Submitted:
20 May 2025
Posted:
20 May 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
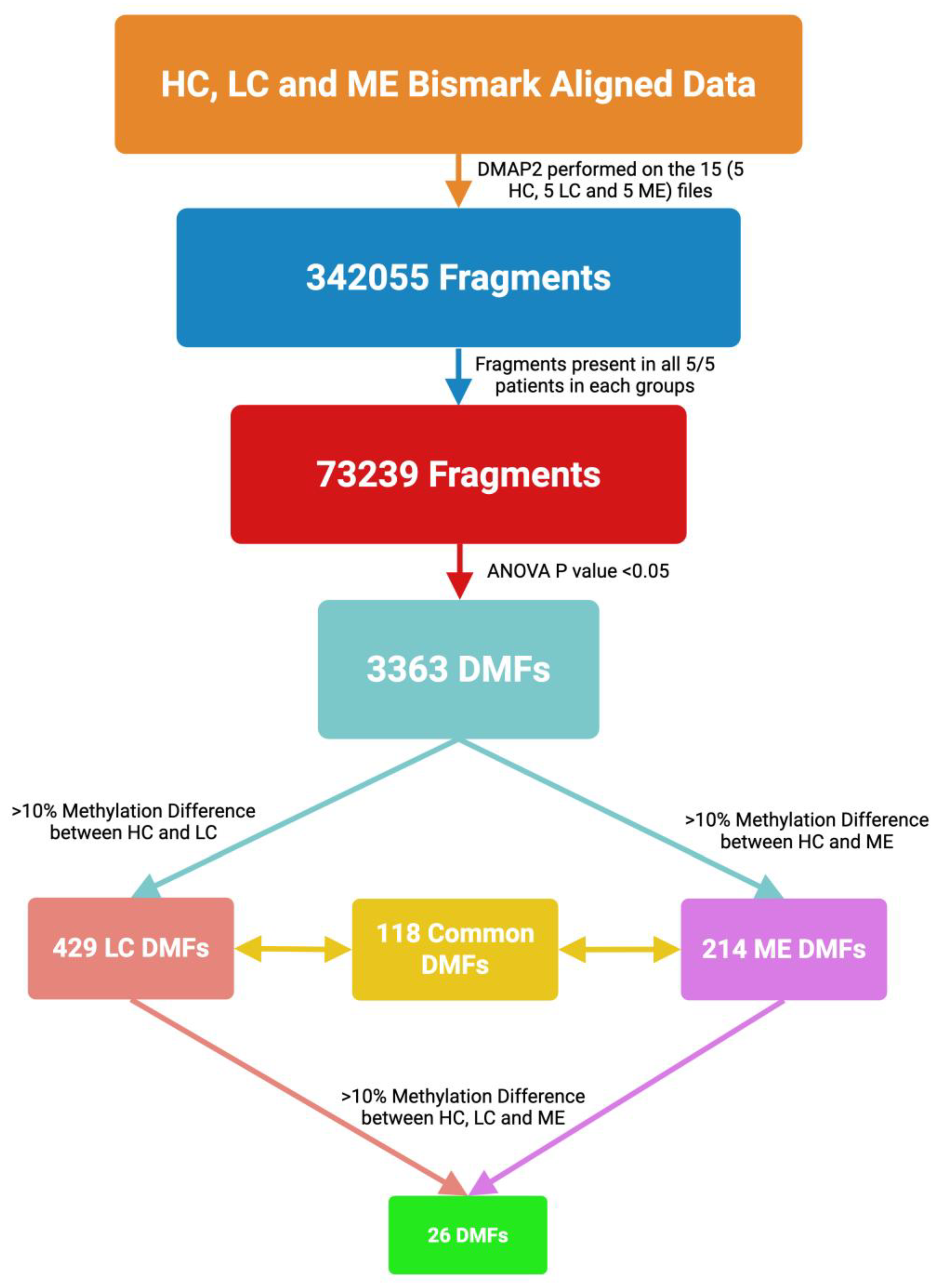
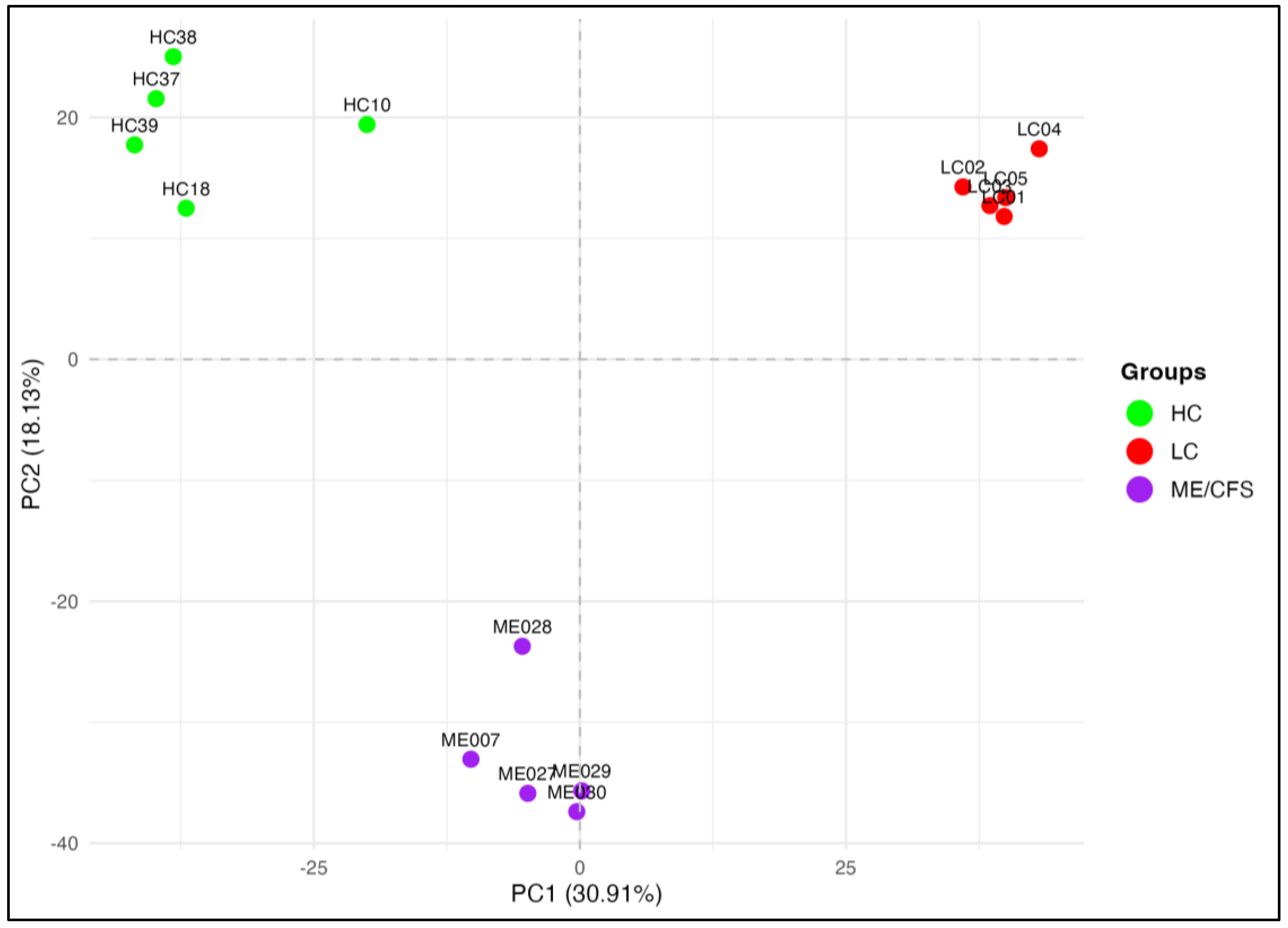
2.1. Whole-Genome DNA Methylation Patterns:
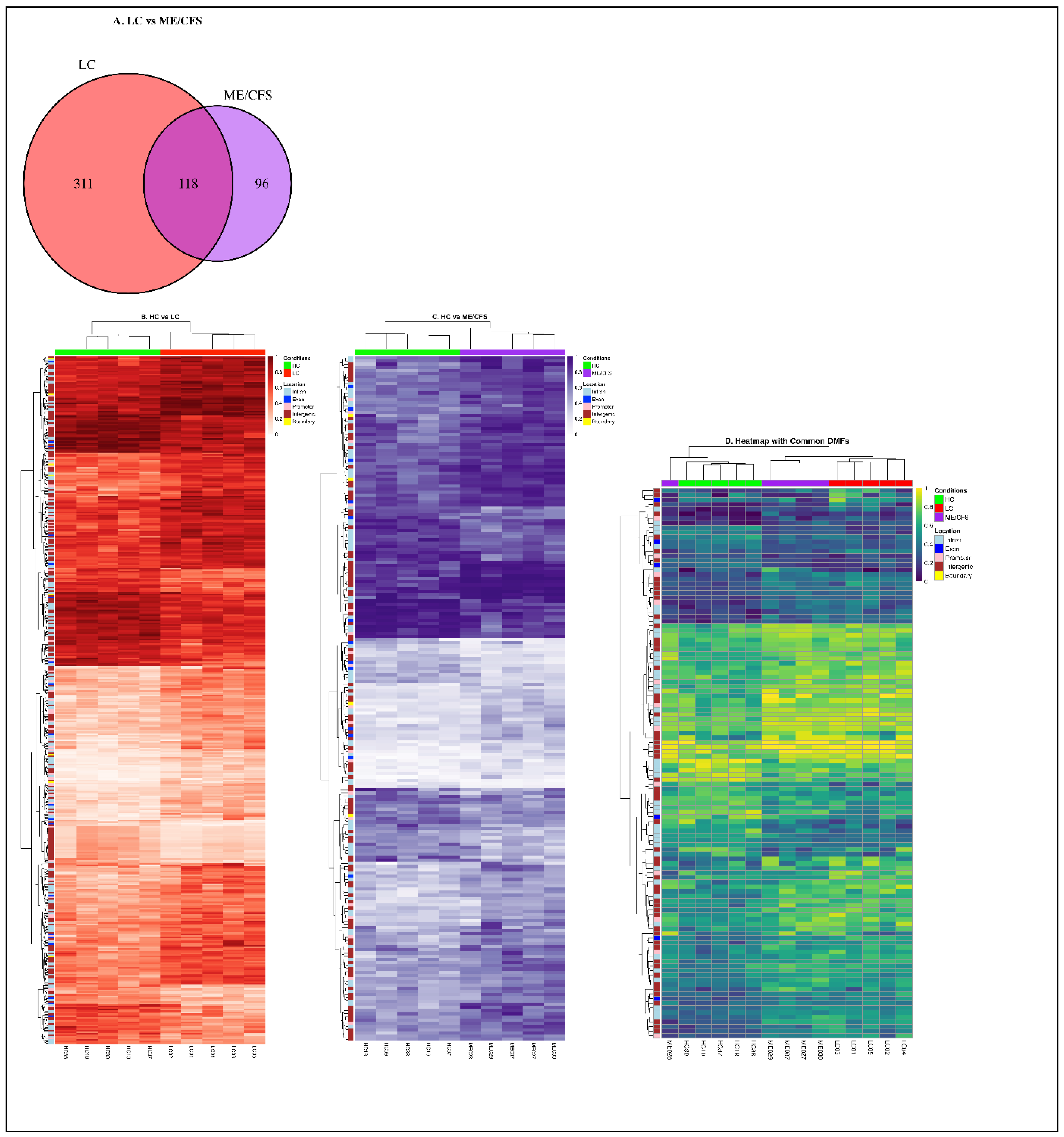
2.2. Characteristics of the Differential Methylation Changes in LC and ME/CFS
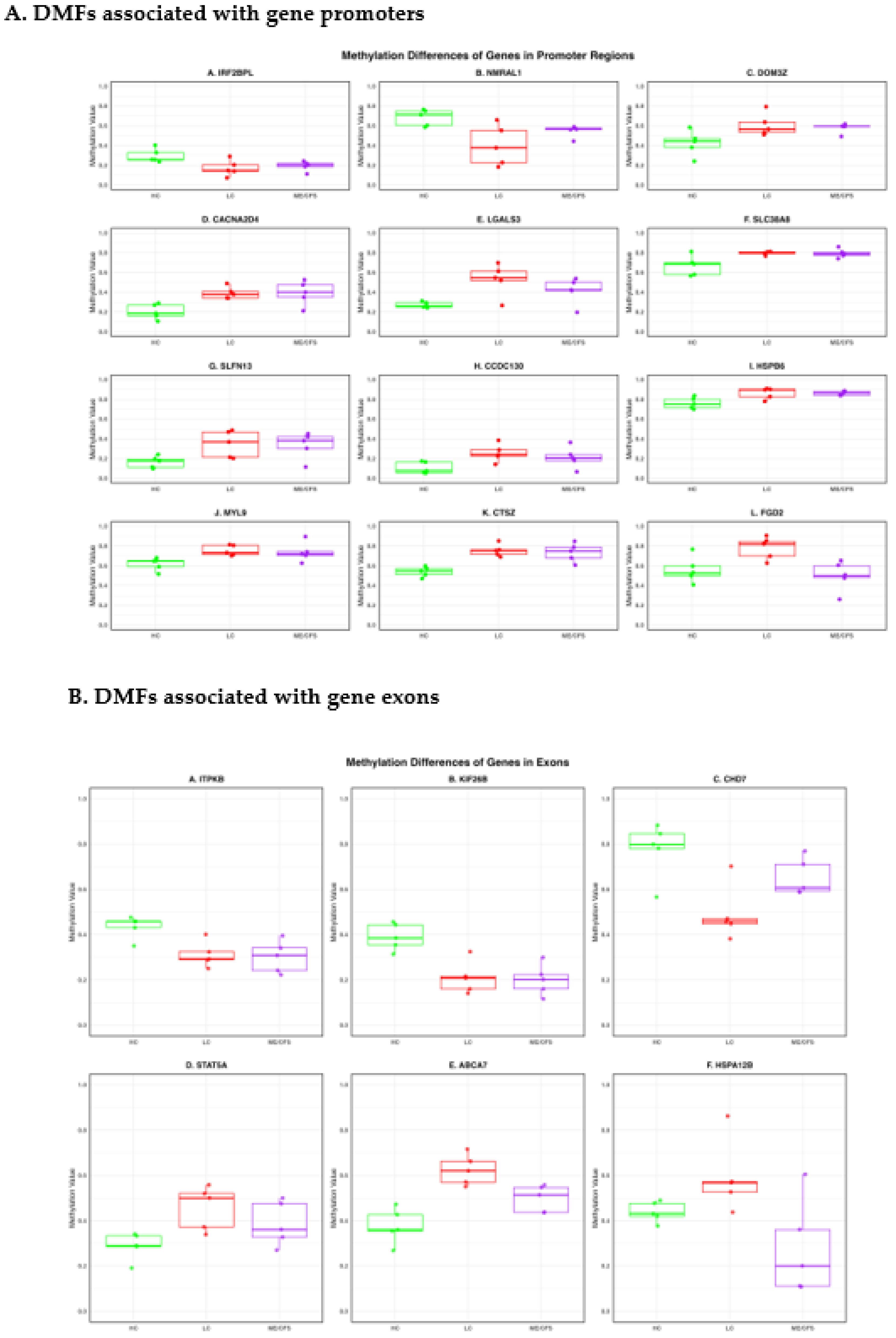
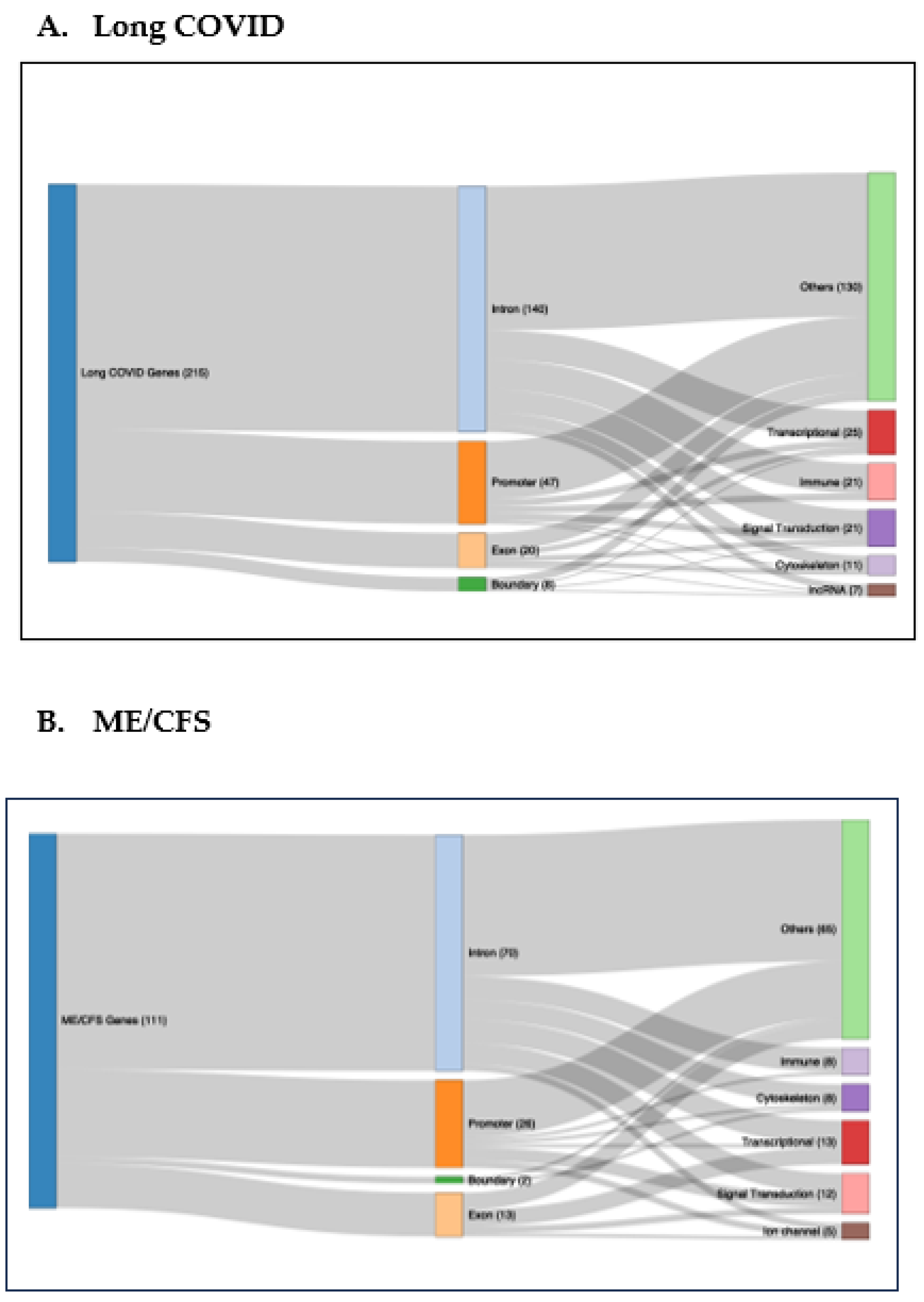
2.3. DMFs Associated with Gene Promoters and Gene Exons.
2.4. Methylation Differences Between Long COVID and ME/CFS
2.4. Functional Pathway Analysis of the DMFs of Long COVID and ME/CFS
3. Discussion
4. Materials and Methods
4.1. The Analysis Cohorts
4.2. PBMC Isolation
4.3. DNA Extraction
4.4. Reduced Representation Bisulphite Sequencing
4.4.1. DNA Sequencing
4.4.2. Statistical Analyses
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO: COVID-19 Cases, World. https://data.who.int/dashboards/covid19/cases (2025). Accessed 3 April 2025.
- Tate WP, Walker MOM, Peppercorn K, Blair ALH, Edgar CD. Towards a Better Understanding of the Complexities of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome and Long COVID. Int J Mol Sci. 2023;24(6):5124. [CrossRef]
- Davis HE, McCorkell L, Vogel JM, Topol EJ. Long COVID: major findings, mechanisms and recommendations. Nature Reviews Microbiology. 2023:1-14. [CrossRef]
- National Academies of Sciences E, Medicine, Health, Medicine D, Board on Global H, Board on Health Sciences P, et al. A Long COVID Definition: A Chronic, Systemic Disease State with Profound Consequences. In: Goldowitz I, Worku T, Brown L, Fineberg HV, editors. Washington (DC): National Academies Press (US) Copyright 2024 by the National Academy of Sciences. All rights reserved.; 2024.
- Collins FS: NIH launches new initiative to study “long COVID”. https://www.nih.gov/about-nih/who-we-are/nih-director/statements/nih-launches-new-initiative-study-long-covid (2021). Accessed 1 April 2025.
- Staff M. AN OUTBREAK of encephalomyelitis in the Royal Free Hospital Group, London, in 1955. Br Med J. 1957;2(5050):895-904.
- Levine PH, Jacobson S, Pocinki AG, Cheney P, Peterson D, Connelly RR, et al. Clinical, epidemiologic, and virologic studies in four clusters of the chronic fatigue syndrome. Arch Intern Med. 1992;152(8):1611-6.
- Shepherd C, Chaudhuri A, Association M, Association ME, Society ANZM. ME/CFS/PVFS: An Exploration of the Key Clinical Issues. ME Association; 2013.
- ME-pedia: Millions Missing protests. https://me-pedia.org/wiki/Millions_Missing_protests (2023). Accessed 10th May 2025.
- Tate W, Walker M, Sweetman E, Helliwell A, Peppercorn K, Edgar C, et al. Molecular Mechanisms of Neuroinflammation in ME/CFS and Long COVID to Sustain Disease and Promote Relapses. Front Neurol. 2022;13:877772. [CrossRef]
- Fang Z, Ahrnsbrak R, Rekito A. Evidence Mounts That About 7% of US Adults Have Had Long COVID. JAMA. 2024;332(1):5-6. [CrossRef]
- Xie Y, Choi T, Al-Aly Z. Postacute Sequelae of SARS-CoV-2 Infection in the Pre-Delta, Delta, and Omicron Eras. N Engl J Med. 2024;391(6):515-25. [CrossRef]
- Das S, Taylor K, Kozubek J, Sardell J, Gardner S. Genetic risk factors for ME/CFS identified using combinatorial analysis. J Transl Med. 2022;20(1):598. [CrossRef]
- Taylor K, Pearson M, Das S, Sardell J, Chocian K, Gardner S. Genetic risk factors for severe and fatigue dominant long COVID and commonalities with ME/CFS identified by combinatorial analysis. Journal of Translational Medicine. 2023;21(1):775. [CrossRef]
- Gentilotti E, Górska A, Tami A, Gusinow R, Mirandola M, Rodríguez Baño J, et al. Clinical phenotypes and quality of life to define post-COVID-19 syndrome: a cluster analysis of the multinational, prospective ORCHESTRA cohort. eClinicalMedicine. 2023;62:102107. [CrossRef]
- Jason LA, Natelson BH, Bonilla H, Sherif ZA, Vernon SD, Verduzco Gutierrez M, et al. What Long COVID investigators can learn from four decades of ME/CFS research. Brain Behavior and Immunity Integrative. 2023;4:100022. [CrossRef]
- Horwitz LI, Thaweethai T, Brosnahan SB, Cicek MS, Fitzgerald ML, Goldman JD, et al. Researching COVID to Enhance Recovery (RECOVER) adult study protocol: Rationale, objectives, and design. PLoS One. 2023;18(6):e0286297. [CrossRef]
- Komaroff AL, Lipkin WI. Insights from myalgic encephalomyelitis/chronic fatigue syndrome may help unravel the pathogenesis of postacute COVID-19 syndrome. Trends in Molecular Medicine. 2021;27(9):895-906. [CrossRef]
- Marshall-Gradisnik S, Eaton-Fitch N. Understanding myalgic encephalomyelitis. Science. 2022;377(6611):1150-1. [CrossRef]
- Komaroff AL: MECFS and Long COVID: Emerging Similarities and Why it Matters. https://massmecfs.org/news-events/823-mecfs-and-long-covid-emerging-similarities-and-why-it-matters (2022). Accessed 12 April 2025.
- Vernon SD, Hartle M, Sullivan K, Bell J, Abbaszadeh S, Unutmaz D, et al. Post-exertional malaise among people with long COVID compared to myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS). Work. 2023;74(4):1179-86. [CrossRef]
- Peppercorn K, Edgar CD, Kleffmann T, Tate WP. A pilot study on the immune cell proteome of long COVID patients shows changes to physiological pathways similar to those in myalgic encephalomyelitis/chronic fatigue syndrome. Sci Rep. 2023;13(1):22068. [CrossRef]
- Eaton-Fitch N, Rudd P, Er T, Hool L, Herrero L, Marshall-Gradisnik S. Immune exhaustion in ME/CFS and long COVID. JCI Insight. 2024;9(20). [CrossRef]
- Helliwell AM, Sweetman EC, Stockwell PA, Edgar CD, Chatterjee A, Tate WP. Changes in DNA methylation profiles of myalgic encephalomyelitis/chronic fatigue syndrome patients reflect systemic dysfunctions. Clin Epigenetics. 2020;12(1):167. [CrossRef]
- Helliwell AM, Stockwell PA, Edgar CD, Chatterjee A, Tate WP. Dynamic Epigenetic Changes during a Relapse and Recovery Cycle in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Int J Mol Sci. 2022;23(19). [CrossRef]
- Brenu EWS, D.R.; Marshall-Gradisnik S.M. . Methylation Profile of CD4+ T Cells in Chronic Fatigue Syndrome/Myalgic Encephalomyelitis. J Clin Cell Immunol. 2014;5. [CrossRef]
- de Vega WC, Vernon SD, McGowan PO. DNA methylation modifications associated with chronic fatigue syndrome. PLoS One. 2014;9(8):e104757. [CrossRef]
- de Vega WC, McGowan PO. The epigenetic landscape of myalgic encephalomyelitis/chronic fatigue syndrome: deciphering complex phenotypes. Epigenomics. 2017;9(11):1337-40. [CrossRef]
- de Vega WC, Erdman L, Vernon SD, Goldenberg A, McGowan PO. Integration of DNA methylation & health scores identifies subtypes in myalgic encephalomyelitis/chronic fatigue syndrome. Epigenomics. 2018;10(5):539-57. [CrossRef]
- Herrera S, de Vega WC, Ashbrook D, Vernon SD, McGowan PO. Genome-epigenome interactions associated with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Epigenetics. 2018;13(12):1174-90. [CrossRef]
- Stockwell PA, Rodger EJ, Gimenez G, Morison IM, Chatterjee A. DMAP2: A Pipeline for Analysis of Whole-Genome-Scale DNA Methylation Sequencing Data. Curr Protoc. 2024;4(9):e70003. [CrossRef]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. [CrossRef]
- Gopalan S, Carja O, Fagny M, Patin E, Myrick JW, McEwen LM, et al. Trends in DNA Methylation with Age Replicate Across Diverse Human Populations. Genetics. 2017;206(3):1659-74. [CrossRef]
- Marcogliese PC, Shashi V, Spillmann RC, Stong N, Rosenfeld JA, Koenig MK, et al. IRF2BPL Is Associated with Neurological Phenotypes. Am J Hum Genet. 2018;103(2):245-60. [CrossRef]
- Zang W, Zheng X. Structure and functions of cellular redox sensor HSCARG/NMRAL1, a linkage among redox status, innate immunity, DNA damage response, and cancer. Free Radic Biol Med. 2020;160:768-74. [CrossRef]
- Di Gregoli K, Somerville M, Bianco R, Thomas AC, Frankow A, Newby AC, et al. Galectin-3 Identifies a Subset of Macrophages With a Potential Beneficial Role in Atherosclerosis. Arterioscler Thromb Vasc Biol. 2020;40(6):1491-509. [CrossRef]
- Kuht HJ, Han J, Maconachie GDE, Park SE, Lee ST, McLean R, et al. SLC38A8 mutations result in arrested retinal development with loss of cone photoreceptor specialization. Hum Mol Genet. 2020;29(18):2989-3002. [CrossRef]
- Yang JY, Deng XY, Li YS, Ma XC, Feng JX, Yu B, et al. Structure of Schlafen13 reveals a new class of tRNA/rRNA- targeting RNase engaged in translational control. Nat Commun. 2018;9(1):1165. [CrossRef]
- Kanno T, Lin WD, Fu JL, Matzke AJM, Matzke M. A genetic screen implicates a CWC16/Yju2/CCDC130 protein and SMU1 in alternative splicing in Arabidopsis thaliana. Rna. 2017;23(7):1068-79. [CrossRef]
- Dreiza CM, Komalavilas P, Furnish EJ, Flynn CR, Sheller MR, Smoke CC, et al. The small heat shock protein, HSPB6, in muscle function and disease. Cell Stress and Chaperones. 2010;15(1):1-11. [CrossRef]
- Krueger S, Bernhardt A, Kalinski T, Baldensperger M, Zeh M, Teller A, et al. Induction of premalignant host responses by cathepsin x/z-deficiency in Helicobacter pylori-infected mice. PLoS One. 2013;8(7):e70242. [CrossRef]
- Kumar CC, Mohan SR, Zavodny PJ, Narula SK, Leibowitz PJ. Characterization and differential expression of human vascular smooth muscle myosin light chain 2 isoform in nonmuscle cells. Biochemistry. 1989;28(9):4027-35. [CrossRef]
- Qin N, Yagel S, Momplaisir ML, Codd EE, D'Andrea MR. Molecular cloning and characterization of the human voltage-gated calcium channel alpha(2)delta-4 subunit. Mol Pharmacol. 2002;62(3):485-96. [CrossRef]
- Huber C, Mårtensson A, Bokoch GM, Nemazee D, Gavin AL. FGD2, a CDC42-specific exchange factor expressed by antigen-presenting cells, localizes to early endosomes and active membrane ruffles. J Biol Chem. 2008;283(49):34002-12. [CrossRef]
- Shen Y, Zhang J, Calarco JA, Zhang Y. EOL-1, the homolog of the mammalian Dom3Z, regulates olfactory learning in C. elegans. J Neurosci. 2014;34(40):13364-70. [CrossRef]
- Apicco DJ, Shlevkov E, Nezich CL, Tran DT, Guilmette E, Nicholatos JW, et al. The Parkinson's disease-associated gene ITPKB protects against α-synuclein aggregation by regulating ER-to-mitochondria calcium release. Proc Natl Acad Sci U S A. 2021;118(1). [CrossRef]
- Uchiyama Y, Sakaguchi M, Terabayashi T, Inenaga T, Inoue S, Kobayashi C, et al. Kif26b, a kinesin family gene, regulates adhesion of the embryonic kidney mesenchyme. Proc Natl Acad Sci U S A. 2010;107(20):9240-5. [CrossRef]
- Reddy NC, Majidi SP, Kong L, Nemera M, Ferguson CJ, Moore M, et al. CHARGE syndrome protein CHD7 regulates epigenomic activation of enhancers in granule cell precursors and gyrification of the cerebellum. Nat Commun. 2021;12(1):5702. [CrossRef]
- Lin JX, Leonard WJ. The role of Stat5a and Stat5b in signaling by IL-2 family cytokines. Oncogene. 2000;19(21):2566-76. [CrossRef]
- Kim WS, Weickert CS, Garner B. Role of ATP-binding cassette transporters in brain lipid transport and neurological disease. J Neurochem. 2008;104(5):1145-66. [CrossRef]
- Dib S, Pahnke J, Gosselet F. Role of ABCA7 in Human Health and in Alzheimer's Disease. Int J Mol Sci. 2021;22(9). [CrossRef]
- Steagall RJ, Rusiñol AE, Truong QA, Han Z. HSPA12B is predominantly expressed in endothelial cells and required for angiogenesis. Arterioscler Thromb Vasc Biol. 2006;26(9):2012-8. [CrossRef]
- Carruthers BM, Jain AK, De Meirleir KL, Peterson DL, Klimas NG, Lerner AM, et al. Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Journal of Chronic Fatigue Syndrome. 2003;11(1):7-115. [CrossRef]
- WHO: A clinical case definition of post COVID-19 condition by a Delphi consensus, 6 October 2021. https://www.who.int/publications/i/item/WHO-2019-nCoV-Post_COVID-19_condition-Clinical_case_definition-2021.1 (2021). Accessed.
- Peppercorn K, ; Edgar, C.; Al Momani, S,; Rodger, E. J.; Tate, W.P.; Chatterjee, A. Application of DNA methylome analysis to patients with ME/CFS. Methods in Molecular Biology. in press: Spinger; 2025.
- Ludgate JL, Wright J, Stockwell PA, Morison IM, Eccles MR, Chatterjee A. A streamlined method for analysing genome-wide DNA methylation patterns from low amounts of FFPE DNA. BMC Med Genomics. 2017;10(1):54. [CrossRef]
- Rodger EJ, Stockwell PA, Almomani S, Eccles MR, Chatterjee A. Protocol for generating high-quality genome-scale DNA methylation sequencing data from human cancer biospecimens. STAR Protoc. 2023;4(4):102714. [CrossRef]
- Stockwell PA, Chatterjee A, Rodger EJ, Morison IM. DMAP: differential methylation analysis package for RRBS and WGBS data. Bioinformatics. 2014;30(13):1814-22. [CrossRef]
- Chatterjee A, Stockwell PA, Rodger EJ, Morison IM. Comparison of alignment software for genome-wide bisulphite sequence data. Nucleic Acids Res. 2012;40(10):e79. Epub 2012 Feb 16. [CrossRef]
|
Chr |
Start | End | Location | GeneID |
%Difference LC v HC |
%Difference ME v HC |
%Difference LC v ME |
| 18 | 34854648 | 34854693 | intron | CELF4 | -13.9 | +15.1 | -29.0 |
| 1 | 90074598 | 90074699 | Intergenic | -36.4 | -15.7 | -20.7 | |
| 16 | 4527540 | 4527641 | Promoter | NMRAL1 | -32.0 | -12.5 | -19.5 |
| 5 | 39185995 | 39186145 | intron | FYB | -33.3 | -14.5 | -18.8 |
| 2 | 240241154 | 240241261 | Intergenic | +13.4 | +32.2 | -18.8 | |
| 7 | 158766236 | 158766379 | Intergenic | +18.4 | +35.4 | -17.0 | |
| 8 | 61778005 | 61778136 | exon | CHD7 | -29.2 | -14.5 | -14.7 |
| 17 | 76129099 | 76129221 | intron | TMC8 | -24.7 | -13.3 | -11.4 |
| 6 | 30854000 | 30854160 | intron | DDR1 | -22.4 | -11.0 | -11.4 |
| 17 | 75429645 | 75429795 | Intergenic | -21.9 | -10.6 | -11.3 | |
| 1 | 59090260 | 59090360 | Intergenic | +12.4 | +22.8 | -10.6 | |
| 2 | 121955379 | 121955533 | Intergenic | +22.3 | +12.2 | +10.1 | |
| 16 | 3137553 | 3137716 | Intergenic | ZNF205 | +26.1 | +14.7 | +11.4 |
| 19 | 1047184 | 1047347 | exon | ABCA7 | +24.6 | +11.8 | +12.8 |
| 7 | 158250978 | 158251159 | Intergenic | +28.1 | +15.3 | +12.8 | |
| 14 | 55587537 | 55587752 | Promoter | LGALS3 | +28.5 | +14.8 | +13.7 |
| 15 | 75336231 | 75336352 | intron | PPCDC | +26.5 | +10.8 | +15.7 |
| 8 | 58055165 | 58055309 | Intergenic | +27.6 | +11.7 | +15.9 | |
| 21 | 46714776 | 46714890 | Intergenic | +31.5 | +13.8 | +17.7 | |
| 8 | 58055310 | 58055463 | Intergenic | +34.2 | +15.7 | +18.5 | |
| 1 | 19110747 | 19110909 | Intergenic | -16.7 | -35.4 | +18.7 | |
| 17 | 76661321 | 76661487 | Intergenic | +11.8 | -11.3 | +23.1 | |
| 10 | 118025165 | 118025303 | Intergenic | +13.1 | -13.1 | +26.2 | |
| 6 | 36969405 | 36969621 | Promoter | FGD2 | +21.7 | -11.1 | +32.8 |
| 20 | 3732943 | 3733092 | exon | HSPA12B | +18.3 | -15.6 | +33.9 |
| 3 | 126945870 | 126946029 | Intergenic | +11.4 | -25.9 | +37.3 |
| A. Promoters | |||||||
| Chromosome | Start | End | Associated Genes |
DM*(%) (LC vs HC) |
DM*(%) (HC vs ME) |
DM* (%) (LC vs ME) |
|
| 19 | 13841885 | 13841989 | CCDC130 | +14.2 | +11.8 | +2.4 | |
| 14 | 77495636 | 77495807 | IRF2BPL | -12.2 | -10.2 | -2.0 | |
| 17 | 33776642 | 33776791 | SLFN13 | +18.1 | +11.7 | +6.4 | |
| 16 | 84076941 | 84077080 | SLC38A8 | +12.7 | +13.0 | -0.3 | |
| 19 | 36249868 | 36250044 | HSPB6 | +11.1 | +10.3 | +0.8 | |
| 16 | 4527540 | 4527641 | NMRAL1 | -32.0 | -12.5 | -19.5 | |
| 14 | 55587537 | 55587752 | LGALS3 | +28.5 | +14.8 | +13.7 | |
| 20 | 57581333 | 57581441 | CTSZ | +21.2 | +18.2 | +2.8 | |
| 20 | 35170171 | 35170286 | MYL9 | +13.2 | +11.6 | +1.6 | |
| 12 | 2027243 | 2027352 | CACNA2D4 | +17.7 | +17.6 | +0.1 | |
| 6 | 36969405 | 36969621 | FGD2 | +21.7 | -11.1 | +32.8 | |
| 6 | 31939186 | 31939321 | DOM3Z | +21.4 | +15.6 | +5.8 | |
| B. Exons | |||||||
| 1 | 245851466 | 245851609 | KIF26B | -19.0 | -18.7 | -0.3 | |
| 8 | 61778005 | 61778136 | CHD7 | -29.2 | -14.5 | -14.7 | |
| 1 | 226821736 | 226821914 | ITPKB | -12.1 | -13.1 | +1.0 | |
| 17 | 40463432 | 40463555 | STAT5A | +16.2 | +11.4 | +4.8 | |
| 20 | 3732943 | 3733092 | HSPA12B | +18.3 | -15.6 | +33.9 | |
| 19 | 1047184 | 1047347 | ABCA7 | +24.6 | +11.8 | +12.8 | |
| Patient | Age | Sex | Patient | Age | Sex | Control | Age | Sex |
| ME030 | 40 | F | LC01 | 43 | F | HC18 | 46 | F |
| ME028 | 19 | F | LC02 | 27 | F | HC39 | 26 | F |
| ME027 | 65 | F | LC03 | 65 | F | HC10 | 59 | F |
| ME029 | 40 | M | LC04 | 42 | M | HC37 | 40 | M |
| ME007 | 27 | F | LC05 | 36 | F | HC38 | 31 | F |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





