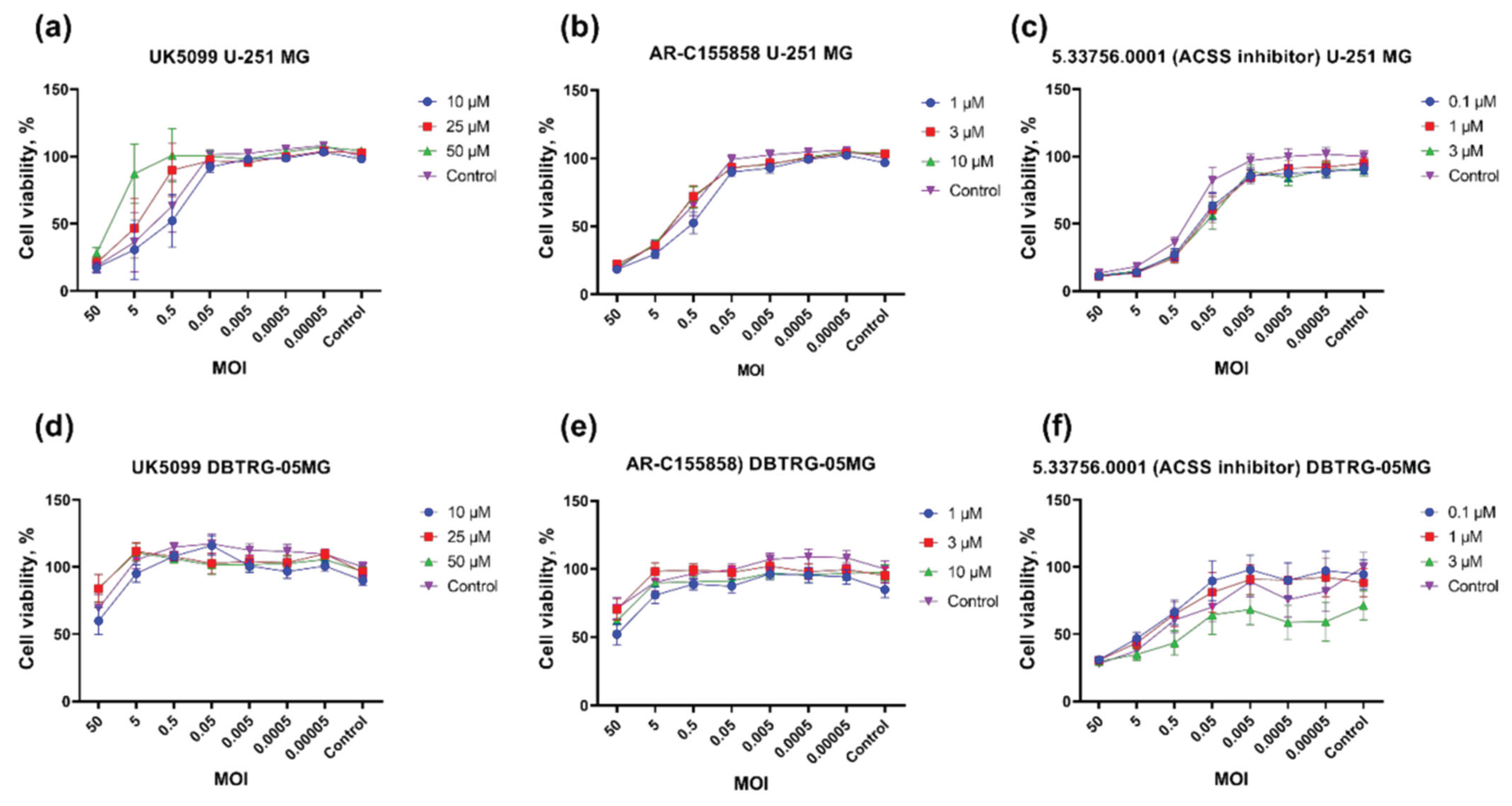
2.7. Import of Pyruvate into Mitochondria Is Essential for Poliovirus Replication
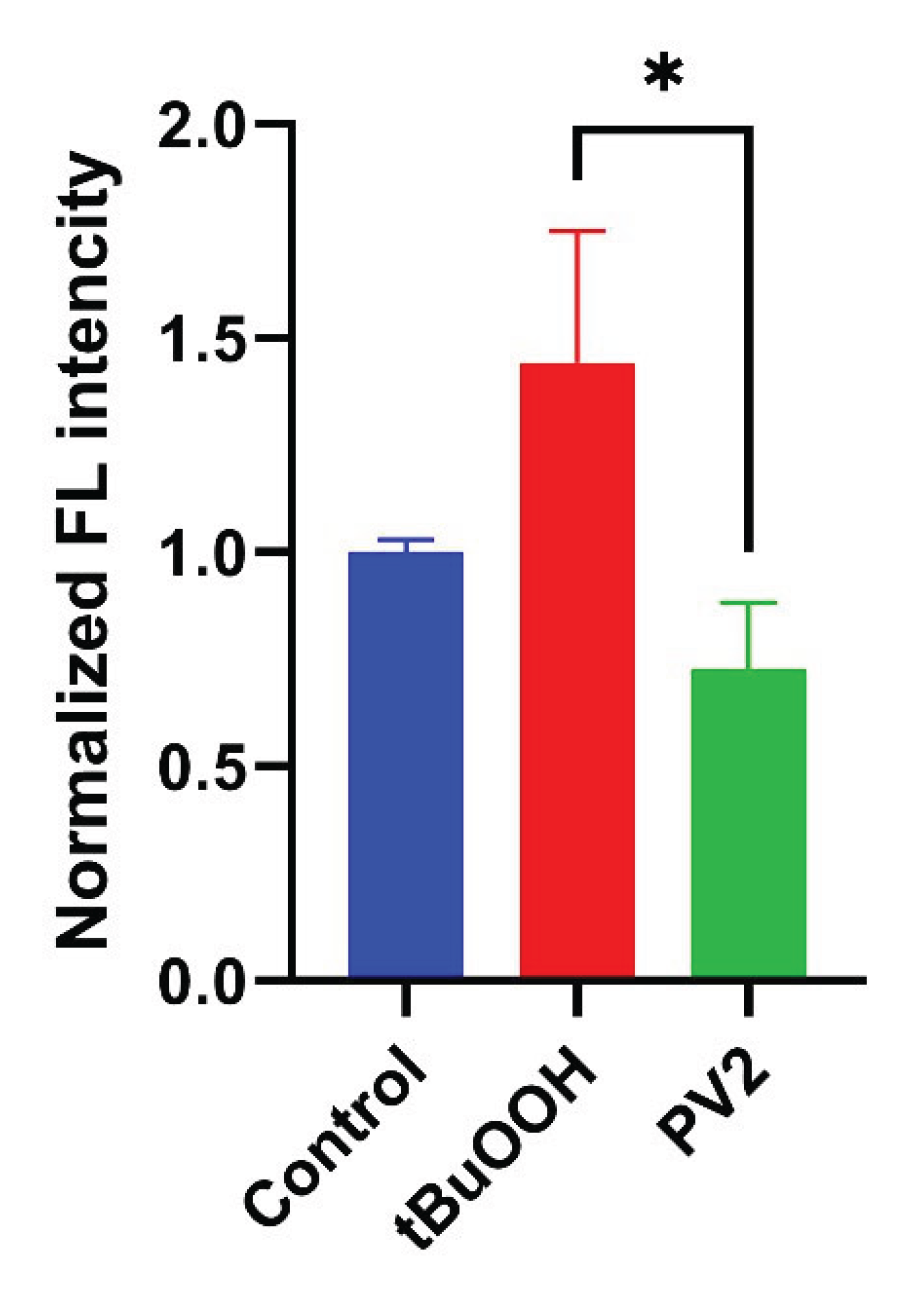
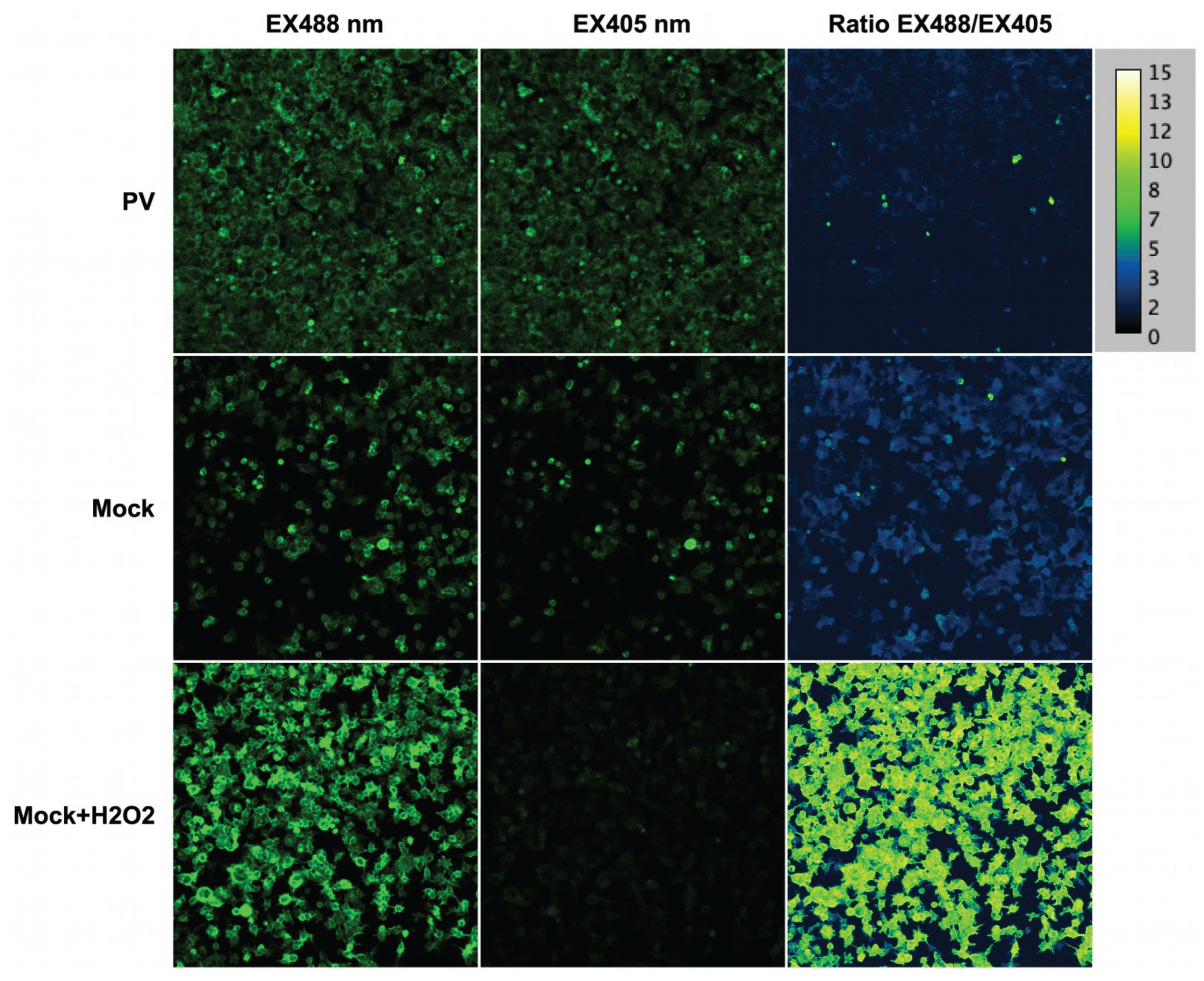
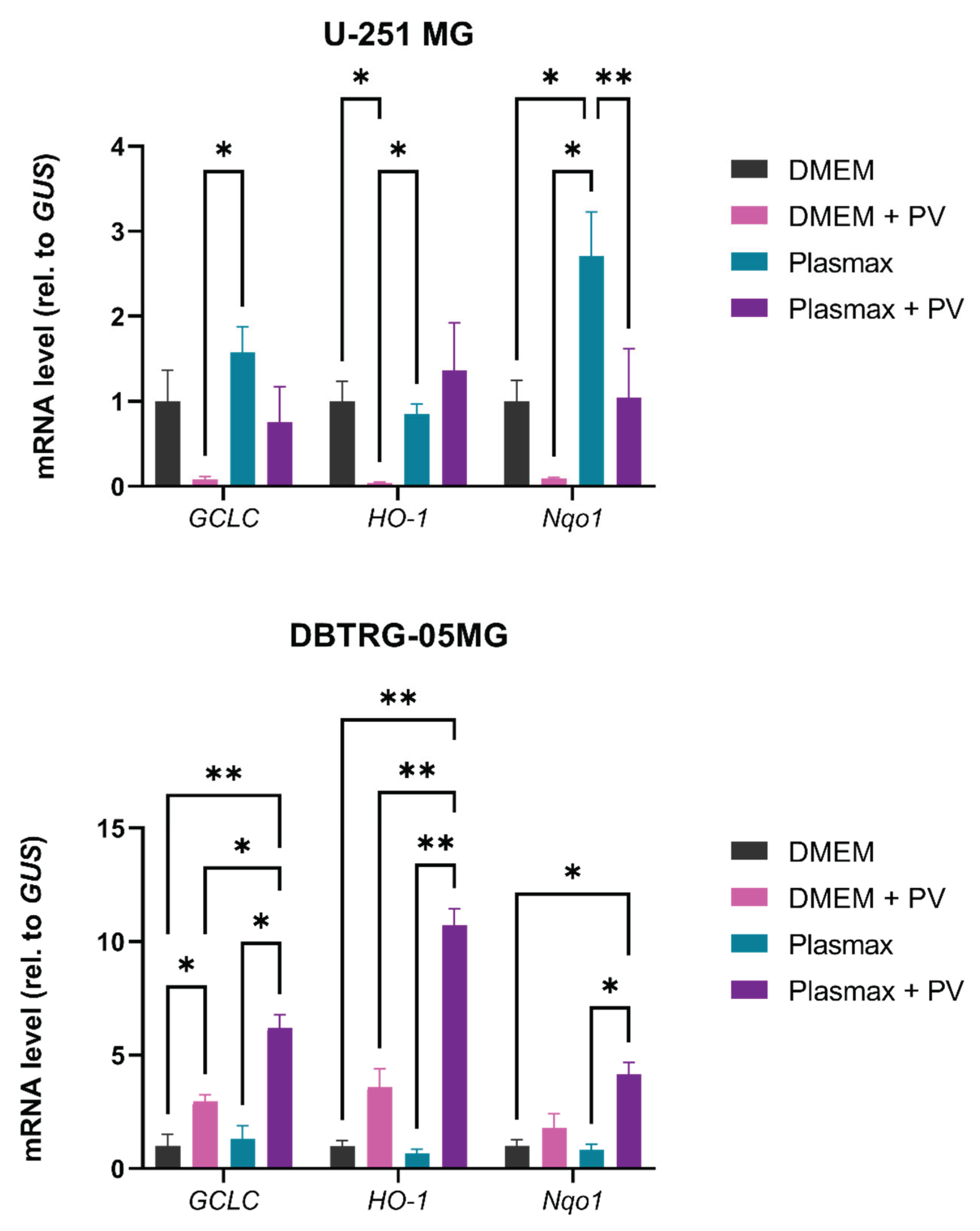
2.8. Poliovirus Infection Does Not Trigger Oxidative Stress
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Cultivation
4.3. Virus
4.4. Cell Morphology Analysis
4.5. Replication Assay
4.6. RT-qPCR
4.7. Cell Viability Assays
4.8. Measurement of Glycolysis and Mitochondrial Respiration
4.9. Immunostaining
4.10. Liquid Chromatography
4.11. Metabolite Quantification by Gas Chromatography-Mass Spectrometry
4.12. Interferon Cell Treatment
4.13. Staining of Neutral Lipids
4.14. Flow Cytometry
4.15. Sensors/Confocal Microscopy
4.16. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ostrom, Q.T.; Gittleman, H.; Farah, P.; Ondracek, A.; Chen, Y.; Wolinsky, Y.; Stroup, N.E.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2006-2010. Neuro Oncol 2013, 15 Suppl 2, ii1-56. [Google Scholar] [CrossRef]
- Tamimi, A.F.; Juweid, M. Epidemiology and Outcome of Glioblastoma. In Glioblastoma; De Vleeschouwer, S., Ed.; Codon Publications: Brisbane (AU), 2017; ISBN 978-0-9944381-2-6. [Google Scholar]
- Ahmadloo, N.; Kani, A.-A.; Mohammadianpanah, M.; Nasrolahi, H.; Omidvari, S.; Mosalaei, A.; Ansari, M. Treatment Outcome and Prognostic Factors of Adult Glioblastoma Multiforme. Journal of the Egyptian National Cancer Institute 2013, 25, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, F.; Ali, H.; Lathia, J.D.; Chen, P. Immunotherapy for Glioblastoma: Current State, Challenges, and Future Perspectives. Cell Mol Immunol 2024, 21, 1354–1375. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, C.; Costa, A.; Osório, L.; Lago, R.C.; Linhares, P.; Carvalho, B.; Caeiro, C. Current Standards of Care in Glioblastoma Therapy. In Glioblastoma; De Vleeschouwer, S., Ed.; Codon Publications: Brisbane (AU), 2017; ISBN 978-0-9944381-2-6. [Google Scholar]
- Weller, M.; van den Bent, M.; Preusser, M.; Le Rhun, E.; Tonn, J.C.; Minniti, G.; Bendszus, M.; Balana, C.; Chinot, O.; Dirven, L.; et al. EANO Guidelines on the Diagnosis and Treatment of Diffuse Gliomas of Adulthood. Nat Rev Clin Oncol 2021, 18, 170–186. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N Engl J Med 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Gesundheit, B.; Ben-David, E.; Posen, Y.; Ellis, R.; Wollmann, G.; Schneider, E.M.; Aigner, K.; Brauns, L.; Nesselhut, T.; Ackva, I.; et al. Effective Treatment of Glioblastoma Multiforme With Oncolytic Virotherapy: A Case-Series. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef]
- Alwithenani, A.; Hengswat, P.; Chiocca, E.A. Oncolytic Viruses as Cancer Therapeutics: From Mechanistic Insights to Clinical Translation. Mol Ther 2025, 33, 2217–2228. [Google Scholar] [CrossRef] [PubMed]
- Alekseeva, O.N.; Hoa, L.T.; Vorobyev, P.O.; Kochetkov, D.V.; Gumennaya, Y.D.; Naberezhnaya, E.R.; Chuvashov, D.O.; Ivanov, A.V.; Chumakov, P.M.; Lipatova, A.V. Receptors and Host Factors for Enterovirus Infection: Implications for Cancer Therapy. Cancers 2024, 16, 3139. [Google Scholar] [CrossRef]
- Shen, Y.; Bai, X.; Zhang, Q.; Liang, X.; Jin, X.; Zhao, Z.; Song, W.; Tan, Q.; Zhao, R.; Jia, W.; et al. Oncolytic Virus VG161 in Refractory Hepatocellular Carcinoma. Nature 2025, 641, 503–511. [Google Scholar] [CrossRef]
- Lawler, S.E.; Speranza, M.-C.; Cho, C.-F.; Chiocca, E.A. Oncolytic Viruses in Cancer Treatment: A Review. JAMA Oncology 2017, 3, 841–849. [Google Scholar] [CrossRef]
- Sosnovtseva, A.O.; Lipatova, A.V.; Grinenko, N.F.; Baklaushev, V.P.; Chumakov, P.M.; Chekhonin, V.P. Sensitivity of C6 Glioma Cells Carrying the Human Poliovirus Receptor to Oncolytic Polioviruses. Bull Exp Biol Med 2016, 161, 821–825. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.C.; Dobrikova, E.Y.; Dobrikov, M.I.; Walton, R.W.; Gemberling, S.L.; Nair, S.K.; Desjardins, A.; Sampson, J.H.; Friedman, H.S.; Friedman, A.H.; et al. Oncolytic Polio Virotherapy of Cancer. [CrossRef]
- Sanchez, E.L.; Lagunoff, M. Viral Activation of Cellular Metabolism. Virology 2015, 479–480, 609–618. [Google Scholar] [CrossRef]
- Gaballah, A.; Bartosch, B. An Update on the Metabolic Landscape of Oncogenic Viruses. Cancers (Basel) 2022, 14, 5742. [Google Scholar] [CrossRef]
- Lévy, P.; Bartosch, B. Metabolic Reprogramming: A Hallmark of Viral Oncogenesis. Oncogene 2016, 35, 4155–4164. [Google Scholar] [CrossRef]
- Lange, P.T.; Lagunoff, M.; Tarakanova, V.L. Chewing the Fat: The Conserved Ability of DNA Viruses to Hijack Cellular Lipid Metabolism. Viruses 2019, 11, 119. [Google Scholar] [CrossRef]
- Chan, R.B.; Tanner, L.; Wenk, M.R. Implications for Lipids during Replication of Enveloped Viruses. Chemistry and Physics of Lipids 2010, 163, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Zakirova, N.F.; Khomich, O.A.; Smirnova, O.A.; Molle, J.; Duponchel, S.; Yanvarev, D.V.; Valuev-Elliston, V.T.; Monnier, L.; Grigorov, B.; Ivanova, O.N.; et al. Hepatitis C Virus Dysregulates Polyamine and Proline Metabolism and Perturbs the Urea Cycle. Cells 2024, 13, 1036. [Google Scholar] [CrossRef]
- Cruz-Pulido, Y.E.; LoMascolo, N.J.; May, D.; Hatahet, J.; Thomas, C.E.; Chu, A.K.W.; Stacey, S.P.; Guzman, M. del M.V.; Aubert, G.; Mounce, B.C. Polyamines Mediate Cellular Energetics and Lipid Metabolism through Mitochondrial Respiration to Facilitate Virus Replication. PLOS Pathogens 2024, 20, e1012711. [Google Scholar] [CrossRef] [PubMed]
- Mastrodomenico, V.; LoMascolo, N.J.; Firpo, M.R.; Villanueva Guzman, M.D.M.; Zaporowski, A.; Mounce, B.C. Persistent Coxsackievirus B3 Infection in Pancreatic Ductal Cells In Vitro Downregulates Cellular Polyamine Metabolism. mSphere 2023, 8, e0003623. [Google Scholar] [CrossRef]
- Wan, Q.; Tavakoli, L.; Wang, T.-Y.; Tucker, A.J.; Zhou, R.; Liu, Q.; Feng, S.; Choi, D.; He, Z.; Gack, M.U.; et al. Hijacking of Nucleotide Biosynthesis and Deamidation-Mediated Glycolysis by an Oncogenic Herpesvirus. Nat Commun 2024, 15, 1442. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, T.; Ramos da Silva, S.; Lee, J.-J.; Lu, C.; Eoh, H.; Jung, J.U.; Gao, S.-J. A Critical Role of Glutamine and Asparagine γ-Nitrogen in Nucleotide Biosynthesis in Cancer Cells Hijacked by an Oncogenic Virus. mBio 2017, 8, e01179-17. [Google Scholar] [CrossRef] [PubMed]
- Verrier, E.R.; Weiss, A.; Bach, C.; Heydmann, L.; Turon-Lagot, V.; Kopp, A.; El Saghire, H.; Crouchet, E.; Pessaux, P.; Garcia, T.; et al. Combined Small Molecule and Loss-of-Function Screen Uncovers Estrogen Receptor Alpha and CAD as Host Factors for HDV Infection and Antiviral Targets. Gut 2020, 69, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Firpo, M.R.; Mastrodomenico, V.; Hawkins, G.M.; Prot, M.; Levillayer, L.; Gallagher, T.; Simon-Loriere, E.; Mounce, B.C. Targeting Polyamines Inhibits Coronavirus Infection by Reducing Cellular Attachment and Entry. ACS Infect Dis 2021, 7, 1423–1432. [Google Scholar] [CrossRef]
- Mastrodomenico, V.; Esin, J.J.; Graham, M.L.; Tate, P.M.; Hawkins, G.M.; Sandler, Z.J.; Rademacher, D.J.; Kicmal, T.M.; Dial, C.N.; Mounce, B.C. Polyamine Depletion Inhibits Bunyavirus Infection via Generation of Noninfectious Interfering Virions. J Virol 2019, 93, e00530-19. [Google Scholar] [CrossRef]
- Bhatt, A.N.; Shenoy, S.; Munjal, S.; Chinnadurai, V.; Agarwal, A.; Vinoth Kumar, A.; Shanavas, A.; Kanwar, R.; Chandna, S. 2-Deoxy-D-Glucose as an Adjunct to Standard of Care in the Medical Management of COVID-19: A Proof-of-Concept and Dose-Ranging Randomised Phase II Clinical Trial. BMC Infect Dis 2022, 22, 669. [Google Scholar] [CrossRef]
- Ivanov, A.V.; Valuev-Elliston, V.T.; Tyurina, D.A.; Ivanova, O.N.; Kochetkov, S.N.; Bartosch, B.; Isaguliants, M.G. Oxidative Stress, a Trigger of Hepatitis C and B Virus-Induced Liver Carcinogenesis. Oncotarget 2016, 8, 3895–3932. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.V.; Valuev-Elliston, V.T.; Ivanova, O.N.; Kochetkov, S.N.; Starodubova, E.S.; Bartosch, B.; Isaguliants, M.G. Oxidative Stress during HIV Infection: Mechanisms and Consequences. [CrossRef]
- Khomich, O.A.; Kochetkov, S.N.; Bartosch, B.; Ivanov, A.V. Redox Biology of Respiratory Viral Infections. Viruses 2018, 10, 392. [Google Scholar] [CrossRef]
- Eagle, H.; Habel, K. THE NUTRITIONAL REQUIREMENTS FOR THE PROPAGATION OF POLIOMYELITIS VIRUS BY THE HELA CELL. Journal of Experimental Medicine 1956, 104, 271–287. [Google Scholar] [CrossRef]
- Mosser, A.G.; Caliguiri, L.A.; Tamm, I. Incorporation of Lipid Precursors into Cytoplasmic Membranes of Poliovirus-Infected HeLa Cells. Virology 1972, 47, 39–47. [Google Scholar] [CrossRef]
- Nchoutmboube, J.; Viktorova, E.; Scott, A.; Ford, L.; Pei, Z.; Watkins, P.; Ernst, R.; Belov, G. Increased Long Chain Acyl-Coa Synthetase Activity and Fatty Acid Import Is Linked to Membrane Synthesis for Development of Picornavirus Replication Organelles. PLoS pathogens 2013, 9, e1003401. [Google Scholar] [CrossRef]
- Vorobyev, P.O.; Kochetkov, D.V.; Chumakov, P.M.; Zakirova, N.F.; Zotova-Nefedorova, S.I.; Vasilenko, K.V.; Alekseeva, O.N.; Kochetkov, S.N.; Bartosch, B.; Lipatova, A.V.; et al. 2-Deoxyglucose, an Inhibitor of Glycolysis, Enhances the Oncolytic Effect of Coxsackievirus. Cancers 2022, 14, 5611. [Google Scholar] [CrossRef] [PubMed]
- Golikov, M.V.; Bartosch, B.; Smirnova, O.A.; Ivanova, O.N.; Ivanov, A.V. Plasma-Like Culture Medium for the Study of Viruses. mBio 2022. [Google Scholar] [CrossRef] [PubMed]
- Golikov, M.V.; Karpenko, I.L.; Lipatova, A.V.; Ivanova, O.N.; Fedyakina, I.T.; Larichev, V.F.; Zakirova, N.F.; Leonova, O.G.; Popenko, V.I.; Bartosch, B.; et al. Cultivation of Cells in a Physiological Plasmax Medium Increases Mitochondrial Respiratory Capacity and Reduces Replication Levels of RNA Viruses. Antioxidants 2022, 11, 97. [Google Scholar] [CrossRef] [PubMed]
- Limpens, R.W.A.L.; van der Schaar, H.M.; Kumar, D.; Koster, A.J.; Snijder, E.J.; van Kuppeveld, F.J.M.; Bárcena, M. The Transformation of Enterovirus Replication Structures: A Three-Dimensional Study of Single- and Double-Membrane Compartments. mBio 2011, 2. [Google Scholar] [CrossRef]
- Mounce, B.C.; Olsen, M.E.; Vignuzzi, M.; Connor, J.H. Polyamines and Their Role in Virus Infection. Microbiol Mol Biol Rev 2017, 81, e00029-17. [Google Scholar] [CrossRef]
- Mastrodomenico, V.; Esin, J.J.; Qazi, S.; Khomutov, M.A.; Ivanov, A.V.; Mukhopadhyay, S.; Mounce, B.C. Virion-Associated Polyamines Transmit with Bunyaviruses to Maintain Infectivity and Promote Entry. ACS Infect. Dis. 2020, 6, 2490–2501. [Google Scholar] [CrossRef]
- Aleksunes, L.M.; Manautou, J.E. Emerging Role of Nrf2 in Protecting against Hepatic and Gastrointestinal Disease. Toxicol Pathol 2007, 35, 459–473. [Google Scholar] [CrossRef]
- Prestwich, R.J.; Errington, F.; Harrington, K.J.; Pandha, H.S.; Selby, P.; Melcher, A. Oncolytic Viruses: Do They Have a Role in Anti-Cancer Therapy? Clin Med Oncol 2008, 2, 83–96. [Google Scholar] [CrossRef]
- Andtbacka, R.H.I.; Kaufman, H.L.; Collichio, F.; Amatruda, T.; Senzer, N.; Chesney, J.; Delman, K.A.; Spitler, L.E.; Puzanov, I.; Agarwala, S.S.; et al. Talimogene Laherparepvec Improves Durable Response Rate in Patients With Advanced Melanoma. J Clin Oncol 2015, 33, 2780–2788. [Google Scholar] [CrossRef]
- Xia, Z.-J.; Chang, J.-H.; Zhang, L.; Jiang, W.-Q.; Guan, Z.-Z.; Liu, J.-W.; Zhang, Y.; Hu, X.-H.; Wu, G.-H.; Wang, H.-Q.; et al. [Phase III randomized clinical trial of intratumoral injection of E1B gene-deleted adenovirus (H101) combined with cisplatin-based chemotherapy in treating squamous cell cancer of head and neck or esophagus]. Ai Zheng 2004, 23, 1666–1670. [Google Scholar]
- Alberts, P.; Tilgase, A.; Rasa, A.; Bandere, K.; Venskus, D. The Advent of Oncolytic Virotherapy in Oncology: The Rigvir® Story. European Journal of Pharmacology 2018, 837, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Frampton, J.E. Teserpaturev/G47Δ: First Approval. BioDrugs 2022, 36, 667–672. [Google Scholar] [CrossRef] [PubMed]
- Romanishin, A.; Vasilev, A.; Khasanshin, E.; Evtekhov, A.; Pusynin, E.; Rubina, K.; Kakotkin, V.; Agapov, M.; Semina, E. Oncolytic Viral Therapy for Gliomas: Advances in the Mechanisms and Approaches to Delivery. Virology 2024, 593, 110033. [Google Scholar] [CrossRef]
- Du, W.; Na, J.; Zhong, L.; Zhang, P. Advances in Preclinical and Clinical Studies of Oncolytic Virus Combination Therapy. Front Oncol 2025, 15, 1545542. [Google Scholar] [CrossRef]
- Burton, C.; Das, A.; McDonald, D.; Vandergrift, W.A.; Patel, S.J.; Cachia, D.; Bartee, E. Oncolytic Myxoma Virus Synergizes with Standard of Care for Treatment of Glioblastoma Multiforme. Oncolytic Virother 2018, 7, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Jiang, H.; Cheng, L.; Ma, B.; Liu, R. Oncolytic Herpes Simplex Virus and Temozolomide Synergistically Inhibit Breast Cancer Cell Tumorigenesis in Vitro and in Vivo. Oncol Lett 2021, 21, 99. [Google Scholar] [CrossRef]
- Romanenko, M.V.; Dolgova, E.V.; Osipov, I.D.; Ritter, G.S.; Sizova, M.S.; Proskurina, A.S.; Efremov, Y.R.; Bayborodin, S.I.; Potter, E.A.; Taranov, O.S.; et al. Oncolytic Effect of Adenoviruses Serotypes 5 and 6 Against U87 Glioblastoma Cancer Stem Cells. Anticancer Research 2019, 39, 6073–6086. [Google Scholar] [CrossRef]
- Cuoco, J.A.; Rogers, C.M.; Mittal, S. The Oncolytic Newcastle Disease Virus as an Effective Immunotherapeutic Strategy against Glioblastoma. 2021. [Google Scholar] [CrossRef]
- Chen, Q.; Wu, J.; Ye, Q.; Ma, F.; Zhu, Q.; Wu, Y.; Shan, C.; Xie, X.; Li, D.; Zhan, X.; et al. Treatment of Human Glioblastoma with a Live Attenuated Zika Virus Vaccine Candidate. mBio 2018, 9. [Google Scholar] [CrossRef]
- Zhu, Z.; Gorman, M.J.; McKenzie, L.D.; Chai, J.N.; Hubert, C.G.; Prager, B.C.; Fernandez, E.; Richner, J.M.; Zhang, R.; Shan, C.; et al. Zika Virus Has Oncolytic Activity against Glioblastoma Stem Cells. Journal of Experimental Medicine 2017, 214, 2843–2857. [Google Scholar] [CrossRef]
- Lipatova, A.V.; Soboleva, A.V.; Gorshkov, V.A.; Bubis, J.A.; Solovyeva, E.M.; Krasnov, G.S.; Kochetkov, D.V.; Vorobyev, P.O.; Ilina, I.Y.; Moshkovskii, S.A.; et al. Multi-Omics Analysis of Glioblastoma Cells’ Sensitivity to Oncolytic Viruses. Cancers 2021, 13, 5268. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.N.S.; Arjona, S.P.; Santerre, M.; Sawaya, B.E. Hallmarks of Metabolic Reprogramming and Their Role in Viral Pathogenesis. Viruses 2022, 14, 602. [Google Scholar] [CrossRef] [PubMed]
- Amatore, D.; Sgarbanti, R.; Aquilano, K.; Baldelli, S.; Limongi, D.; Civitelli, L.; Nencioni, L.; Garaci, E.; Ciriolo, M.R.; Palamara, A.T. Influenza Virus Replication in Lung Epithelial Cells Depends on Redox-sensitive Pathways Activated by NOX4-derived ROS. [CrossRef]
- Vancura, A.; Bu, P.; Bhagwat, M.; Zeng, J.; Vancurova, I. Metformin as an Anticancer Agent. Trends in Pharmacological Sciences 2018, 39, 867–878. [Google Scholar] [CrossRef]
- Benjanuwattra, J.; Chaiyawat, P.; Pruksakorn, D.; Koonrungsesomboon, N. Therapeutic Potential and Molecular Mechanisms of Mycophenolic Acid as an Anticancer Agent. European Journal of Pharmacology 2020, 887, 173580. [Google Scholar] [CrossRef] [PubMed]
- Cantor, J.R. The Rise of Physiologic Media. Trends in Cell Biology 2019, 29, 854–861. [Google Scholar] [CrossRef]
- Voorde, J.V.; Ackermann, T.; Pfetzer, N.; Sumpton, D.; Mackay, G.; Kalna, G.; Nixon, C.; Blyth, K.; Gottlieb, E.; Tardito, S. Improving the Metabolic Fidelity of Cancer Models with a Physiological Cell Culture Medium. Science Advances 2019. [Google Scholar] [CrossRef]
- Cantor, J.R.; Abu-Remaileh, M.; Kanarek, N.; Freinkman, E.; Gao, X.; Louissaint, A.; Lewis, C.A.; Sabatini, D.M. Physiologic Medium Rewires Cellular Metabolism and Reveals Uric Acid as an Endogenous Inhibitor of UMP Synthase. Cell 2017, 169, 258–272.e17. [Google Scholar] [CrossRef]
- Rawat, V.; DeLear, P.; Prashanth, P.; Ozgurses, M.E.; Tebeje, A.; Burns, P.A.; Conger, K.O.; Solís, C.; Hasnain, Y.; Novikova, A.; et al. Drug Screening in Human Physiologic Medium Identifies Uric Acid as an Inhibitor of Rigosertib Efficacy. JCI Insight 2024, 9, e174329. [Google Scholar] [CrossRef]
- Flickinger, K.M.; Wilson, K.M.; Rossiter, N.J.; Hunger, A.L.; Vishwasrao, P.V.; Lee, T.D.; Fritz, C.A.M.; Richards, R.M.; Hall, M.D.; Cantor, J.R. Conditional Lethality Profiling Reveals Anticancer Mechanisms of Action and Drug-Nutrient Interactions. Science Advances 2024. [Google Scholar] [CrossRef]
- Khadka, S.; Arthur, K.; Barekatain, Y.; Behr, E.; Washington, M.; Ackroyd, J.; Crowley, K.; Suriyamongkol, P.; Lin, Y.-H.; Pham, C.-D.; et al. Impaired Anaplerosis Is a Major Contributor to Glycolysis Inhibitor Toxicity in Glioma. Cancer Metab 2021, 9, 27. [Google Scholar] [CrossRef]
- Lee, M.-J.; Chen, Y.; Huang, Y.-P.; Hsu, Y.-C.; Chiang, L.-H.; Chen, T.-Y.; Wang, G.-J. Exogenous Polyamines Promote Osteogenic Differentiation by Reciprocally Regulating Osteogenic and Adipogenic Gene Expression. J Cell Biochem 2013, 114, 2718–2728. [Google Scholar] [CrossRef] [PubMed]
- Quemener, V.; Blanchard, Y.; Lescoat, D.; Havouis, R.; Moulinoux, J.P. Depletion in Nuclear Spermine during Human Spermatogenesis, a Natural Process of Cell Differentiation. Am J Physiol 1992, 263, C343–347. [Google Scholar] [CrossRef]
- Pegg, A.E. Mammalian Polyamine Metabolism and Function. IUBMB Life 2009, 61, 880–894. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, K.; Murozumi, K.; Shirahata, A.; Park, M.H.; Kashiwagi, K.; Igarashi, K. Independent Roles of eIF5A and Polyamines in Cell Proliferation. Biochem J 2005, 385, 779–785. [Google Scholar] [CrossRef] [PubMed]
- Casero, R.A.; Murray Stewart, T.; Pegg, A.E. Polyamine Metabolism and Cancer: Treatments, Challenges and Opportunities. Nat Rev Cancer 2018, 18, 681–695. [Google Scholar] [CrossRef]
- Lewis, E.C.; Kraveka, J.M.; Ferguson, W.; Eslin, D.; Brown, V.I.; Bergendahl, G.; Roberts, W.; Wada, R.K.; Oesterheld, J.; Mitchell, D.; et al. A Subset Analysis of a Phase II Trial Evaluating the Use of DFMO as Maintenance Therapy for High-Risk Neuroblastoma. Int J Cancer 2020, 147, 3152–3159. [Google Scholar] [CrossRef]
- Sholler, G.L.S.; Ferguson, W.; Bergendahl, G.; Bond, J.P.; Neville, K.; Eslin, D.; Brown, V.; Roberts, W.; Wada, R.K.; Oesterheld, J.; et al. Maintenance DFMO Increases Survival in High Risk Neuroblastoma. Sci Rep 2018, 8, 14445. [Google Scholar] [CrossRef]
- Saulnier Sholler, G.L.; Gerner, E.W.; Bergendahl, G.; MacArthur, R.B.; VanderWerff, A.; Ashikaga, T.; Bond, J.P.; Ferguson, W.; Roberts, W.; Wada, R.K.; et al. A Phase I Trial of DFMO Targeting Polyamine Addiction in Patients with Relapsed/Refractory Neuroblastoma. PLoS One 2015, 10, e0127246. [Google Scholar] [CrossRef]
- Duke, E.S.; Bradford, D.; Sinha, A.K.; Mishra-Kalyani, P.S.; Lerro, C.C.; Rivera, D.; Wearne, E.; Miller, C.P.; Leighton, J.; Sabit, H.; et al. US Food and Drug Administration Approval Summary: Eflornithine for High-Risk Neuroblastoma After Prior Multiagent, Multimodality Therapy. J Clin Oncol 2024, 42, 3047–3057. [Google Scholar] [CrossRef]
- Romão, L.; do Canto, V.P.; Netz, P.A.; Moura-Neto, V.; Pinto, Â.C.; Follmer, C. Conjugation with Polyamines Enhances the Antitumor Activity of Naphthoquinones against Human Glioblastoma Cells. Anti-Cancer Drugs 2018, 29, 520. [Google Scholar] [CrossRef]
- Redgate, E.S.; Boggs, S.; Grudziak, A.; Deutsch, M. Polyamines in Brain Tumor Therapy. J Neuro-Oncol 1995, 25, 167–179. [Google Scholar] [CrossRef] [PubMed]
- A, A.G.; D, L.G.; Vassileios, R.; Vasiliki, G.; P, K.A. Difluoromethylornithine in Cancer: New Advances. Future Oncology 2017. [Google Scholar] [CrossRef]
- McBenedict, B.; Hauwanga, W.N.; Pogodina, A.; Singh, G.; Thomas, A.; Ibrahim, A.M.A.; Johnny, C.; Pessôa, B.L.; McBenedict, B.; Hauwanga, W.N.; et al. Approaches in Adult Glioblastoma Treatment: A Systematic Review of Emerging Therapies. Cureus 2024, 16. [Google Scholar] [CrossRef] [PubMed]
- Kuemmerle, A.; Schmid, C.; Bernhard, S.; Kande, V.; Mutombo, W.; Ilunga, M.; Lumpungu, I.; Mutanda, S.; Nganzobo, P.; Tete, D.N.; et al. Effectiveness of Nifurtimox Eflornithine Combination Therapy (NECT) in T. b. Gambiense Second Stage Sleeping Sickness Patients in the Democratic Republic of Congo: Report from a Field Study. PLoS Negl Trop Dis 2021, 15, e0009903. [Google Scholar] [CrossRef]
- Olsen, M.E.; Filone, C.M.; Rozelle, D.; Mire, C.E.; Agans, K.N.; Hensley, L.; Connor, J.H. Polyamines and Hypusination Are Required for Ebolavirus Gene Expression and Replication. mBio 2016, 7, e00882-16. [Google Scholar] [CrossRef] [PubMed]
- Fiches, G.N.; Wu, Z.; Zhou, D.; Biswas, A.; Li, T.-W.; Kong, W.; Jean, M.; Santoso, N.G.; Zhu, J. Polyamine Biosynthesis and eIF5A Hypusination Are Modulated by the DNA Tumor Virus KSHV and Promote KSHV Viral Infection. PLoS Pathog 2022, 18, e1010503. [Google Scholar] [CrossRef]
- Gibson, W.; van Breemen, R.; Fields, A.; LaFemina, R.; Irmiere, A. D,L-Alpha-Difluoromethylornithine Inhibits Human Cytomegalovirus Replication. J Virol 1984, 50, 145–154. [Google Scholar] [CrossRef]
- Rojas-Luna, L.; Posadas-Modragón, A.; Avila-Trejo, A.M.; Alcántara-Farfán, V.; Rodríguez-Páez, L.I.; Santiago-Cruz, J.A.; Pastor-Alonso, M.O.; Aguilar-Faisal, J.L. Inhibition of Chikungunya Virus Replication by N-ω-Chloroacetyl-L-Ornithine in C6/36, Vero Cells and Human Fibroblast BJ. Antiviral Therapy 2023. [Google Scholar] [CrossRef]
- Mounce, B.C.; Cesaro, T.; Vlajnić, L.; Vidiņa, A.; Vallet, T.; Weger-Lucarelli, J.; Passoni, G.; Stapleford, K.A.; Levraud, J.-P.; Vignuzzi, M. Chikungunya Virus Overcomes Polyamine Depletion by Mutation of nsP1 and the Opal Stop Codon To Confer Enhanced Replication and Fitness. J Virol 2017, 91, e00344-17. [Google Scholar] [CrossRef]
- Mounce, B.C.; Cesaro, T.; Moratorio, G.; Hooikaas, P.J.; Yakovleva, A.; Werneke, S.W.; Smith, E.C.; Poirier, E.Z.; Simon-Loriere, E.; Prot, M.; et al. Inhibition of Polyamine Biosynthesis Is a Broad-Spectrum Strategy against RNA Viruses. Journal of Virology 2016, 90, 9683–9692. [Google Scholar] [CrossRef]
- Mao, B.; Wang, Z.; Pi, S.; Long, Q.; Chen, K.; Cui, J.; Huang, A.; Hu, Y. Difluoromethylornithine, a Decarboxylase 1 Inhibitor, Suppresses Hepatitis B Virus Replication by Reducing HBc Protein Levels. Front Cell Infect Microbiol 2020, 10, 158. [Google Scholar] [CrossRef] [PubMed]
- Mounce, B.C.; Poirier, E.Z.; Passoni, G.; Simon-Loriere, E.; Cesaro, T.; Prot, M.; Stapleford, K.A.; Moratorio, G.; Sakuntabhai, A.; Levraud, J.-P.; et al. Interferon-Induced Spermidine-Spermine Acetyltransferase and Polyamine Depletion Restrict Zika and Chikungunya Viruses. Cell Host Microbe 2016, 20, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Olsen, M.E.; Cressey, T.N.; Mühlberger, E.; Connor, J.H. Differential Mechanisms for the Involvement of Polyamines and Hypusinated eIF5A in Ebola Virus Gene Expression. J Virol 2018, 92, e01260-18. [Google Scholar] [CrossRef] [PubMed]
- Gibson, W.; Roizman, B. Compartmentalization of Spermine and Spermidine in the Herpes Simplex Virion. Proc Natl Acad Sci U S A 1971, 68, 2818–2821. [Google Scholar] [CrossRef]
- Mastrodomenico, V.; LoMascolo, N.J.; Cruz-Pulido, Y.E.; Cunha, C.R.; Mounce, B.C. Polyamine-Linked Cholesterol Incorporation in Rift Valley Fever Virus Particles Promotes Infectivity. ACS Infect Dis 2022, 8, 1439–1448. [Google Scholar] [CrossRef]
- Firpo, M.R.; LoMascolo, N.J.; Petit, M.J.; Shah, P.S.; Mounce, B.C. Polyamines and eIF5A Hypusination Facilitate SREBP2 Synthesis and Cholesterol Production Leading to Enhanced Enterovirus Attachment and Infection. PLoS Pathog 2023, 19, e1011317. [Google Scholar] [CrossRef]
- Kicmal, T.M.; Tate, P.M.; Dial, C.N.; Esin, J.J.; Mounce, B.C. Polyamine Depletion Abrogates Enterovirus Cellular Attachment. Journal of Virology 2019, 93, e01054-19. [Google Scholar] [CrossRef]
- Gassen, N.C.; Papies, J.; Bajaj, T.; Emanuel, J.; Dethloff, F.; Chua, R.L.; Trimpert, J.; Heinemann, N.; Niemeyer, C.; Weege, F.; et al. SARS-CoV-2-Mediated Dysregulation of Metabolism and Autophagy Uncovers Host-Targeting Antivirals. Nat Commun 2021, 12, 3818. [Google Scholar] [CrossRef]
- Darnell, J.E.; Eagle, H. Glucose and Glutamine in Poliovirus Production by HeLa Cells. Virology 1958, 6, 556–566. [Google Scholar] [CrossRef]
- Zhang, Z.; Hu, H.; Luo, Q.; Yang, K.; Zou, Z.; Shi, M.; Liang, W. Dihydroxyacetone Phosphate Accumulation Leads to Podocyte Pyroptosis in Diabetic Kidney Disease. J Cell Mol Med 2024, 28, e18073. [Google Scholar] [CrossRef]
- Hernandez, A.; Sonavane, M.; Smith, K.R.; Seiger, J.; Migaud, M.E.; Gassman, N.R. Dihydroxyacetone Suppresses mTOR Nutrient Signaling and Induces Mitochondrial Stress in Liver Cells. PLoS One 2022, 17, e0278516. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.R.; Hayat, F.; Andrews, J.F.; Migaud, M.E.; Gassman, N.R. Dihydroxyacetone Exposure Alters NAD(P)H and Induces Mitochondrial Stress and Autophagy in HEK293T Cells. Chem Res Toxicol 2019, 32, 1722–1731. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.V.; Bartosch, B.; Isaguliants, M.G. Oxidative Stress in Infection and Consequent Disease. Oxid Med Cell Longev 2017, 2017, 3496043. [Google Scholar] [CrossRef] [PubMed]
- Kavouras, J.H.; Prandovszky, E.; Valyi-Nagy, K.; Kovacs, S.K.; Tiwari, V.; Kovacs, M.; Shukla, D.; Valyi-Nagy, T. Herpes Simplex Virus Type 1 Infection Induces Oxidative Stress and the Release of Bioactive Lipid Peroxidation By-Products in Mouse P19N Neural Cell Cultures. J Neurovirol 2007, 13, 416–425. [Google Scholar] [CrossRef]
- Chernyak, B.V.; Popova, E.N.; Prikhodko, A.S.; Grebenchikov, O.A.; Zinovkina, L.A.; Zinovkin, R.A. COVID-19 and Oxidative Stress. Biochemistry (Mosc) 2020, 85, 1543–1553. [Google Scholar] [CrossRef]
- Tung, W.-H.; Hsieh, H.-L.; Lee, I.-T.; Yang, C.-M. Enterovirus 71 Induces Integrin Β1/EGFR-Rac1-Dependent Oxidative Stress in SK-N-SH Cells: Role of HO-1/CO in Viral Replication. J Cell Physiol 2011, 226, 3316–3329. [Google Scholar] [CrossRef]
- Cheng, M.-L.; Weng, S.-F.; Kuo, C.-H.; Ho, H.-Y. Enterovirus 71 Induces Mitochondrial Reactive Oxygen Species Generation That Is Required for Efficient Replication. PLoS One 2014, 9, e113234. [Google Scholar] [CrossRef]
- Xie, B.; Zhou, J.-F.; Lu, Q.; Li, C.-J.; Chen, P. Oxidative Stress in Patients with Acute Coxsackie Virus Myocarditis. Biomed Environ Sci 2002, 15, 48–57. [Google Scholar]
- Sciacovelli, M.; Frezza, C. Oncometabolites: Unconventional Triggers of Oncogenic Signalling Cascades. Free Radical Biology and Medicine 2016, 100, 175–181. [Google Scholar] [CrossRef]
- Le, T.H.; Lipatova, A.V.; Volskaya, M.A.; Tikhonova, O.A.; Chumakov, P.M. The State of The Jak/Stat Pathway Affects the Sensitivity of Tumor Cells to Oncolytic Enteroviruses. Mol Biol 2020, 54, 570–577. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A SIMPLE METHOD OF ESTIMATING FIFTY PER CENT ENDPOINTS l’2.
- Fiehn, O. Metabolomics by Gas Chromatography-Mass Spectrometry: Combined Targeted and Untargeted Profiling. Curr Protoc Mol Biol 2016, 114, 30.4.1–30.4.32. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, O.N.; Krasnov, G.S.; Snezhkina, A.V.; Kudryavtseva, A.V.; Fedorov, V.S.; Zakirova, N.F.; Golikov, M.V.; Kochetkov, S.N.; Bartosch, B.; Valuev-Elliston, V.T.; et al. Transcriptome Analysis of Redox Systems and Polyamine Metabolic Pathway in Hepatoma and Non-Tumor Hepatocyte-like Cells. Biomolecules 2023, 13, 714. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.V.; Smirnova, O.A.; Petrushanko, I.Y.; Ivanova, O.N.; Karpenko, I.L.; Alekseeva, E.; Sominskaya, I.; Makarov, A.A.; Bartosch, B.; Kochetkov, S.N.; et al. HCV Core Protein Uses Multiple Mechanisms to Induce Oxidative Stress in Human Hepatoma Huh7 Cells. Viruses 2015, 7, 2745–2770. [Google Scholar] [CrossRef] [PubMed]
- Abubaker, A.A.; Vara, D.; Eggleston, I.; Canobbio, I.; Pula, G. A Novel Flow Cytometry Assay Using Dihydroethidium as Redox-Sensitive Probe Reveals NADPH Oxidase-Dependent Generation of Superoxide Anion in Human Platelets Exposed to Amyloid Peptide β. Platelets 2019, 30, 181–189. [Google Scholar] [CrossRef]
- Pak, V.V.; Ezeriņa, D.; Lyublinskaya, O.G.; Pedre, B.; Tyurin-Kuzmin, P.A.; Mishina, N.M.; Thauvin, M.; Young, D.; Wahni, K.; Gache, S.A.M.; et al. Ultrasensitive Genetically Encoded Indicator for Hydrogen Peroxide Identifies Roles for the Oxidant in Cell Migration and Mitochondrial Function. Cell Metabolism 2020, 31, 642–653.e6. [Google Scholar] [CrossRef]
- Smirnova, O.A.; Ivanova, O.N.; Fedyakina, I.T.; Yusubalieva, G.M.; Baklaushev, V.P.; Yanvarev, D.V.; Kechko, O.I.; Mitkevich, V.A.; Vorobyev, P.O.; Fedorov, V.S.; et al. SARS-CoV-2 Establishes a Productive Infection in Hepatoma and Glioblastoma Multiforme Cell Lines. Cancers 2023, 15, 632. [Google Scholar] [CrossRef]
| Compound | Target | Metabolic pathway | Sourse |
|---|---|---|---|
| 5.33756.0001 | ACSS | Fatty acid biosynthesis | Sigma |
| APCHA | Spermine synthase (SMS) | Polyamine biosynthesis | Sigma |
| DFMO | Ornithine decarboxylase (ODC) | Polyamine biosynthesis | MedChemExpress |
| Deferiprone (DFP) | Hypusination (DOHH inhibitor) | eIF5a hypusination | Sigma |
| Diethylnorspermine (DENSpm) | Spermidine/spermine-N1-acetyltransferase (SSAT) | Inducer of polyamine catabolism | Santa-Cruz Biotechnologies |
| GC7 | Hypusination (DOHH inhibitor) | eIF5a hypusination | Santa-Cruz Biotechnologies |
| MCHA | Spermidine synthase (SRM) | Polyamine biosynthesis | Sigma |
| AR-C155858 | MCT1 and MCT2 inhibitor | Glycolysis | MedChemExpress |
| MDL72.527 | Polyamine oxidases (PAOX, SMOX) | Polyamine catabolism | Sigma |
| Sardomozide (SAM-486a) | AdoMetDC (ADM1) | Polyamine biosynthesis | MedChemExpress |
| UK-5099 (PF-1005023) | Inhibitor of the mitochondrial pyruvate carrier (MPC) | Link between glycolysis and TCA cycle | Sigma |
| Name | 5'-3' Sequence | 3'-5' Sequence |
|---|---|---|
| AGPS | GCGCGAGCTACGGGTCTG | CTCTCCGCGCTTTGCACT |
| Aldo | GGCCCCACGATAGTGTGAAT | TCTCTAATGACCCCTGCCCT |
| DGAT | TATTGCGGCCAATGTCTTTGC | CACTGGAGTGATAGACTCAACCA |
| FASN | TACAAGCTGCGTGCCGCTGA | ACCCTCGATGACGTGGACGGAT |
| GCLC | GGATTTGGAAATGGGCAATTG | CTCAGATATACTGCAGGCTTGGAA |
| HO1 | CCAGCAACAAAGTGCAAGATTC | TCACATGGCATAAAGCCCTACAG |
| IGS15 | GCCGATCTTCTGGGTGATCTG | ATGGGCTGGGACCTGACG |
| MxA | GGTGGTCCCCAGTAATGTGG | CGTCAAGATTCCGATGGTCCT |
| Nqo1 | CCGTGGATCCCTTGCAGAGA | AGGACCCTTCCGGAGTAAGA |
| OAS-1 | ACAACCAGGTCAGCGTCAGAT | AGGTGGTAAAGGGTGGCTCC |
| TPI | AGCTCATCGGCACTCTGAAC | CCGGGCGAAGTCGATATAGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





